Association Between InDel and CNV Variation in the FBLN1 Gene and Slaughter Traits in Cattle
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Collection of Animal Samples
2.3. Extraction of Genomic DNA
2.4. Primer Design
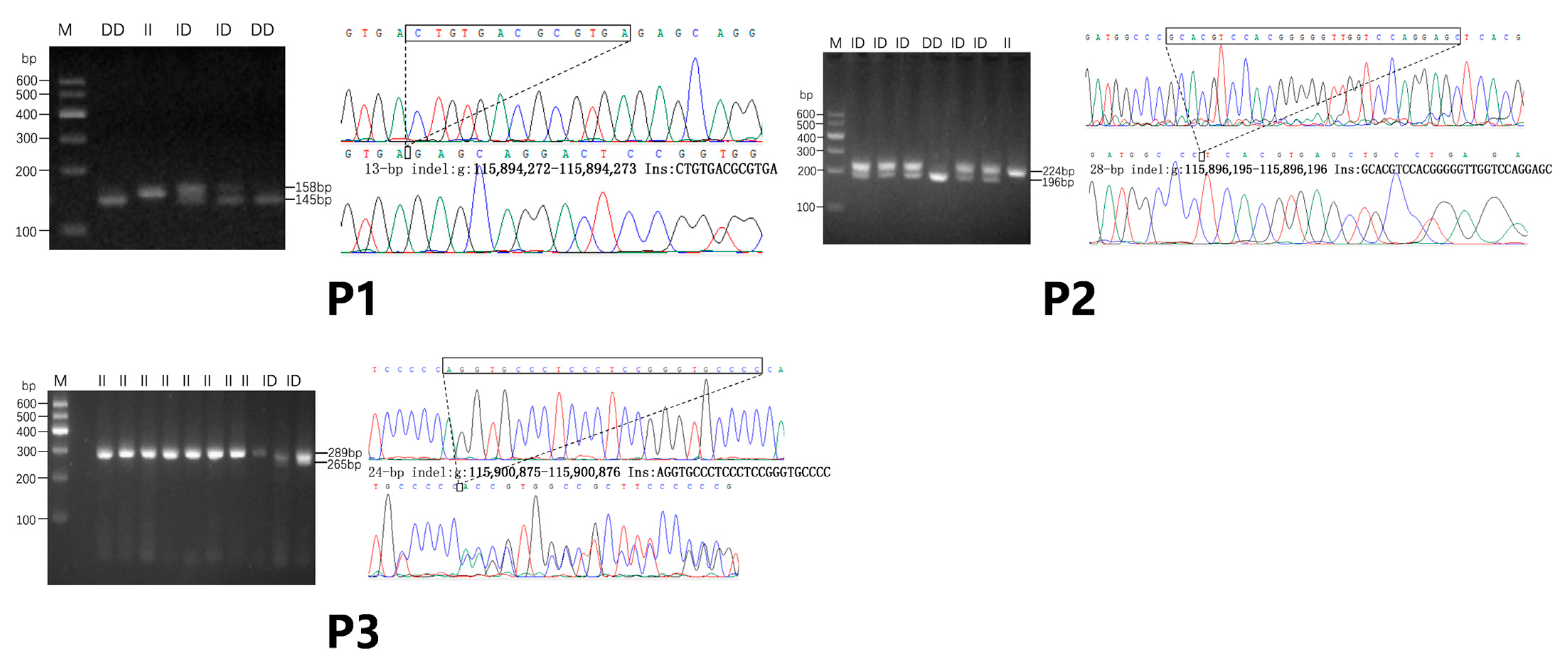
2.5. Identification and Genotyping of InDel Variants
2.6. Statistical Analysis
3. Results
3.1. Identification and Analysis of Bovine FBLN1 Gene
3.2. Estimation of InDel (P1, P2, P3) Polymorphism Parameters of FBLN1
3.3. Analysis of Linkage Disequilibrium (LD)
3.4. CNV Identification: Distribution of Genotypes in the Bovine FBLN1 Gene
3.5. Association Analysis of FBLN1 with Slaughter Traits
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| InDel | Insertion–deletion |
| CNV | Copy number variation |
| ECM | Extracellular matrix |
| NGS | Next-generation sequencing |
| RNA-seq | RNA sequencing |
| DEG | Differentially expressed gene |
| ALF | Adlibitum feeding |
| RF | Restricted feeding |
| GSEA | Gene set enrichment analysis |
| LD | Linkage disequilibrium |
| 5′ UTR | 5′ Untranslated region |
References
- Pałka, S.E.; Drąg-Kozak, E.; Migdał, Ł.; Kmiecik, M. Effect of a Diet Supplemented with Nettle (Urtica dioica L.) or Fenugreek (Trigonella foenum-graecum L.) on the Content of Selected Heavy Metals in Liver and Rabbit Meat. Animals 2022, 12, 827. [Google Scholar] [CrossRef] [PubMed]
- Barro, A.G.; Marestone, B.S.; dos S, E.R.; Ferreira, G.A.; Vero, J.G.; Terto, D.K.; de SD Muniz, C.A.; Bridi, A.M. Genetic Parameters for Frame Size and Carcass Traits in Nellore Cattle. Trop. Anim. Health. Prod. 2023, 55, 71. [Google Scholar] [CrossRef] [PubMed]
- da Silva Neto, J.B.; Peripoli, E.; Pereira, A.S.C.; Stafuzza, N.B.; Lôbo, R.B.; Fukumasu, H.; Ferraz, J.B.S.; Baldi, F. Weighted Genomic Prediction for Growth and Carcass-Related Traits in Nelore Cattle. Anim. Genet. 2023, 54, 271–283. [Google Scholar] [CrossRef] [PubMed]
- Deng, T.; Liang, M.; Du, L.; Li, K.; Li, J.; Qian, L.; Xue, Q.; Qiu, S.; Xu, L.; Zhang, L.; et al. Transcriptome Analysis of Compensatory Growth and Meat Quality Alteration after Varied Restricted Feeding Conditions in Beef Cattle. Int. J. Mol. Sci. 2024, 25, 2704. [Google Scholar] [CrossRef]
- Debeer, P.; Schoenmakers, E.F.P.M.; Twal, W.O.; Argraves, W.S.; De Smet, L.; Fryns, J.P.; Van De Ven, W.J.M. The Fibulin-1 Gene (FBLN1) Is Disrupted in a t (12;22) Associated with a Complex Type of Synpolydactyly. J. Med. Genet. 2002, 39, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Karamanos, N.K.; Theocharis, A.D.; Piperigkou, Z.; Manou, D.; Passi, A.; Skandalis, S.S.; Vynios, D.H.; Orian-Rousseau, V.; Ricard-Blum, S.; Schmelzer, C.E.H.; et al. A Guide to the Composition and Functions of the Extracellular Matrix. FEBS J. 2021, 288, 6850–6912. [Google Scholar] [CrossRef]
- Yang, H.; Wang, W.; Liu, H.; Zhang, C.; Cao, Y.; Long, L.; Han, X.; Wang, Y.; Yan, F.; Li, G.; et al. miR615-3p Inhibited FBLN1 and Osteogenic Differentiation of Umbilical Cord Mesenchymal Stem Cells by Associated with YTHDF2 in a m6A-miRNA Interaction Manner. Cell Prolif. 2024, 57, e13607. [Google Scholar] [CrossRef] [PubMed]
- Singh, U.; Sun, T.; Larsson, T.; Elliott, R.W.; Kostka, G.; Fundele, R.H. Expression and Functional Analysis of Fibulin-1 (Fbln1) during Normal and Abnormal Placental Development of the Mouse. Placenta 2006, 27, 1014–1021. [Google Scholar] [CrossRef]
- McCoy, M.D.; Sarasua, S.M.; DeLuca, J.M.; Davis, S.; Phelan, K.; Rogers, R.C.; Boccuto, L. State of the Science for Kidney Disorders in Phelan-McDermid Syndrome: UPK3A, FBLN1, WNT7B, and CELSR1 as Candidate Genes. Genes 2022, 13, 1042. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Ye, M.; Chen, X.; Zhao, H.; Hasim, A.; Guo, X. Discovery and Validation of FBLN1 and ANT3 as Potential Biomarkers for Early Detection of Cervical Cancer. Cancer Cell Int. 2021, 21, 125. [Google Scholar] [CrossRef]
- Pan, Y.; Fei, L.; Wang, S.; Chen, H.; Jiang, C.; Li, H.; Wang, C.; Yang, Y.; Zhang, Q.; Chen, Y. Integrated Analysis of Single-Cell, Spatial and Bulk RNA-Sequencing Identifies a Cell-Death Signature for Predicting the Outcomes of Head and Neck Cancer. Front. Immunol. 2024, 15, 1487966. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A Revolutionary Tool for Transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- MacArthur Clark, J.A.; Sun, D. Guidelines for the Ethical Review of Laboratory Animal Welfare People’s Republic of China National Standard GB/T 35892-2018 [Issued 6 February 2018 Effective from 1 September 2018]. Anim. Models Exp. Med. 2020, 3, 103–113. [Google Scholar] [CrossRef] [PubMed]
- GB/T 27643-2011; Beef Carcass and Cuts. The Standardization Administration of the People’s Republic of China: Beijing, China, 2011.
- Aljanabi, S.M.; Martinez, I. Universal and Rapid Salt-Extraction of High Quality Genomic DNA for PCR-Based Techniques. Nucleic Acids. Res. 1997, 25, 4692–4693. [Google Scholar] [CrossRef] [PubMed]
- Berim, G.O.; Ruckenstein, E. Calculation of Nanodrop Profile from Fluid Density Distribution. Adv. Colloid Interface Sci. 2016, 231, 15–22. [Google Scholar] [CrossRef]
- Gao, Y.; Jiang, J.; Yang, S.; Hou, Y.; Liu, G.E.; Zhang, S.; Zhang, Q.; Sun, D. CNV Discovery for Milk Composition Traits in Dairy Cattle Using Whole Genome Resequencing. BMC Genom. 2017, 18, 265. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Bi, Y.; Wang, Z.; Zhu, H.; Liu, M.; Wu, X.; Pan, C. Goat SNX29: mRNA Expression, InDel and CNV Detection, and their associations with litter Size. Front. Vet. Sci. 2022, 9, 981315. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing Real-Time PCR Data by the Comparative CT Method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Nei, M. Analysis of Gene Diversity in Subdivided Populations. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3323. [Google Scholar] [CrossRef]
- Graffelman, J.; Weir, B.S. The Transitivity of the Hardy-Weinberg Law. Forensic. Sci. Int. Genet. 2022, 58, 102680. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Guo, X.; Yongfu, L.; Wang, T.; Bao, P.; Chu, M.; Wu, X.; Yan, P.; Liang, C. Identification of circRNA-Associated ceRNA Networks in the Longissimus Dorsi of Yak under Different Feeding Systems. BMC Vet. Res. 2024, 20, 67. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.; Sun, Y.; Han, Y.; Liu, Y.; Jin, S. Transcriptome Comparison Revealed the Difference in Subcutaneous Fat Metabolism of Qinghai Yak under Different Feeding Conditions. PLoS ONE 2024, 19, e0311224. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Jiang, E.; Zhang, K.; Zhang, S.; Jiang, F.; Song, E.; Chen, H.; Guo, P.; Lan, X. Genetic Variations within the Bovine CRY2 Gene Are Significantly Associated with Carcass Traits. Animals 2022, 12, 1616. [Google Scholar] [CrossRef] [PubMed]
- Kenny, D.; Murphy, C.P.; Sleator, R.D.; Evans, R.D.; Berry, D.P. Contribution of Herd Characteristics to Best Linear Unbiased Estimates of Slaughter Traits in Beef Cattle. Animal 2021, 15, 100321. [Google Scholar] [CrossRef]
- Kelly, D.N.; Sleator, R.D.; Murphy, C.P.; Conroy, S.B.; Berry, D.P. Phenotypic and Genetic Associations between Feeding Behavior and Carcass Merit in Crossbred Growing Cattle. J. Anim. Sci. 2021, 99, skab285. [Google Scholar] [CrossRef] [PubMed]
- Cantarero-Aparicio, M.Á.; Angón, E.; González-Esquivel, C.; Peña, F.; Caballero-Villalobos, J.; Ryan, E.G.; Perea, J.M. Carcass and Meat Quality Traits in Female Lidia Cattle Slaughtered at Different Ages. Animals 2024, 14, 850. [Google Scholar] [CrossRef]
- de Carvalho Porto Barbosa, M.; Fioravanti, M.C.S.; Peripolli, V.; do Egito, A.A.; Juliano, R.S.; Ramos, A.F.; Cardoso, D.; Laudares, K.M.; Feijó, G.L.D.; Prado, C.S.; et al. Performance, Carcass, and Meat Traits of Locally Adapted Brazilian Cattle Breeds under Feedlot Conditions. Trop. Anim. Health. Prod. 2023, 55, 243. [Google Scholar] [CrossRef] [PubMed]
- Tran, H.; VanDusen, W.J.; Argraves, W.S. The Self-Association and Fibronectin-Binding Sites of Fibulin-1 Map to Calcium-Binding Epidermal Growth Factor-like Domains. J. Biol. Chem. 1997, 272, 22600–22606. [Google Scholar] [CrossRef]
- Fujiwara, H.; Ferreira, M.; Donati, G.; Marciano, D.K.; Linton, J.M.; Sato, Y.; Hartner, A.; Sekiguchi, K.; Reichardt, L.F.; Watt, F.M. The Basement Membrane of Hair Follicle Stem Cells Is a Muscle Cell Niche. Cell 2011, 144, 577–589. [Google Scholar] [CrossRef]
- Mattei, M.G.; Pan, T.C.; Zhang, R.Z.; Timpl, R.; Chu, M.L. The Fibulin-1 Gene (FBLN1) Is Located on Human Chromosome 22 and on Mouse Chromosome 15. Genomics 1994, 22, 437–438. [Google Scholar] [CrossRef]
- Tang, R.; Jian, Y.; Hu, Z.; Li, L.; Wang, H.; Miao, P.; Yang, Z.; Tang, M. A Bioinformatics-Based Study on Methylation Alterations of the FBLN1 Gene in Hippocampal Tissue of Alzheimer’s Disease Model DKO and DTG Mice. Int. J. Mol. Sci. 2024, 25, 9036. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Geng, X.; Yang, F.; Zhang, H. FBLN1 Promotes Chondrocyte Proliferation by Increasing Phosphorylation of Smad2. J. Orthop. Sci. 2022, 27, 242–248. [Google Scholar] [CrossRef]
- Raman, R.; Antony, M.; Nivelle, R.; Lavergne, A.; Zappia, J.; Guerrero-Limón, G.; Caetano da Silva, C.; Kumari, P.; Sojan, J.M.; Degueldre, C.; et al. The Osteoblast Transcriptome in Developing Zebrafish Reveals Key Roles for Extracellular Matrix Proteins Col10a1a and Fbln1 in Skeletal Development and Homeostasis. Biomolecules 2024, 14, 139. [Google Scholar] [CrossRef]
- Shen, S.; Zhu, L.; Yang, Y.; Bi, Y.; Li, J.; Wang, Y.; Pan, C.; Wang, S.; Lan, X. Exploration of the Polymorphism Distribution of Bovine HMGA2 Gene in Worldwide Breeds and Its Associations with Ovarian Traits. Animals 2024, 14, 796. [Google Scholar] [CrossRef]
- Mattick, J.S.; Gagen, M.J. The Evolution of Controlled Multitasked Gene Networks: The Role of Introns and Other Noncoding RNAs in the Development of Complex Organisms. Mol. Biol. Evol. 2001, 18, 1611–1630. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Liu, P.; Zhang, M.; Kang, Y.; Lv, L.; Xu, H.; Zhang, Q.; Li, R.; Pan, C.; Lan, X. The Detection of a Functional 168 Bp Deletion of the HOXB13 Gene Determining Short Tail and Its Association with Senior Growth Traits in Sheep Breeds Worldwide. Animals 2024, 14, 1617. [Google Scholar] [CrossRef]
- Rose, A.B. Introns as Gene Regulators: A Brick on the Accelerator. Front. Genet. 2019, 9, 672. [Google Scholar] [CrossRef] [PubMed]



| Primer Names | Primer Sequences(5′-3′) | Region | Product Sizes (bp) | Polymorphisms |
|---|---|---|---|---|
| P1 | F:TGAGAGTAAGCTCAGAAACGGA R:TGCTGCTAACCTCTGAGTTCC | Intron1 | 145/158 | Yes |
| P2 | F:GCTTCAGTTTCCAAAGGCCG R:CCCTGAGTAGGTGACGAGA | Intron1 | 196/224 | Yes |
| P3 | F:CTGCAATTGAAGCACCTGGAT R:GGGCTCAGAGACGGTTTGTC | Intron1 | 265/289 | Yes |
| P4 | F:CGGATGCGCTAACAAGAAGTC R:CAGTATTATGGCCCCCTGCC | Intron1 | 95/107 | No |
| P5 | F:AGTGACCTCTCAGCAAGGGT R:AGGGAGGGACAGCCTAGTTT | Intron4 | 234/252 | No |
| Primers | Chromosome | Start | End | Length | Location |
|---|---|---|---|---|---|
| CNV1 | 5 | 115,918,196 | 115,920,996 | 2800 | Exon7 |
| CNV2 | 5 | 115,937,723 | 115,940,523 | 2800 | Intron14 |
| Primers | Primer Sequences (5′–3′) | Sizes (bp) |
|---|---|---|
| CNV1 | F:CGCATGTGCTTTCTAGTCCC R:TCATGCTTTTTACGCAGCGG | 127 |
| CNV2 | F:CGAACCTTGGTTTGCTGACG R:CTTGAGAGGCACATTGGGGG | 140 |
| BTF3 | F:AACCAGGAGAAACTCGCCAA R:TTCGGTGAAATGCCCTCTCG | 166 |
| Loci | Sample Sizes | Genotypic Frequencies | Allelic Frequencies | HWE p-Value | Population Parameter Estimates | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| II | ID | DD | I | D | Ho | He | Ne | PIC | |||
| P1 | 641 | 0.168 (n = 108) | 0.557 (n = 357) | 0.275 (n = 176) | 0.447 | 0.553 | 0.0136 | 0.506 | 0.494 | 1.978 | 0.372 |
| P2 | 638 | 0.404 (n = 258) | 0.528 (n = 337) | 0.067 (n = 43) | 0.668 | 0.332 | 1.27 × 10−6 | 0.557 | 0.443 | 1.796 | 0.345 |
| P3 | 417 | 0.866 (n = 361) | 0.134 (n = 56) | 0 (n = 0) | 0.933 | 0.067 | 0.142 | 0.875 | 0.125 | 1.143 | 0.117 |
| Loci | Sizes (bp) | Genotypic Frequencies | ||
|---|---|---|---|---|
| Loss | Medium | Gain | ||
| CNV1 | 127 | 0.031 | 0.055 | 0.913 |
| CNV2 | 140 | 0.362 | 0.378 | 0.260 |
| Loci | Gender | Traits (kg) | Sample Size | Observed Genotypes (Mean ± SE) | p-Value | ||
|---|---|---|---|---|---|---|---|
| II | ID | DD | |||||
| P1 | Male | Three-rib S-cut abdomen | 94 | 1.38 ± 0.08 a (n = 19) | 1.35 ± 0.04 b (n = 59) | 1.12 ± 0.07 a (n = 16) | 0.040 |
| Female | Three-rib S-cut abdomen | 382 | 1.26 ± 0.04 A (n = 64) | 1.27 ± 0.03 A (n = 204) | 1.13 ± 0.03 B (n = 114) | 0.001 | |
| Left limbs weight | 388 | 213.36 ± 3.90 A (n = 65) | 210.15 ± 2.25 A (n = 207) | 199.75 ± 3.13 B (n = 116) | 0.007 | ||
| Right limbs weight | 388 | 214.32 ± 4.04 A (n = 65) | 210.68 ± 2.31 A (n = 207) | 199.71 ± 3.19 B (n = 116) | 0.005 | ||
| Entry weight | 388 | 329.14 ± 5.80 A (n = 65) | 329.39 ± 3.47 A (n = 207) | 306.09 ± 4.53 B (n = 116) | 0.000134 | ||
| Beef neck edge | 386 | 1.28 ± 0.03 A (n = 65) | 1.26 ± 0.02 A (n = 206) | 1.15 ± 0.03 B (n = 115) | 0.000450 | ||
| Short rib | 379 | 3.21 ± 0.08 ab (n = 62) | 3.25 ± 0.05 a (n = 204) | 3.01 ± 0.06 b (n = 113) | 0.012 | ||
| Beef slices | 384 | 2.04 ± 0.05 A (n = 65) | 2.05 ± 0.03 A (n = 205) | 1.87 ± 0.04 B (n = 114) | 0.001 | ||
| Triangular brisket | 395 | 6.47 ± 0.14 A (n = 65) | 6.19 ± 0.09 A (n = 213) | 5.80 ± 0.13 B (n = 117) | 0.003 | ||
| High rib | 397 | 16.65 ± 0.36 A (n = 67) | 16.41 ± 0.22 A (n = 214) | 15.38 ± 0.30 B (n = 116) | 0.007 | ||
| Sirloin | 393 | 13.04 ± 0.26 A (n = 67) | 12.78 ± 0.16 A (n = 210) | 11.73 ± 0.24 B (n = 116) | 0.000139 | ||
| P2 | Male | Right limbs weight | 94 | 223.15 ± 5.20 b (n = 31) | 230.57 ± 3.79 b (n = 59) | 262.00 ± 21.83 a (n = 4) | 0.048 |
| Triangular brisket | 106 | 6.72 ± 0.19 B (n = 42) | 7.35 ± 0.16 A (n = 60) | 8.63 ± 0.62 A (n = 4) | 0.003 | ||
| Three-rib S-cut abdomen | 97 | 1.21 ± 0.07 b (n = 33) | 1.37 ± 0.04 ab (n = 60) | 1.56 ± 0.29 a (n = 4) | 0.047 | ||
| High rib | 107 | 16.88 ± 0.84 b (n = 42) | 19.78 ± 0.52 ab (n = 61) | 22.60 ± 1.98 a (n = 4) | 0.012 | ||
| Sirloin | 103 | 12.43 ± 0.36 b (n = 38) | 13.51 ± 0.30 ab (n = 61) | 15.50 ± 1.34 a (n = 4) | 0.012 | ||
| Female | Entry weight | 385 | 313.34 ± 3.83 B (n = 161) | 329.91 ± 3.60 A (n = 204) | 315.75 ± 7.82 AB (n = 20) | 0.006 | |
| Left limbs weight | 385 | 201.98 ± 2.76 b (n = 161) | 211.02 ± 2.16 a (n = 204) | 213.03 ± 8.17 a (n = 20) | 0.026 | ||
| Right limbs weight | 385 | 202.27 ± 2.82 b (n = 161) | 211.38 ± 2.22 a (n = 204) | 215.10 ± 8.43 a (n = 20) | 0.024 | ||
| Beef neck edge | 383 | 1.18 ± 0.02 b (n = 159) | 1.25 ± 0.02 a (n = 204) | 1.26 ± 0.08 a (n = 20) | 0.047 | ||
| Beef slices | 382 | 1.93 ± 0.03 b (n = 159) | 2.05 ± 0.03 a (n = 203) | 1.94 ± 0.09 ab (n = 20) | 0.026 | ||
| Triangular brisket | 393 | 5.93 ± 0.12 b (n = 164) | 6.28 ± 0.09 a (n = 208) | 6.05 ± 0.22 ab (n = 21) | 0.041 | ||
| High rib | 394 | 15.79 ± 0.26 b (n = 164) | 16.52 ± 0.21 a (n = 208) | 15.27 ± 0.79 b (n = 22) | 0.038 | ||
| Sirloin | 389 | 12.16 ± 0.21 b (n = 161) | 12.82 ± 0.15 a (n = 206) | 11.89 ± 0.52 b (n = 22) | 0.017 | ||
| P3 | Female | Short rib | 291 | 3.16 ± 0.05 b (n = 251) | 3.50 ± 0.18 a (n = 40) | 0.012 | |
| Meat tendon | 218 | 4.88 ± 0.36 b (n = 188) | 7.80 ± 1.01 a (n = 30) | 0.01 | |||
| Variation | Gender | Traits (kg) | Sample Size | Observed Genotypes (Mean ± SE) | p-Value | ||
|---|---|---|---|---|---|---|---|
| Loss | Medium | Gain | |||||
| CNV1 | Female | Carcass fat | 113 | 84.10 ± 1.11 B (n = 5) | 91.45 ± 1.55 A (n = 108) | 0.001 | |
| CNV2 | Female | Skirt steak | 115 | 1.95 ± 0.06 b (n = 42) | 2.18 ± 0.06 a (n = 41) | 2.14 ± 0.06 ab (n = 32) | 0.019 |
| Chunky II | 115 | 38.46 ± 1.36 B (n = 42) | 43.71 ± 1.21 A (n = 41) | 44.70 ± 1.36 A (n = 32) | 0.002 | ||
| Beef plate | 113 | 2.70 ± 0.07 b (n = 41) | 2.92 ± 0.06 a (n = 40) | 2.73 ± 0.08 ab (n = 32) | 0.049 | ||
| Beef short plate | 114 | 6.12 ± 0.17 (n = 41) | 6.48 ± 0.19 (n = 41) | 6.77 ± 0.19 (n = 32) | 0.05 | ||
| Three-rib S-cut abdomen | 113 | 1.29 ± 0.05 b (n = 41) | 1.46 ± 0.05 a (n = 41) | 1.30 ± 0.04 b (n = 31) | 0.02 | ||
| High rib | 115 | 16.19 ± 0.42 B (n = 42) | 17.85 ± 0.36 A (n = 41) | 17.51 ± 0.45 A (n = 32) | 0.009 | ||
| Oxtail | 111 | 0.68 ± 0.04 b (n = 41) | 0.75 ± 0.03 ab (n = 39) | 0.83 ± 0.04 a (n = 31) | 0.013 | ||
| Knuckle tendon | 109 | 1.18 ± 0.05 b (n = 41) | 1.25 ± 0.05 ab (n = 39) | 1.39 ± 0.08 a (n = 29) | 0.043 | ||
| Ribeye | 115 | 11.20 ± 0.33 b (n = 42) | 12.23 ± 0.21 a (n = 41) | 12.10 ± 0.35 a (n = 32) | 0.026 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, H.; Zhu, Q.; Li, Y.; Zhang, Y.; Zhang, C.; Mao, C.; Jiang, F.; Pan, C.; Lan, X.; Deng, T. Association Between InDel and CNV Variation in the FBLN1 Gene and Slaughter Traits in Cattle. Agriculture 2025, 15, 518. https://doi.org/10.3390/agriculture15050518
Gu H, Zhu Q, Li Y, Zhang Y, Zhang C, Mao C, Jiang F, Pan C, Lan X, Deng T. Association Between InDel and CNV Variation in the FBLN1 Gene and Slaughter Traits in Cattle. Agriculture. 2025; 15(5):518. https://doi.org/10.3390/agriculture15050518
Chicago/Turabian StyleGu, Hongye, Qihui Zhu, Yafang Li, Yuli Zhang, Chiyuan Zhang, Cui Mao, Fugui Jiang, Chuanying Pan, Xianyong Lan, and Tianyu Deng. 2025. "Association Between InDel and CNV Variation in the FBLN1 Gene and Slaughter Traits in Cattle" Agriculture 15, no. 5: 518. https://doi.org/10.3390/agriculture15050518
APA StyleGu, H., Zhu, Q., Li, Y., Zhang, Y., Zhang, C., Mao, C., Jiang, F., Pan, C., Lan, X., & Deng, T. (2025). Association Between InDel and CNV Variation in the FBLN1 Gene and Slaughter Traits in Cattle. Agriculture, 15(5), 518. https://doi.org/10.3390/agriculture15050518





