3.1. Analysis of Yield Components by Treatment
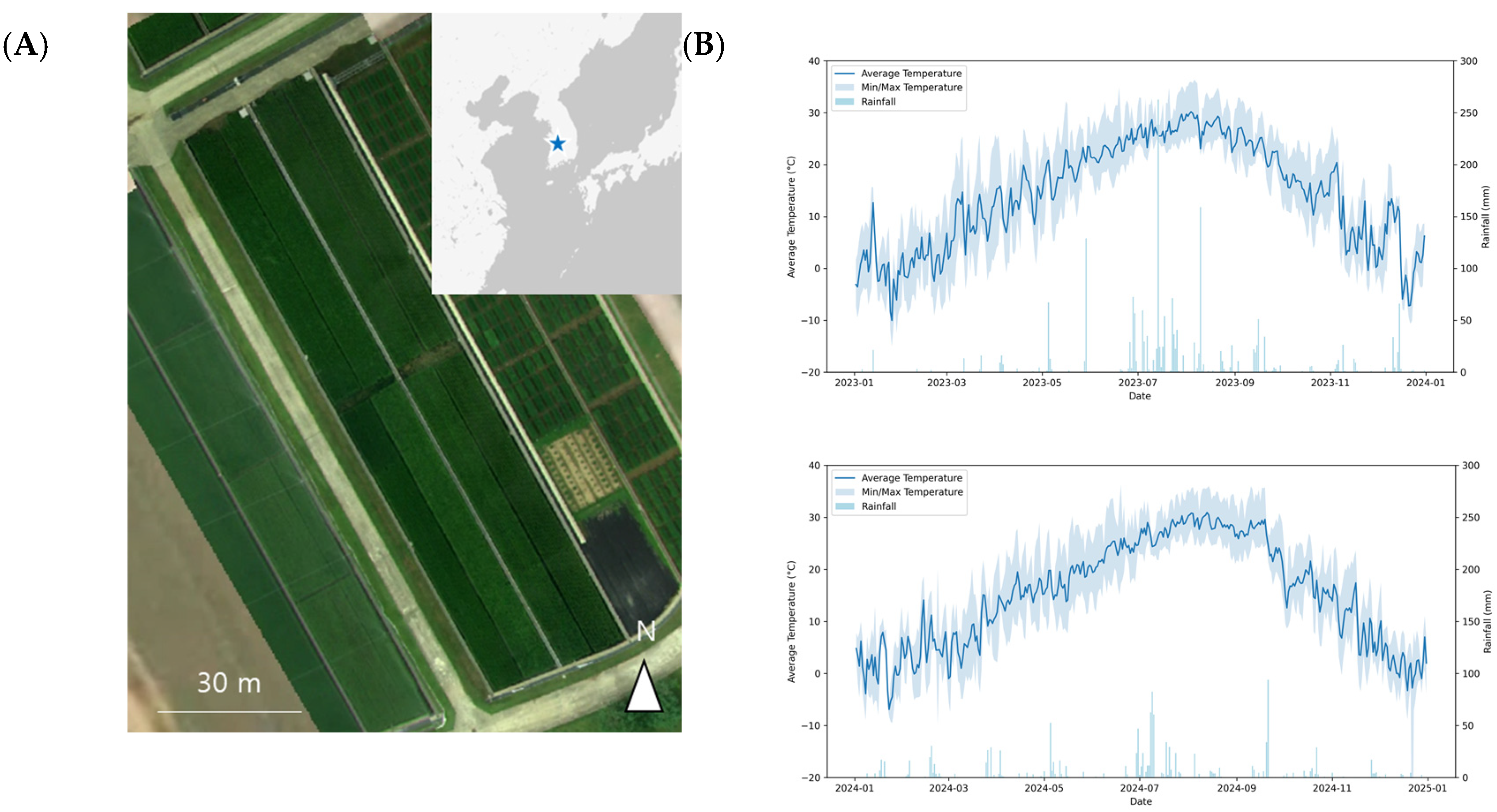
The yield and yield components of rice harvested in 2023 and 2024 at the National Institute of Crop Science in Wanju, South Korea, were analyzed (
Table 4). In the 2023 data, the high-nitrogen treatment (23-F-9N) recorded the highest yield (875.83 g/m
2), grain number (GN: 38,111.0 grains/m
2), and panicle number (PN: 384.6 panicles/m
2), highlighting the effect of nitrogen fertilizer. In contrast, treatments with low nitrogen levels (23-F-0N) or late transplantation (23-F-0N-LT) showed lower yield and GN values. FGR remained stable above 0.88 across most treatments but was the highest in 23-F-0N-LT (0.95). TGW varied depending on the treatment, with relatively lower values observed for 23-9N-LT.
In the 2024 dataset, the effects of nitrogen levels and rice variety combinations on the yield components were evident. Treatments 24-F-9N-DJ and 24-F-9N-SI had the highest values for yield (826.82 and 818.24 g/m2, respectively) and GN (32,821.3 and 32,710.3 grains/m2, respectively), indicating a positive effect of nitrogen fertilizer on yield. Conversely, the low-nitrogen treatment (24-F-0N-SD) resulted in the lowest values for all yield components. TGW was the highest in 24-F-0N-SD (32.19 g) and lowest in 24-F-9N-NP (25.38 g). Although the FGR was generally stable, it showed a decreasing trend in 24-F-0N-SD, decreasing to 0.76.
Overall, nitrogen levels significantly affected key yield components, such as yield, GN, and PN, with high-nitrogen treatments consistently producing superior results under different conditions. Differences between field and soil bin conditions were observed across the years, with a notable decrease in yield in the soil bin treatments in 2024. These findings evidently demonstrate the influence of nitrogen treatments and environmental conditions on yield components, which is supported by the statistically significant differences observed among treatments.
3.3. Log-Normal Parameter Analysis of VIs
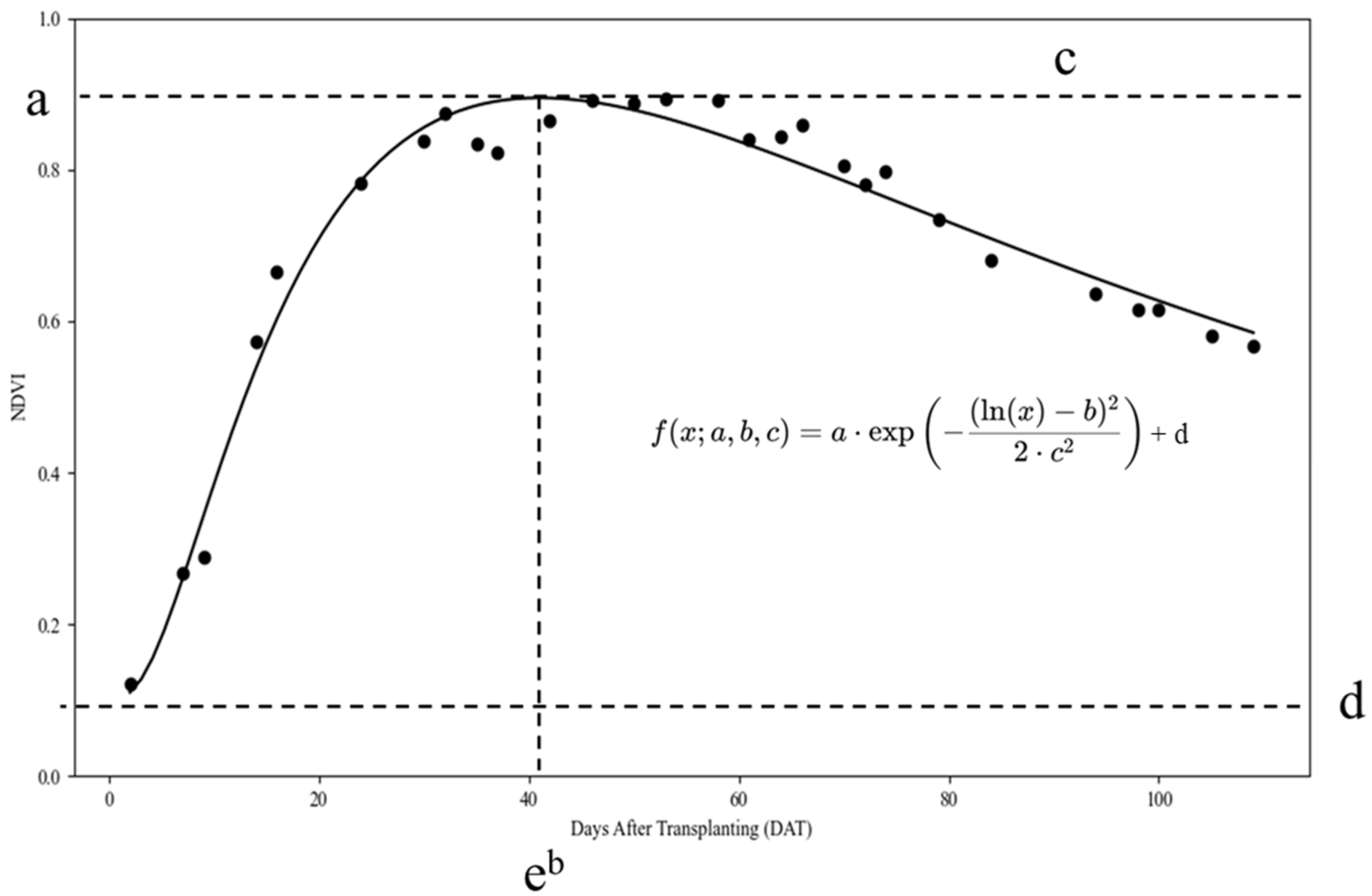
Time-series VIs measured within a 1 m
2 area for each sample were analyzed using a four-parameter log-normal model.
Table 5 presents the log-normal model fitting parameters (
a,
b,
c, and
d) for the NDVI data across various treatment conditions for 2023 and 2024. As nitrogen treatment levels increased, parameter
a, representing the peak value of the NDVI curve, generally exhibited an increasing trend. This indicates that higher nitrogen levels contribute to the elevation of maximum NDVI values. Parameter
b, which indicated the timing of the NDVI peak, tended to decrease with delayed transplantation dates. This implies that the late transplantation may have caused the NDVI peak to occur slightly earlier. This trend was particularly evident in the 2023 field data, where late transplanting treatments had lower
b values than standard transplanting treatments.
Parameter c, representing the spread or distribution of the NDVI curve, varied with the treatment conditions. Higher nitrogen levels typically correspond to higher c values, indicating a broader distribution of the NDVI curve. In contrast, parameter d, representing the baseline or minimum NDVI value, showed minimal variation across treatments, suggesting that the baseline NDVI values remained relatively stable regardless of nitrogen level or transplanting time.
These findings provide valuable insights into how NDVI profiles respond to variations in nitrogen levels and transplantation time. They enhance our understanding of how these factors affect the temporal behavior of VIs. Overall, the R
2 values for the fitted models were consistently high across different treatments, indicating that the four-parameter log-normal model effectively captured variations in NDVI over time and across treatment conditions.
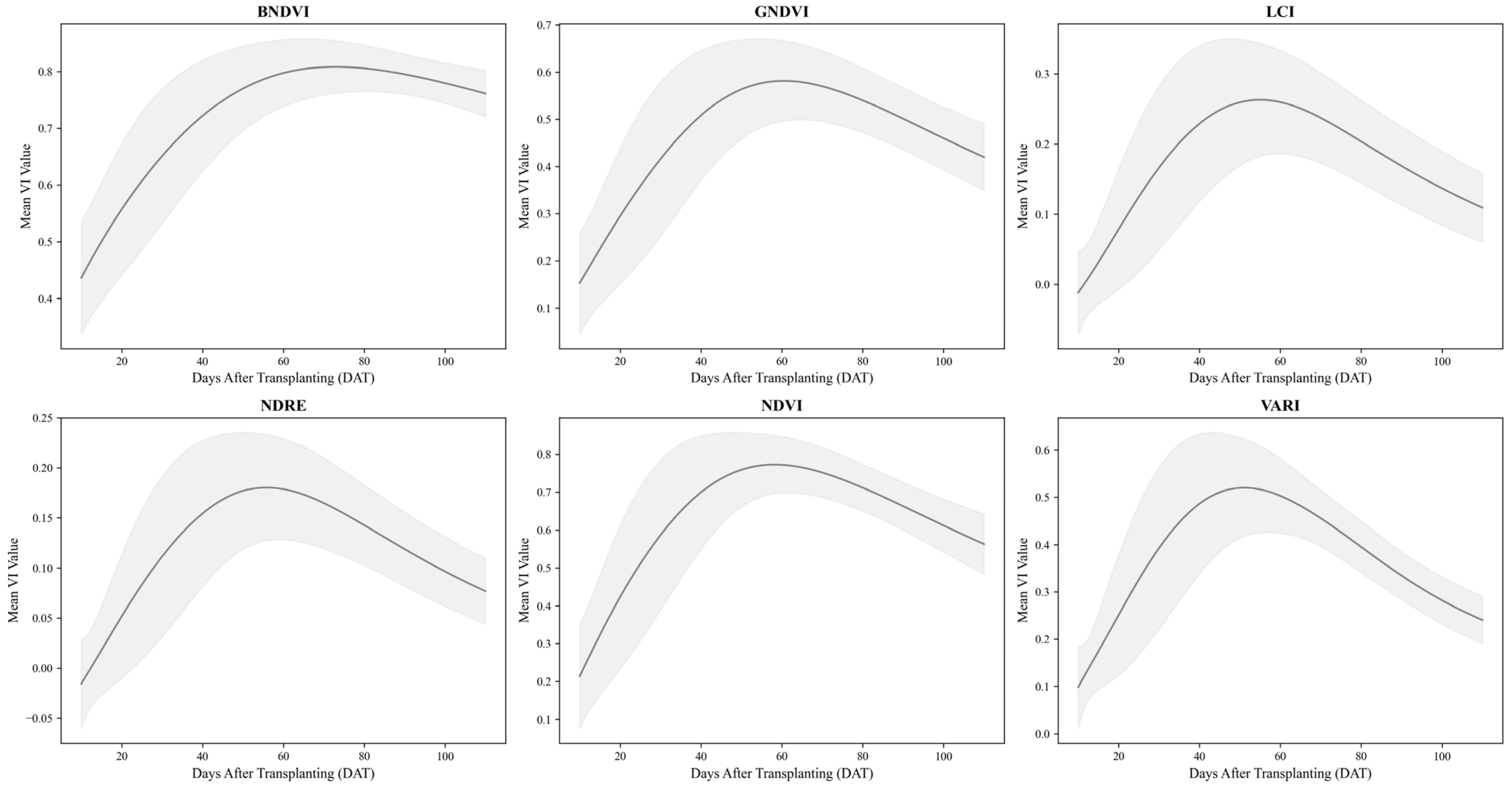
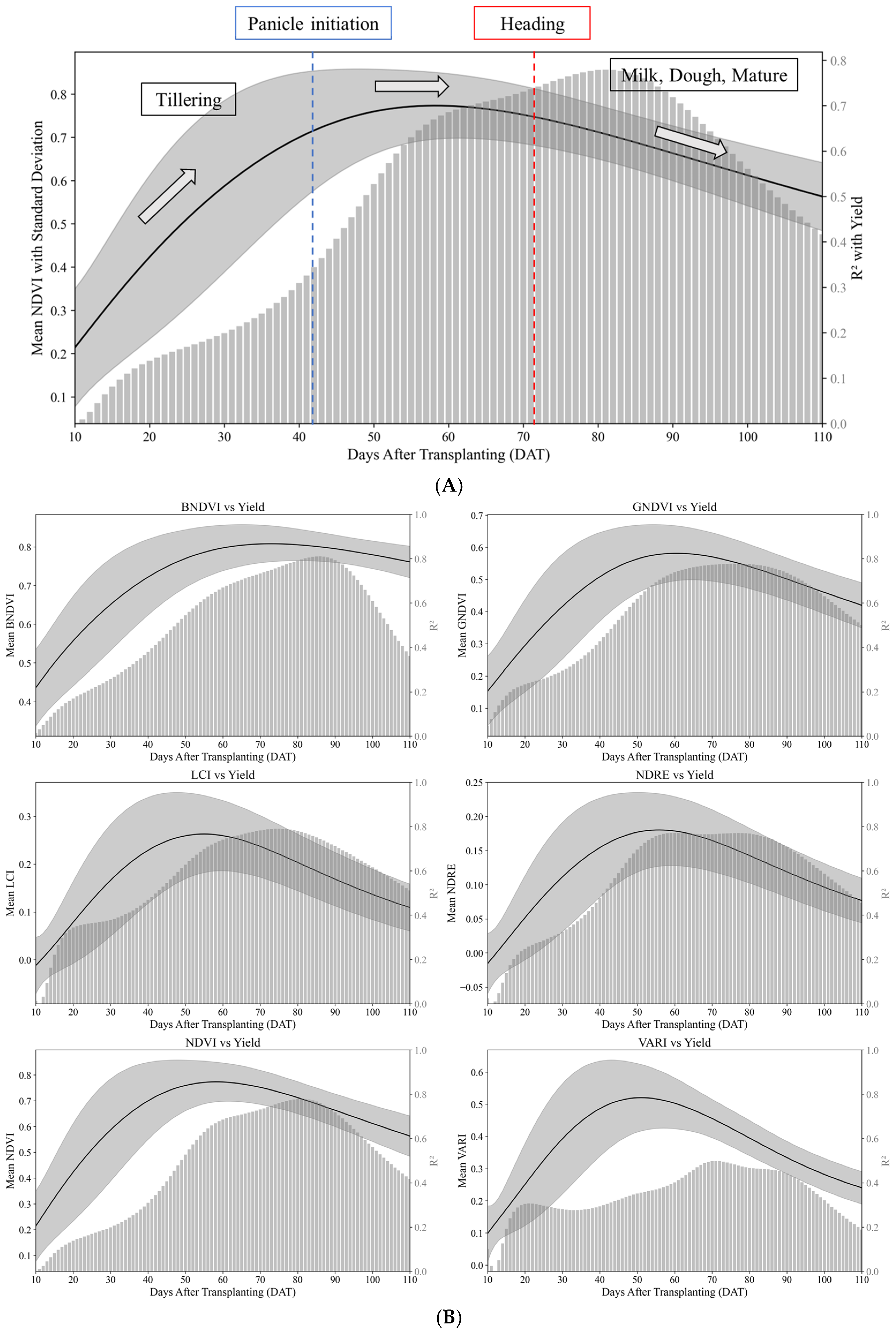
Figure 5 shows the deviations in the log-normal curves for each VI. The shaded regions represent the standard deviation around the mean log-normal model fit, visually representing how each index varied across the treatments.
Table 6 presents the average log-normal model parameters and R
2 values for each VI. The VIs included the BNDVI, GNDVI, LCI, NDRE, NDVI, and VARI. The consistently high R
2 values across the indices demonstrated the robustness of the log-normal model in fitting the time-series data. Differences in the parameter values among the VIs highlight their unique responses to environmental and treatment factors, further validating the applicability of this approach for assessing vegetation dynamics.
3.4. Comparison of Log-Normal Graphs of VIs by Treatment and Variety
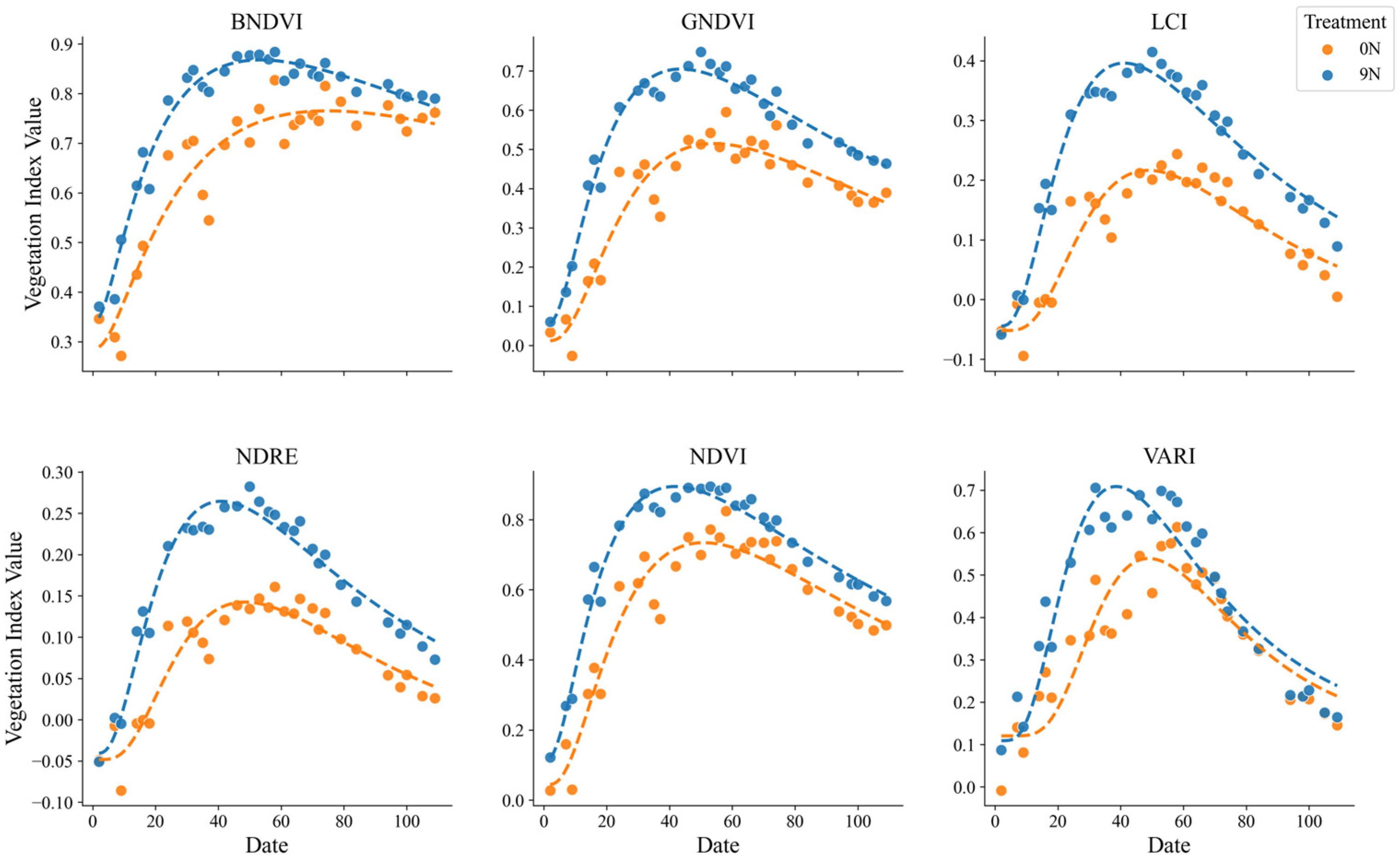
Graphical comparisons were conducted to examine differences among treatments and varieties for each VI fitted as time-series data. Nitrogen application rates influenced the VI curves, with higher nitrogen levels generally resulting in increased values across most indices, indicating improvements in plant health and biomass accumulation. BNDVI, GNDVI, and NDVI exhibited more pronounced peaks and higher values with increasing nitrogen levels.
Furthermore, nitrogen application advanced the timing of VI peaks, suggesting that nitrogen treatments accelerated the time to reach maximum biomass. This pattern, characterized by a rapid peak followed by a decline, reflects the growth dynamics observed under the nitrogen treatment. This highlights the critical role of N in shaping the timing and patterns of plant growth (
Figure 6).
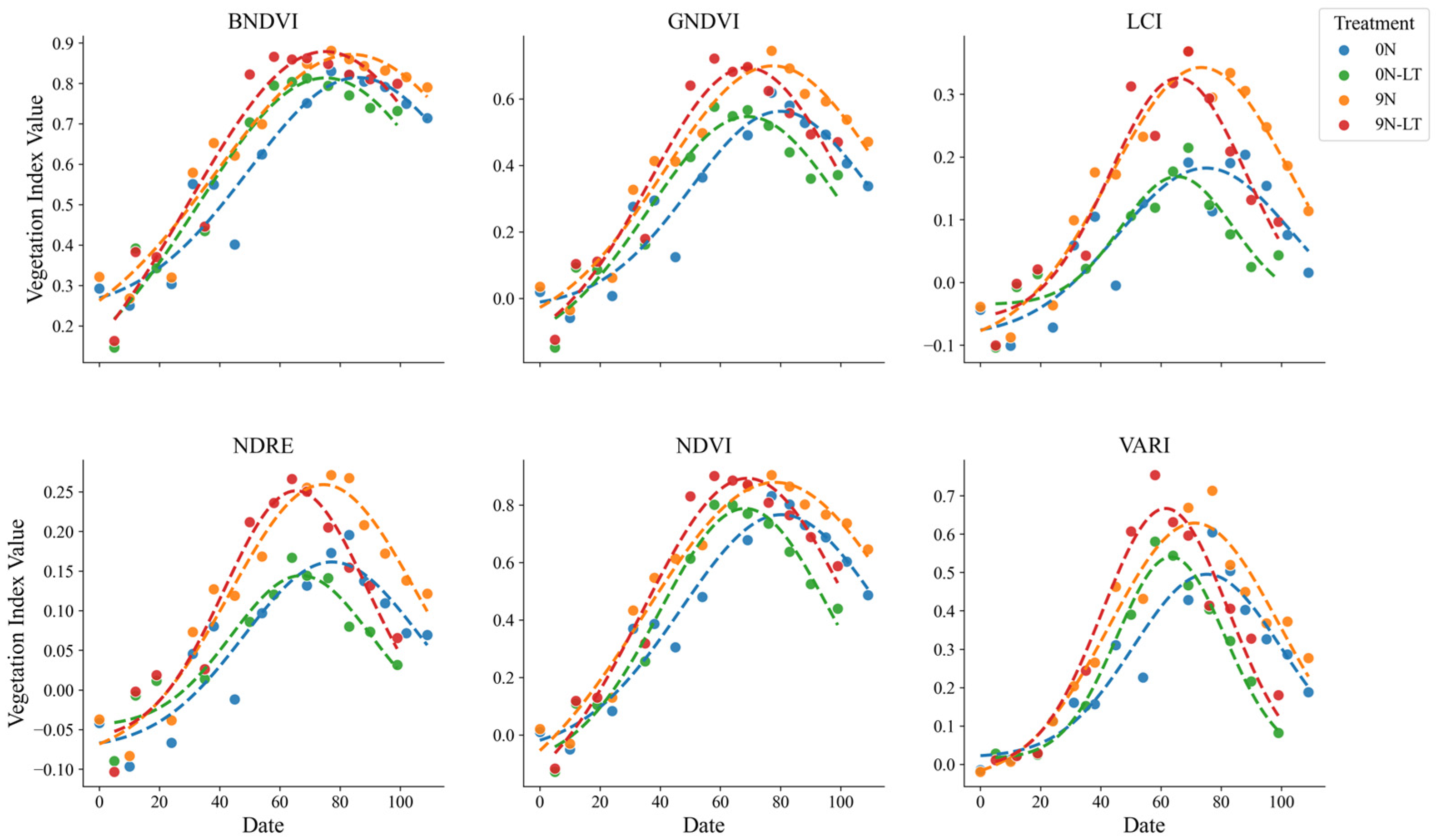
The results were used to analyze the effects of transplanting time and nitrogen treatment on VIs (
Figure 7). A longer transplantation time shortened the number of days required to reach the peak VI. Furthermore, a delay of approximately 20 days in transplanting had minimal effect on the peak VI values under nitrogen-treated conditions. However, under nitrogen-untreated conditions, transplanting reduced the overall VI values. This indicates that delayed transplanting considerably affected growth and VIs in the nitrogen-untreated plots.
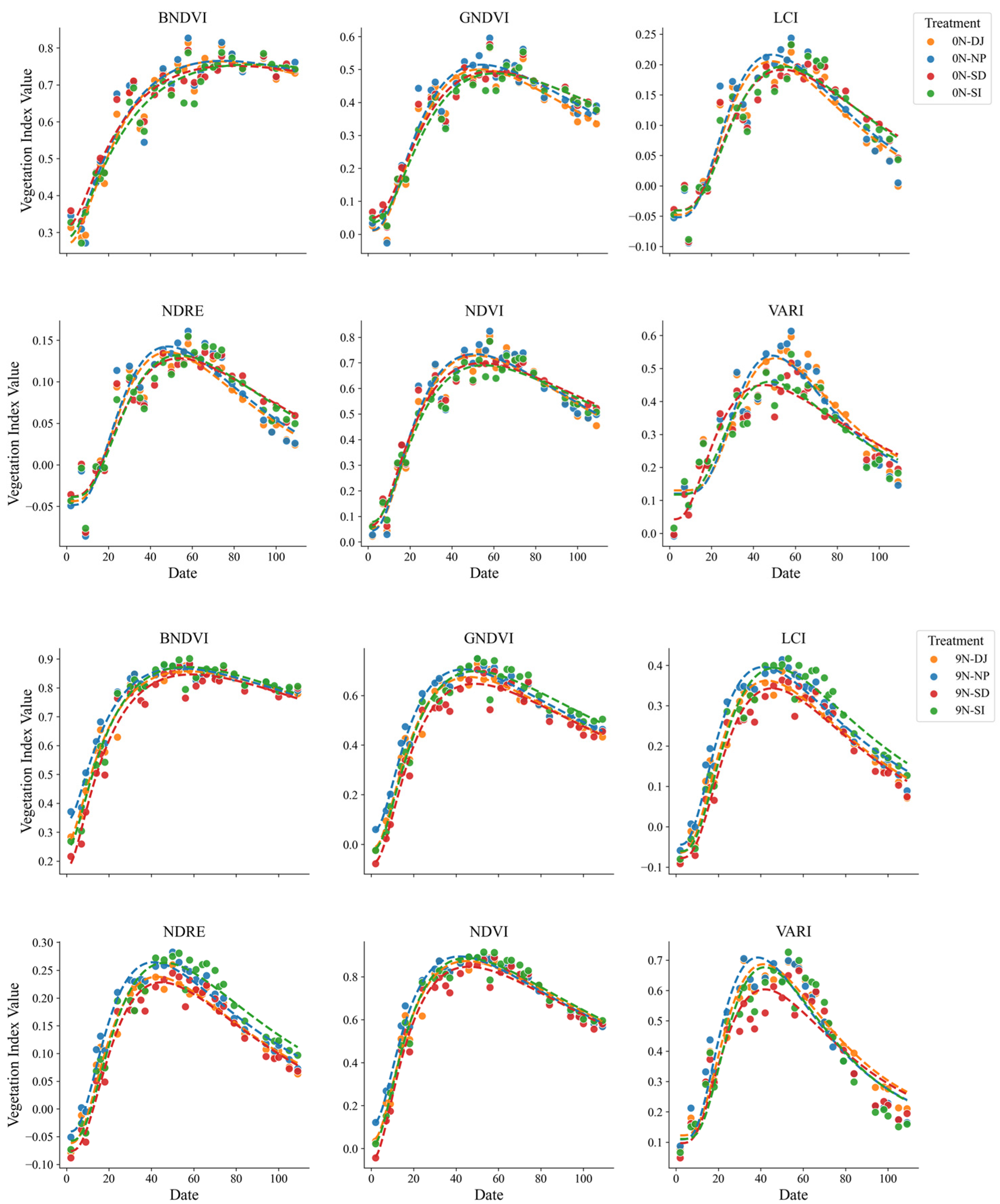
Minimal differences were observed in the VI patterns under the nitrogen treatment among the four rice varieties (Shindongjin, Dongjin-1, Nampyeong, and Saeilmi), indicating that ecological or phenotypic differences were not evident. This consistency suggests that the log-normal model can be broadly applied to varieties with similar traits, thus providing a reliable framework for analyzing time-series VIs under diverse conditions (
Figure 8).
3.5. Effect of VI Patterns on Yield and Yield Components
The correlations between the time-series VI values fitted to a log-normal model and the yield are shown in
Figure 9. The R
2 values were consistently higher during the mid-to-late growth stages than during the early tillering stage for all VIs. This indicates that variations in VIs during the early growth phase have a limited influence on yield formation. The gradual increase in R
2 values during the early growth phase can be attributed to biomass accumulation and its relationship with yield [
35].
A detailed analysis focusing on the heading stage revealed that the R
2 values peaked during or immediately after the heading stage for most VIs. This indicates that the heading stage is closely related to yield formation and that changes in VIs after heading are crucial in yield prediction. Specifically, the increase in R
2 values after heading can be attributed to the physical canopy of rice leaves being overshadowed by panicles, which affects the VI values [
36]. This suggests that the rate and extent of VI decline after heading may be determined by the amount and speed of panicle emergence following nitrogen treatment. These findings highlight that VIs indicate biomass as well as structural changes, such as panicle formation, during the post-heading phase.
The timing of peak R2 values varied among VIs, with NDRE showing the earliest and strongest correlation with yield. This suggests that NDRE is particularly sensitive to early growth and nutrient conditions, forming a relationship with yield more rapidly than the other VIs. Conversely, the peak value of VI had less influence on yield than expected, and high standard deviations in VI values did not necessarily correlate with yield differences. The strongest correlations between VIs and yield were observed during the post-peak decline phase, indicating that decreases in VI values during the late growth stages significantly influenced yield-related factors.
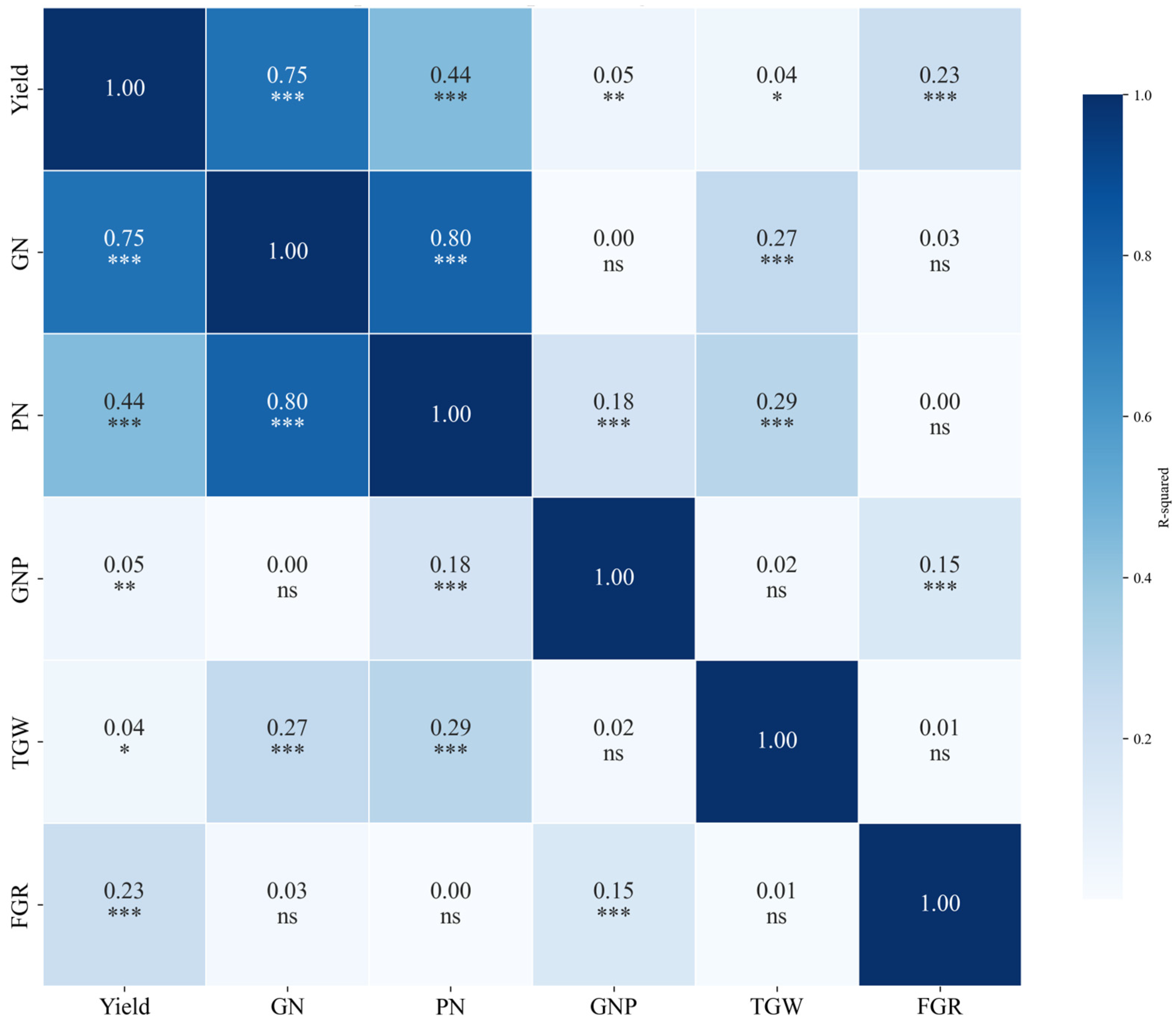
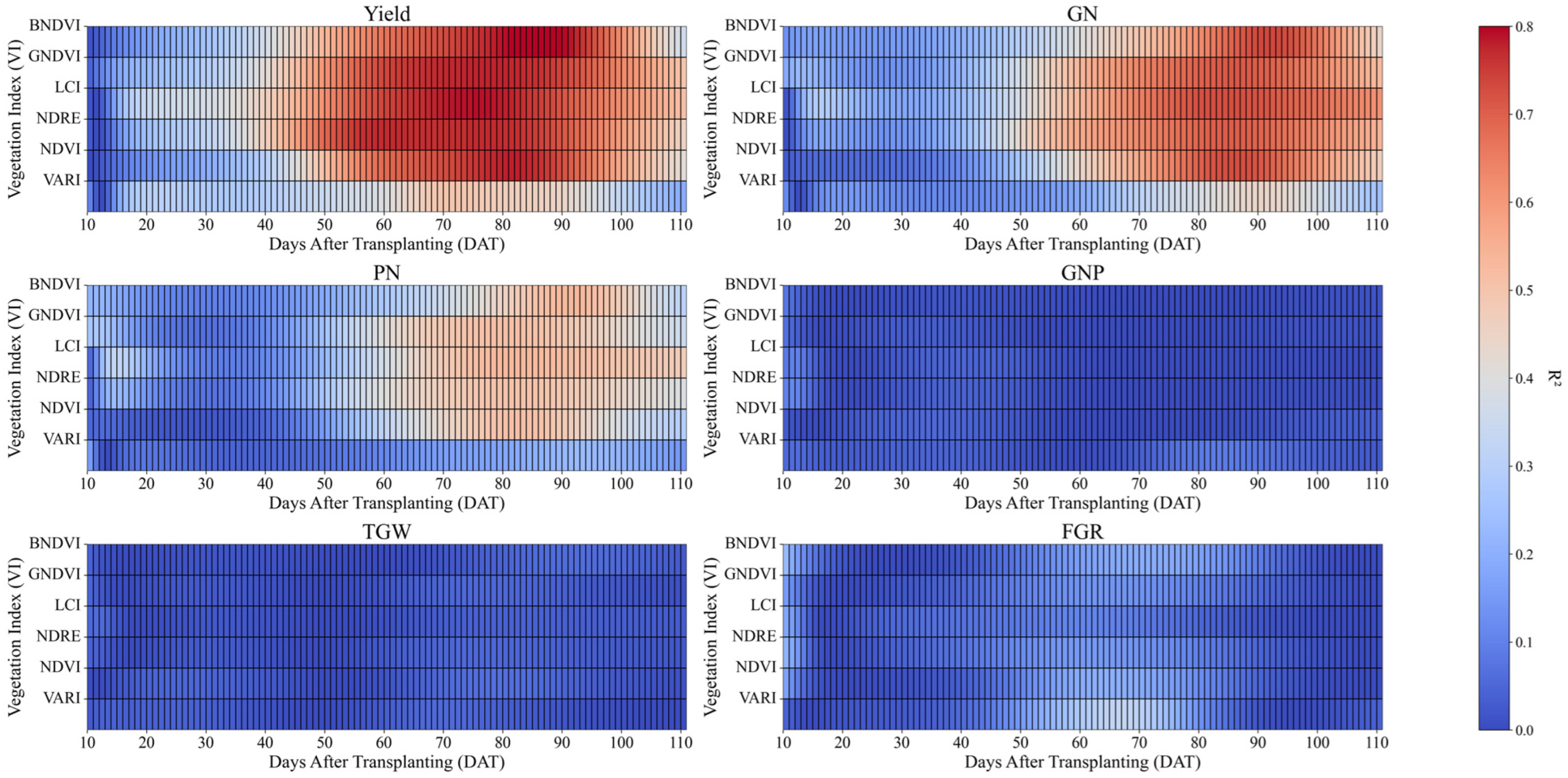
Additionally,
Figure 10 shows a heatmap of the R
2 values for each VI in relation to rice yield, GN, PN, GNP, TGW, and FGR. The overall correlation patterns between the VIs and yield components were similar, with R
2 values peaking earliest for yield, followed by GN and PN. The magnitude of R
2 was highest for yield, followed by GN and PN. This indicates that VIs are strongly linked to yield as well as to GN and PN, although the strength of these relationships varies by index. However, GNP, TGW, and FGR were not significantly correlated with yield or major yield components, such as GN and PN, at any growth stage. This suggests that these components have a limited influence on yield variability compared with other yield-related factors.
Table 7 summarizes the DAT at which each VI peaked and the maximum R
2 values for the yield, GN, and PN. It also includes the differences between the DAT at the peak VI value and DAT at the maximum R
2. R
2max typically occurred later than the peak VI values for all VIs, with this lag particularly pronounced for GN and PN. This suggests that vegetation growth patterns influenced GN and PN over an extended period, whereas the relationship between the VIs and yield was relatively immediate.
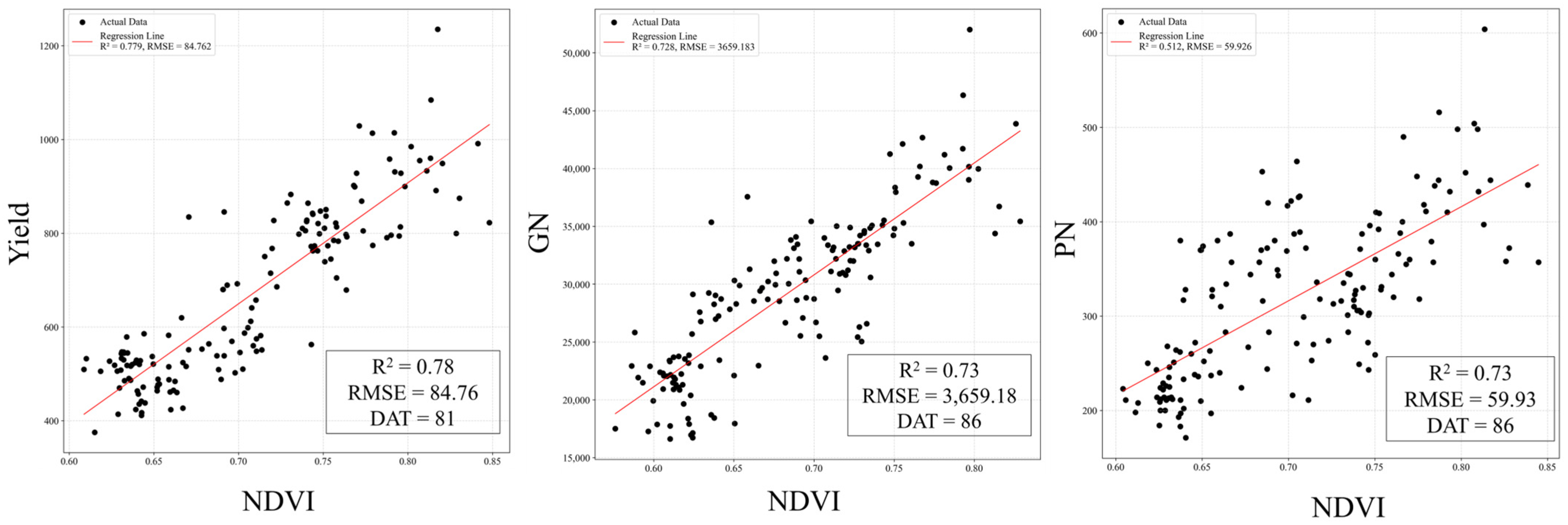
Figure 11 shows the regression analysis of the relationships between the VI values, highlighting the R
2 and RMSE values. For NDVI, the maximum R
2 with yield was observed at DAT 81 (R
2 = 0.78) with an RMSE of 84.76. For GN, the maximum R
2 was recorded at DAT 86 (R
2 = 0.73) with an RMSE of 3659.18. For PN, the maximum R
2 occurred at DAT 82 (R
2 = 0.51) with an RMSE of 59.95. These results indicate that VI data from specific time points can effectively explain the relationship between yield and its components. However, they also suggest that using VI data from inappropriate time points may lead to errors in yield estimation, underscoring the importance of selecting the correct timing for VI data in predictive analyses.
3.7. Performance Evaluation of Yield Estimation Models
The parameters of four types (
a,
b,
c, and
d) derived from individual samples fitted to the model were used as variables for each yield component in the multivariate regression analysis. The analysis employed a linear regression model, PLSR, which effectively handled multicollinearity while objectively identifying the variable characteristics and importance. Additionally, nonlinear machine learning models, such as RFR, GBR, and XGBR, were used to evaluate model performance. R
2 values were evaluated and determined using the LOOCV method. Regression graphs for the NDVI, models, and each yield component are shown in
Figure 13.
Among the VIs, LCI, NDRE, and NDVI consistently demonstrated high predictive performance across all models (
Table 8 and
Table 9). On average, LCI and NDRE were better predictors of GN and PN, respectively. LCI showed strong performance in nonlinear models such as RFR, XGBR, and GBR while maintaining a relatively high accuracy in PLSR. NDRE, with its sensitivity to early and mid-growth stages, achieved high predictive performance for GN and PN as well as for yield in XGBR and GBR. Similarly, NDVI exhibited excellent performance in yield prediction, with R
2 values of 0.82 in PLSR and RFR, comparable to NDRE.
NDVI showed moderate accuracy for GN prediction, with R2 values ranging from 0.58 to 0.76, depending on the model, demonstrating its utility in predicting grain production under diverse conditions. The NDVI performed reliably for PN prediction, achieving a peak R2 value of 0.74 RFR, indicating its effectiveness in representing panicle density. However, NDVI exhibited relatively lower predictive power for GNP, TGW, and FGR, with R2 values below 0.5 for most models, revealing its limitations in capturing meticulous details related to grain quality and weight.
For yield component prediction, GN and PN consistently recorded higher R2 values than GNP, TGW, and FGR for all VIs and models, thereby achieving greater predictive accuracy. In contrast, BNDVI showed moderate predictive performance for most yield components but relatively lower accuracy for PN compared to other indices. LCI and NDRE were the most stable and reliable VIs for predicting GN and PN.
Overall, the nonlinear models (XGBR, RFR, and GBR) outperformed PLSR in predicting complex traits such as GN and PN. However, the observed differences in performance among the regression models may stem from both the features of the data and the inherent capabilities of the models themselves. For example, nonlinear models might better capture complex relationships in the data, leading to higher accuracy in predicting traits like GN and PN compared to the linear PLSR model. However, the performance of each model can also be influenced by factors such as the size and quality of the training data, the choice of hyperparameters, and the specific evaluation metrics used. Further investigation is needed to disentangle the effects of data features and model capabilities on the observed performance differences. GNP, TGW, and FGR consistently recorded lower R2 values for all VIs, indicating their limited contributions to yield variability. Despite these limitations, NDVI remains a dependable indicator for predicting key yield components, such as GN and PN, reaffirming its significance in yield estimation models.
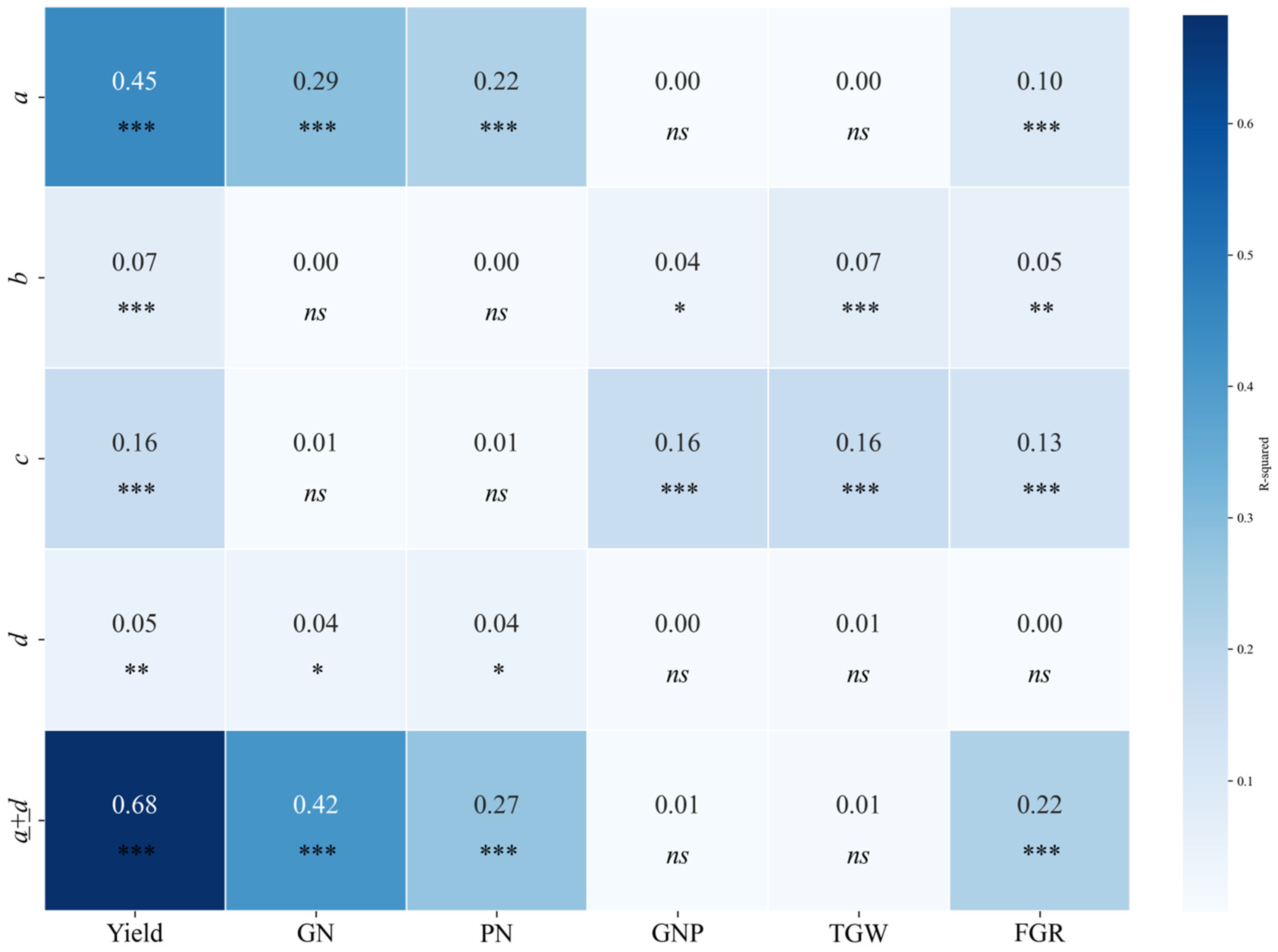
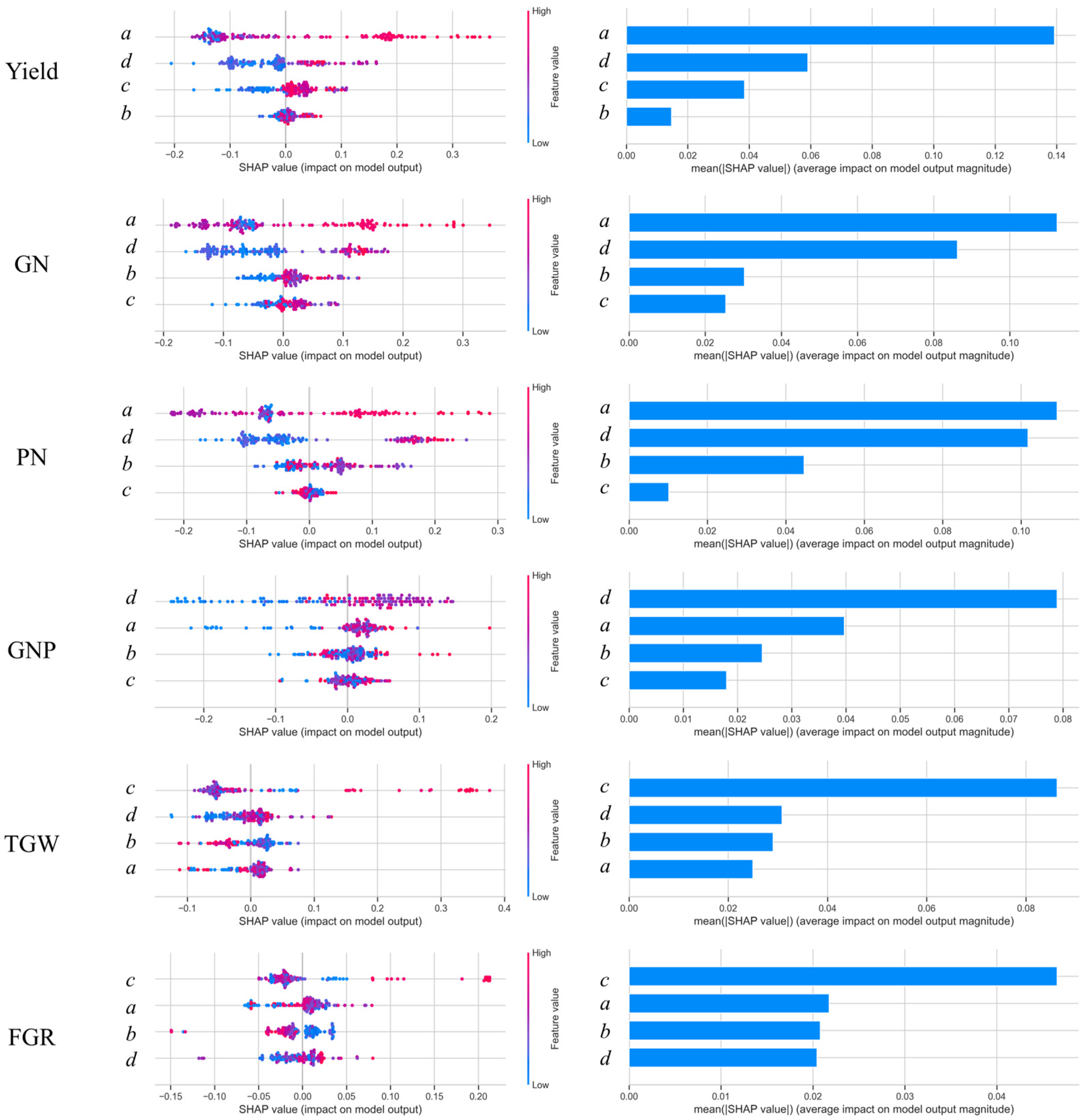
The SHAP value analysis for the NDVI model parameters highlighted the relative importance of four parameters (
a,
b,
c, and
d) in predicting the yield components (
Figure 14). Parameter
a, representing the peak value of the NDVI curve, consistently exhibited the highest contribution to the model outputs across all yield components, reaffirming its strong association with maximum vegetation vigor and yield. Parameter
c, indicative of the spread of the curve, also played a notable role, particularly for PN and GN, where broader distributions in the NDVI curves reflected extended growth activity critical for yield formation.
In contrast, parameters b and d showed varying effects depending on the yield component. For GN and PN, b was moderately important, suggesting that the timing of the peak NDVI was relevant for determining reproductive development. However, for secondary yield components, such as GNP and TGW, both b and d had comparatively lower contributions, indicating their reduced influence on finer yield traits.
Overall, the SHAP value analysis underscores the dominant roles of a and c in predicting yield-related traits, underscoring their importance in NDVI-based models. This finding highlights the critical effect of maximum VIs and growth distribution patterns on yield estimation.