The m6A Methylation Profile Identified That OsHMT9.1 Deregulates Chromium Toxicity in Rice (Oryza sativa L.) Through Negative Regulatory Functions
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Phenotypic and Ion Concentration Measurements
2.3. Semi-Thin Section of Rice Root Tip
2.4. m6A Immunoprecipitation (IP) and cDNA Library Construction
2.5. High-Throughput MeRIP-Seq and RNA-Seq
2.6. Vector Construction and Transgenic Rice Production
2.7. Expression Pattern Analysis of OsHMT9.1
2.8. Subcellular Localization of OsHMT9.1
3. Results
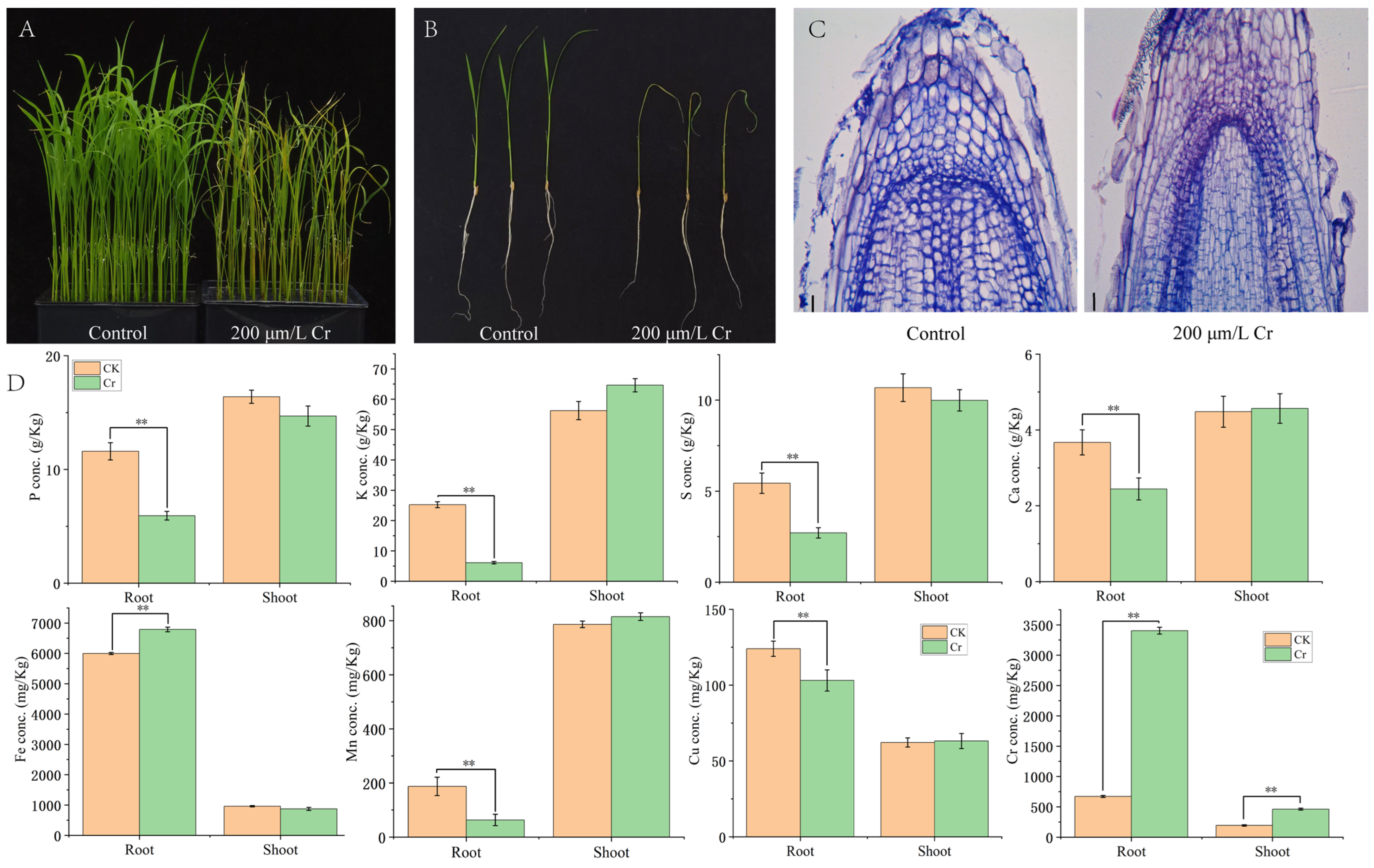
3.1. Phenotypic and Ionic Responses of Rice Seedlings to Cr Stress
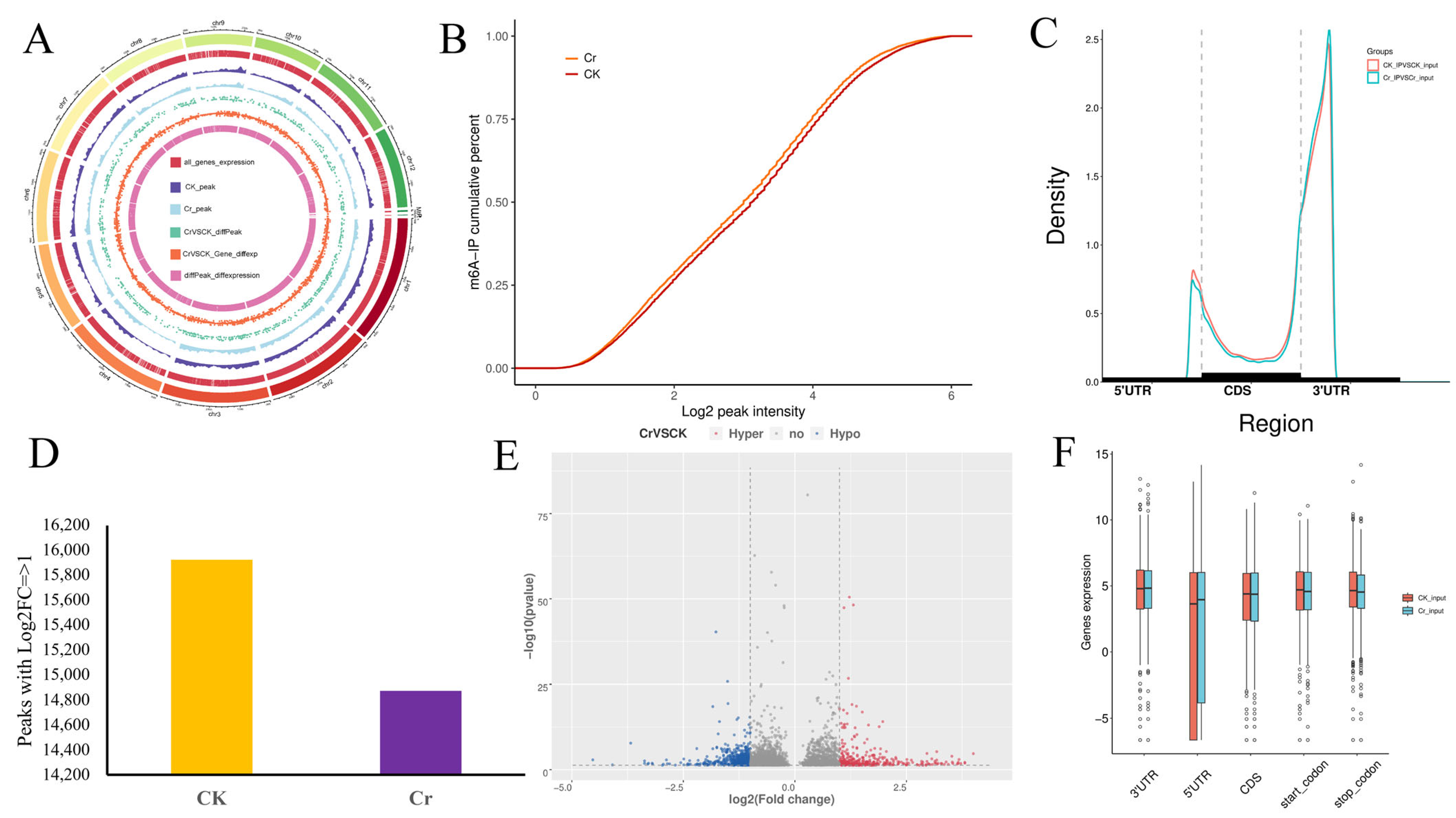
3.2. The Overview of Transcriptome-Wide m6A Methylation in the Roots of Rice Under Cr Stress
3.3. Differentially m6A-Modified Peaks and Genes in Response to Cr
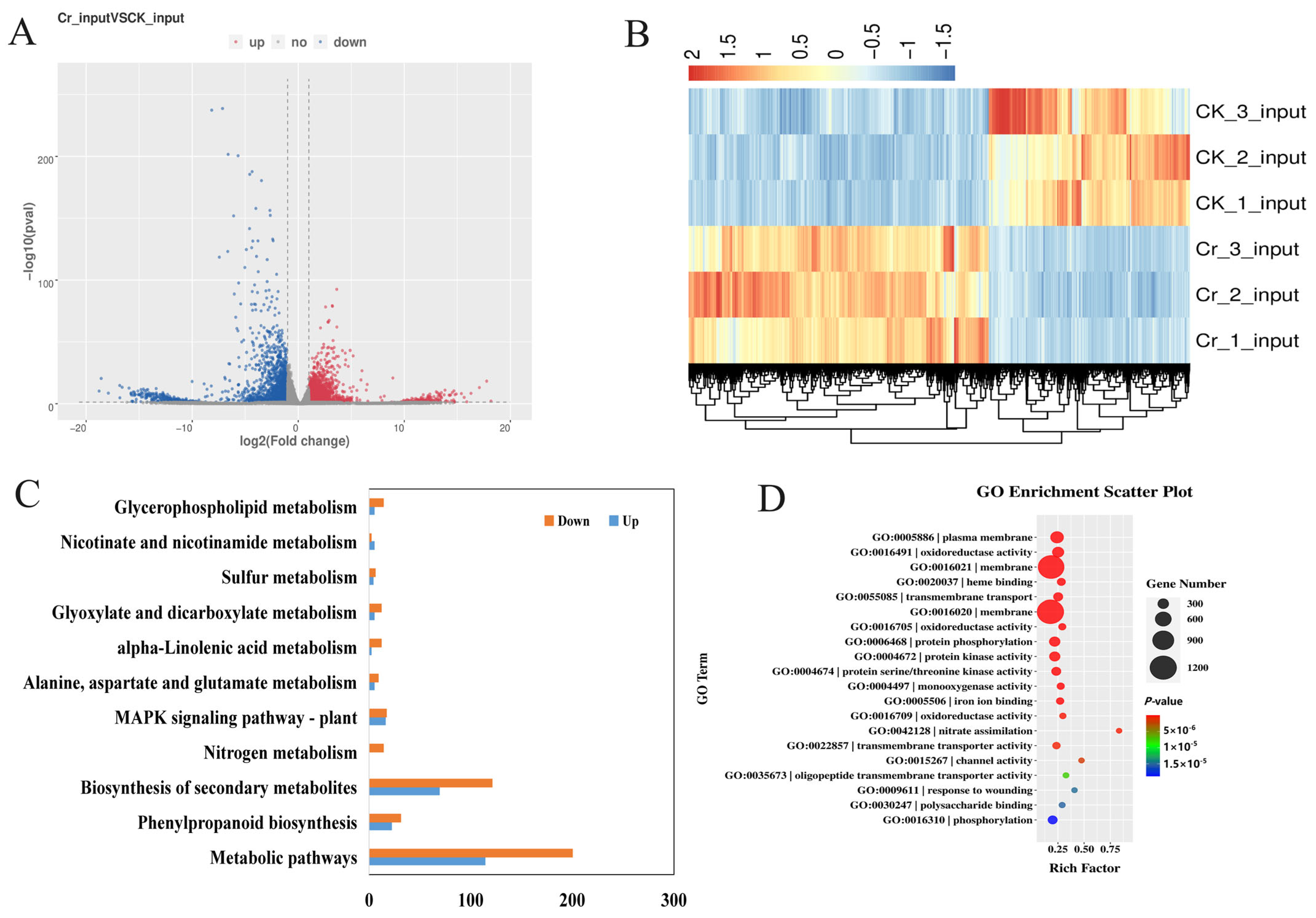
3.4. Differentially Expressed Genes in Rice Roots Under Cr Stress
3.5. Differentially m6A-Modified DEGs in Rice Roots Under Cr Stress
3.6. DEGs and DMPs Play Roles of Cr Stress Response in Rice Roots
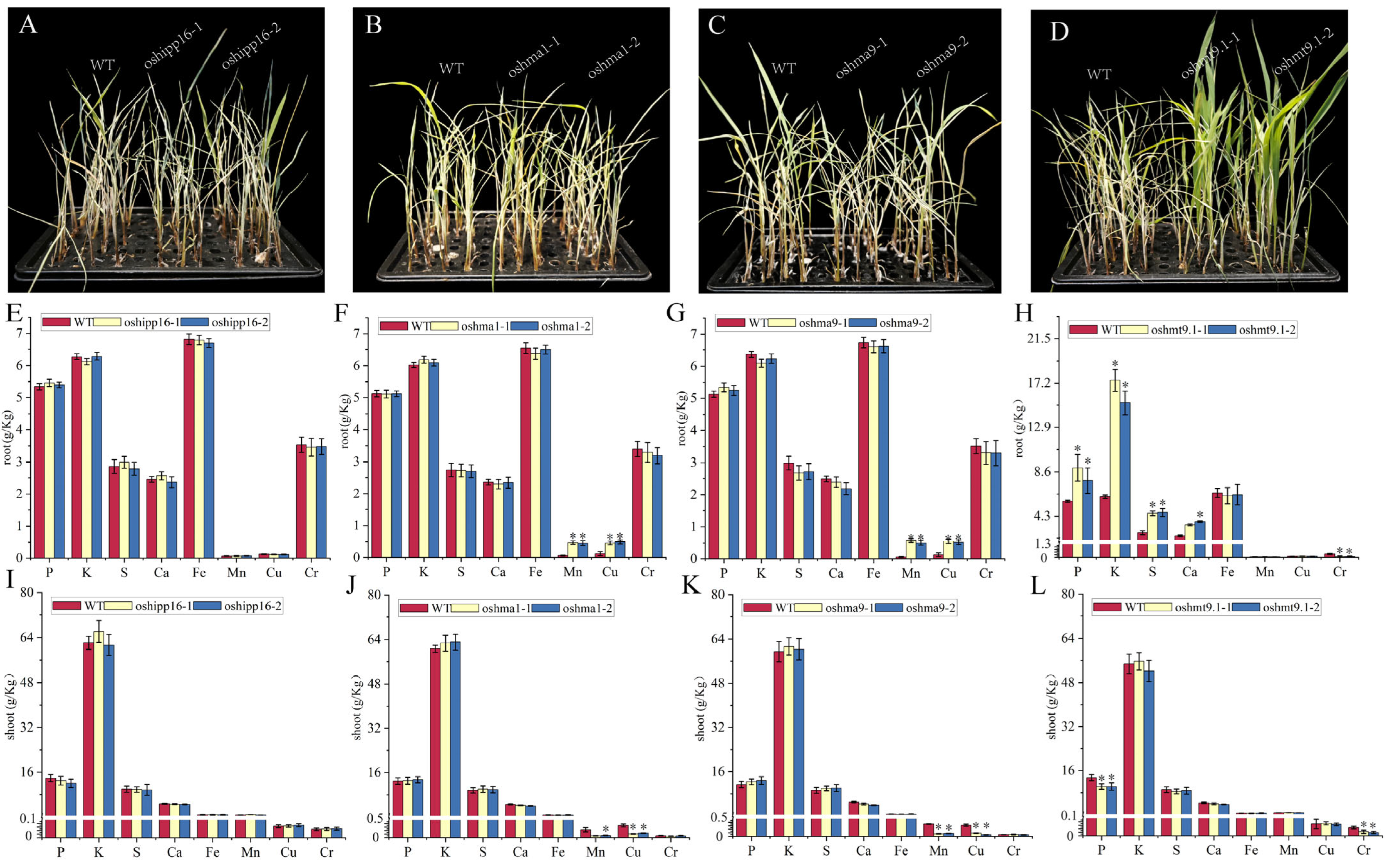
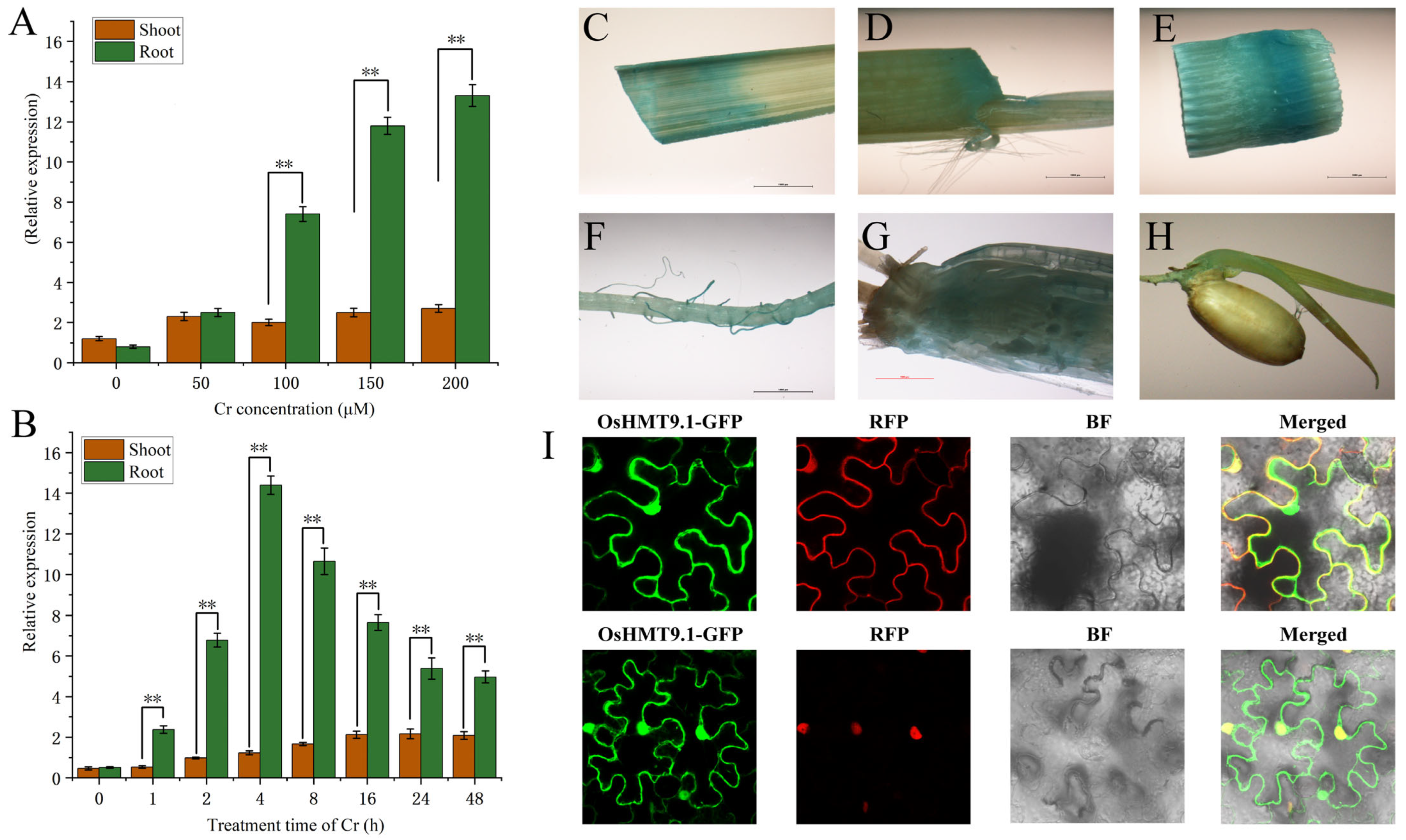
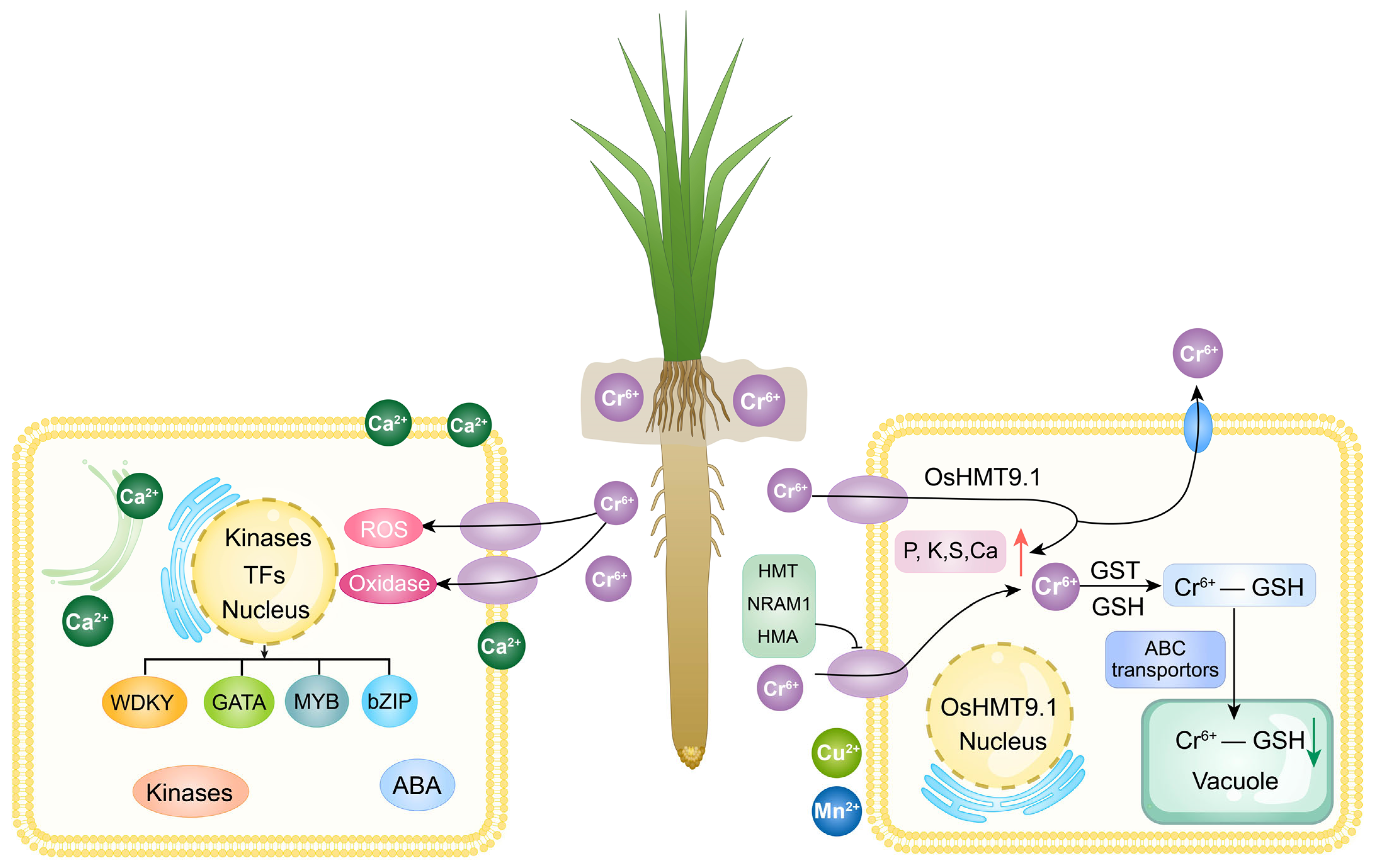
3.7. OsHMT9.1 Negatively Regulates Chromium Stress in Rice
3.8. Expression Characteristics of OsHMT9.1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Ali, S.; Mir, R.A.; Tyagi, A.; Manzar, N.; Kashyap, A.S.; Mushtaq, M.; Raina, A.; Park, S.; Sharma, S.; Mir, Z.A.; et al. Chromium Toxicity in Plants: Signaling, Mitigation, and Future Perspectives. Plants 2023, 12, 1502. [Google Scholar] [CrossRef] [PubMed]
- Rahman, Z.; Singh, V.P. The relative impact of toxic heavy metals (THMs) (arsenic (As), cadmium (Cd), chromium (Cr)(VI), mercury (Hg), and lead (Pb)) on the total environment: An overview. Environ. Monit. Assess. 2019, 191, 419. [Google Scholar] [CrossRef] [PubMed]
- Shanker, A.K.; Cervantes, C.; Loza-Tavera, H.; Avudainayagam, S. Chromium toxicity in plants. Environ. Int. 2005, 31, 739–753. [Google Scholar] [CrossRef] [PubMed]
- Panda, S.K. Chromium-mediated oxidative stress and ultrastructural changes in root cells of developing rice seedlings. J. Plant Physiol. 2007, 164, 1419–1428. [Google Scholar] [CrossRef]
- Sudo, E.; Itouga, M.; Yoshida-Hatanaka, K.; Ono, Y.; Sakakibara, H. Gene expression and sensitivity in response to copper stress in rice leaves. J. Exp. Bot. 2008, 59, 3465–3474. [Google Scholar] [CrossRef]
- Wei, L.H.; Song, P.; Wang, Y.; Lu, Z.; Tang, Q.; Yu, Q.; Xiao, Y.; Zhang, X.; Duan, H.C.; Jia, G. The m(6)A Reader ECT2 Controls Trichome Morphology by Affecting mRNA Stability in Arabidopsis. Plant Cell 2018, 30, 968–985. [Google Scholar] [CrossRef]
- Arribas-Hernández, L.; Bressendorff, S.; Hansen, M.H.; Poulsen, C.; Erdmann, S.; Brodersen, P. An m(6)A-YTH Module Controls Developmental Timing and Morphogenesis in Arabidopsis. Plant Cell 2018, 30, 952–967. [Google Scholar] [CrossRef]
- Duan, H.C.; Wei, L.H.; Zhang, C.; Wang, Y.; Chen, L.; Lu, Z.; Chen, P.R.; He, C.; Jia, G. ALKBH10B Is an RNA N(6)-Methyladenosine Demethylase Affecting Arabidopsis Floral Transition. Plant Cell 2017, 29, 2995–3011. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Liang, Z.; Gu, X.; Chen, Y.; Teo, Z.W.; Hou, X.; Cai, W.M.; Dedon, P.C.; Liu, L.; Yu, H. N(6)-Methyladenosine RNA Modification Regulates Shoot Stem Cell Fate in Arabidopsis. Dev. Cell 2016, 38, 186–200. [Google Scholar] [CrossRef]
- Yue, H.; Nie, X.; Yan, Z.; Weining, S. N6-methyladenosine regulatory machinery in plants: Composition, function and evolution. Plant Biotechnol. J. 2019, 17, 1194–1208. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Zhang, Y.C.; Liao, J.Y.; Yu, Y.; Zhou, Y.F.; Feng, Y.Z.; Yang, Y.W.; Lei, M.Q.; Bai, M.; Wu, H.; et al. The subunit of RNA N6-methyladenosine methyltransferase OsFIP regulates early degeneration of microspores in rice. PLoS Genet. 2019, 15, e1008120. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Wang, H.; Wang, J.; Liu, X.; Lin, F.; Lu, J. N6-methyladenosine RNA methylation is involved in virulence of the rice blast fungus Pyricularia oryzae (syn. Magnaporthe oryzae). FEMS Microbiol. Lett. 2019, 366, fny286. [Google Scholar]
- Tian, S.; Wu, N.; Zhang, L.; Wang, X. RNA N(6) -methyladenosine modification suppresses replication of rice black streaked dwarf virus and is associated with virus persistence in its insect vector. Mol. Plant Pathol. 2021, 22, 1070–1081. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Wang, J.; Zhang, Y.; Kong, Y.; Dong, H.; Feng, X.; Li, T.; Zhou, C.; Yu, J.; Xin, D.; et al. Changes in the m6A RNA methylome accompany the promotion of soybean root growth by rhizobia under cadmium stress. J. Hazard. Mater. 2023, 441, 129843. [Google Scholar] [CrossRef] [PubMed]
- Su, T.; Fu, L.; Kuang, L.; Chen, D.; Zhang, G.; Shen, Q.; Wu, D. Transcriptome-wide m6A methylation profile reveals regulatory networks in roots of barley under cadmium stress. J. Hazard. Mater. 2022, 423, 127140. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Ma, S.; Yu, Y.; Feng, H.; Wang, Y.; Liu, C.; He, S.; Yang, M.; Chen, Q.; Xin, D.; et al. Transcriptome-wide m(6)A methylation profiling identifies GmAMT1;1 as a promoter of lead and cadmium tolerance in soybean nodules. J. Hazard. Mater. 2024, 465, 133263. [Google Scholar] [CrossRef]
- Basit, F.; Chen, M.; Ahmed, T.; Shahid, M.; Noman, M.; Liu, J.; An, J.; Hashem, A.; Fahad Al-Arjani, A.B.; Alqarawi, A.A.; et al. Seed Priming with Brassinosteroids Alleviates Chromium Stress in Rice Cultivars via Improving ROS Metabolism and Antioxidant Defense Response at Biochemical and Molecular Levels. Antioxidants 2021, 10, 1089. [Google Scholar] [CrossRef]
- Cheng, Q.; Wang, P.; Wu, G.; Wang, Y.; Tan, J.; Li, C.; Zhang, X.; Liu, S.; Huang, S.; Huang, T.; et al. Coordination of m(6)A mRNA methylation and gene transcriptome in rice response to cadmium stress. Rice 2021, 14, 62. [Google Scholar] [CrossRef]
- Shahid, M.; Shamshad, S.; Rafiq, M.; Khalid, S.; Bibi, I.; Niazi, N.K.; Dumat, C.; Rashid, M.I. Chromium speciation, bioavailability, uptake, toxicity and detoxification in soil-plant system: A review. Chemosphere 2017, 178, 513–533. [Google Scholar] [CrossRef]
- Trinh, N.N.; Huang, T.L.; Chi, W.C.; Fu, S.F.; Chen, C.C.; Huang, H.J. Chromium stress response effect on signal transduction and expression of signaling genes in rice. Physiol. Plant. 2014, 150, 205–224. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.L.; Huang, L.Y.; Fu, S.F.; Trinh, N.N.; Huang, H.J. Genomic profiling of rice roots with short- and long-term chromium stress. Plant Mol. Biol. 2014, 86, 157–170. [Google Scholar] [CrossRef]
- Bukhari, S.A.; Shang, S.; Zhang, M.; Zheng, W.; Zhang, G.; Wang, T.Z.; Shamsi, I.H.; Wu, F. Genome-wide identification of chromium stress-responsive micro RNAs and their target genes in tobacco (Nicotiana tabacum) roots. Environ. Toxicol. Chem. 2015, 34, 2573–2582. [Google Scholar] [CrossRef] [PubMed]
- Dubey, S.; Misra, P.; Dwivedi, S.; Chatterjee, S.; Bag, S.K.; Mantri, S.; Asif, M.H.; Rai, A.; Kumar, S.; Shri, M.; et al. Transcriptomic and metabolomic shifts in rice roots in response to Cr (VI) stress. BMC Genom. 2010, 11, 648. [Google Scholar] [CrossRef]
- Zeng, F.; Qiu, B.; Wu, X.; Niu, S.; Wu, F.; Zhang, G. Glutathione-mediated alleviation of chromium toxicity in rice plants. Biol. Trace Elem. Res. 2012, 148, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Lei, L.; Wang, J.; Zheng, H.; Xin, W.; Liu, H.; Zou, D. qCTB7 positively regulates cold tolerance at booting stage in rice. Theor. Appl. Genet. 2023, 136, 135. [Google Scholar] [CrossRef]
- Siahpoosh, M.R.; Sanchez, D.H.; Schlereth, A.; Scofield, G.N.; Furbank, R.T.; van Dongen, J.T.; Kopka, J. Modification of OsSUT1 gene expression modulates the salt response of rice Oryza sativa cv. Taipei 309. Plant Sci. 2012, 182, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Gao, Z.; Xiao, G.; Huang, R.; Zhang, H. Leucine-Rich Repeat Receptor-Like Kinase FON1 Regulates Drought Stress and Seed Germination by Activating the Expression of ABA-Responsive Genes in Rice. Plant Mol. Biol. Report. 2014, 32, 1158–1168. [Google Scholar] [CrossRef]
- Islam, M.A.; Du, H.; Ning, J.; Ye, H.; Xiong, L. Characterization of Glossy1-homologous genes in rice involved in leaf wax accumulation and drought resistance. Plant Mol. Biol. 2009, 70, 443–456. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.M.; Lin, W.R.; Kao, C.H.; Hong, C.Y. Gene knockout of glutathione reductase 3 results in increased sensitivity to salt stress in rice. Plant Mol. Biol. 2015, 87, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.J.; Zhao, B.C.; Ge, W.N.; Zhang, Y.F.; Song, Y.; Sun, D.Y.; Guo, Y. An apoplastic h-type thioredoxin is involved in the stress response through regulation of the apoplastic reactive oxygen species in rice. Plant Physiol. 2011, 157, 1884–1899. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.W.; Zhao, Y.N.; Liu, X.S.; Rono, J.K.; Yang, Z.M. A metal chaperone OsHIPP16 detoxifies cadmium by repressing its accumulation in rice crops. Environ. Pollut. 2022, 311, 120058. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Xiong, S. OsHIPP24 is a Copper Metallochaperone Which Affects Rice Growth. J. Plant Biol. 2021, 64, 145–153. [Google Scholar] [CrossRef]
- Lee, S.; Kim, Y.Y.; Lee, Y.; An, G. Rice P1B-type heavy-metal ATPase, OsHMA9, is a metal efflux protein. Plant Physiol. 2007, 145, 831–842. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, R.; Bashir, K.; Ishimaru, Y.; Nishizawa, N.K.; Nakanishi, H. The role of heavy-metal ATPases, HMAs, in zinc and cadmium transport in rice. Plant Signal. Behav. 2012, 7, 1605–1607. [Google Scholar] [CrossRef]
- Chen, Z.; Fujii, Y.; Yamaji, N.; Masuda, S.; Takemoto, Y.; Kamiya, T.; Yusuyin, Y.; Iwasaki, K.; Kato, S.; Maeshima, M.; et al. Mn tolerance in rice is mediated by MTP8.1, a member of the cation diffusion facilitator family. J. Exp. Bot. 2013, 64, 4375–4387. [Google Scholar] [CrossRef] [PubMed]
- Lan, H.X.; Wang, Z.F.; Wang, Q.H.; Wang, M.M.; Bao, Y.M.; Huang, J.; Zhang, H.S. Characterization of a vacuolar zinc transporter OZT1 in rice (Oryza sativa L.). Mol. Biol. Rep. 2013, 40, 1201–1210. [Google Scholar] [CrossRef] [PubMed]
- Kavitha, P.G.; Kuruvilla, S.; Mathew, M.K. Functional characterization of a transition metal ion transporter, OsZIP6 from rice (Oryza sativa L.). Plant Physiol. Biochem. 2015, 97, 165–174. [Google Scholar]
- Zhang, H.; Li, Y.; Pu, M.; Xu, P.; Liang, G.; Yu, D. Oryza sativa POSITIVE REGULATOR OF IRON DEFICIENCY RESPONSE 2 (OsPRI2) and OsPRI3 are involved in the maintenance of Fe homeostasis. Plant Cell Environ. 2020, 43, 261–274. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, H.; Ogawa, I.; Ishimaru, Y.; Mori, S.; Nishizawa, N.K. Iron deficiency enhances cadmium uptake and translocation mediated by the Fe2+ transporters OsIRT1 and OsIRT2 in rice. Soil Sci. Plant Nutr. 2010, 52, 464–469. [Google Scholar] [CrossRef]
- Tan, L.; Qu, M.; Zhu, Y.; Peng, C.; Wang, J.; Gao, D.; Chen, C. ZINC TRANSPORTER5 and ZINC TRANSPORTER9 Function Synergistically in Zinc/Cadmium Uptake. Plant Physiol. 2020, 183, 1235–1249. [Google Scholar] [CrossRef]
- Rono, J.K.; Le Wang, L.; Wu, X.C.; Cao, H.W.; Zhao, Y.N.; Khan, I.U.; Yang, Z.M. Identification of a new function of metallothionein-like gene OsMT1e for cadmium detoxification and potential phytoremediation. Chemosphere 2021, 265, 129136. [Google Scholar] [CrossRef]
- Mittal, D.; Chakrabarti, S.; Sarkar, A.; Singh, A.; Grover, A. Heat shock factor gene family in rice: Genomic organization and transcript expression profiling in response to high temperature, low temperature and oxidative stresses. Plant Physiol. Biochem. 2009, 47, 785–795. [Google Scholar] [CrossRef]
- Uraguchi, S.; Kamiya, T.; Sakamoto, T.; Kasai, K.; Sato, Y.; Nagamura, Y.; Yoshida, A.; Kyozuka, J.; Ishikawa, S.; Fujiwara, T. Low-affinity cation transporter (OsLCT1) regulates cadmium transport into rice grains. Proc. Natl. Acad. Sci. USA 2011, 108, 20959–20964. [Google Scholar] [CrossRef]
- Peris-Peris, C.; Serra-Cardona, A.; Sánchez-Sanuy, F.; Campo, S.; Ariño, J.; San Segundo, B. Two NRAMP6 Isoforms Function as Iron and Manganese Transporters and Contribute to Disease Resistance in Rice. Mol. Plant-Microbe Interact. 2017, 30, 385–398. [Google Scholar] [CrossRef]
- Ishimaru, Y.; Bashir, K.; Nakanishi, H.; Nishizawa, N.K. OsNRAMP5, a major player for constitutive iron and manganese uptake in rice. Plant Signal. Behav. 2012, 7, 763–766. [Google Scholar] [CrossRef]
- Barceló, J.; Poschenrieder, C.; Gunsé, B. Water Relations of Chromium VI Treated Bush Bean Plants (Phaseolus vulgaris L. cv. Contender) under both Normal and Water Stress Conditions. J. Exp. Bot. 1986, 37, 178–187. [Google Scholar] [CrossRef]
- Panda, S.K.; Patra, H.K. Effect of salicylic acid potentiates cadmium-induced oxidative damage in Oryza sativa L. leaves. Acta Physiol. Plant. 2007, 29, 567–575. [Google Scholar] [CrossRef]
- Yang, D.; Li, S.; Xiao, Y.; Lu, L.; Zheng, Z.; Tang, D.; Cui, H. Transcriptome analysis of rice response to blast fungus identified core genes involved in immunity. Plant Cell Environ. 2021, 44, 3103–3121. [Google Scholar] [CrossRef] [PubMed]
- Abe, T.; Ito, M.; Takahashi, R.; Honma, T.; Kuramata, M.; Ishikawa, S. QTL Pyramiding and Its Use in Breeding for Increasing the Phytoextraction Efficiency of Soil Cd via High-Cd-Accumulating Rice. Plants 2022, 11, 2178. [Google Scholar] [CrossRef]
- Ali, M.K.; Sun, Z.H.; Yang, X.M.; Pu, X.Y.; Duan, C.L.; Li, X.; Wang, L.X.; Yang, J.Z.; Zeng, Y.W. NILs of Cold Tolerant Japonica Cultivar Exhibited New QTLs for Mineral Elements in Rice. Front. Genet. 2021, 12, 789645. [Google Scholar] [CrossRef]
- Pan, X.; Li, Y.; Liu, W.; Liu, S.; Min, J.; Xiong, H.; Dong, Z.; Duan, Y.; Yu, Y.; Li, X. QTL mapping and candidate gene analysis of cadmium accumulation in polished rice by genome-wide association study. Sci. Rep. 2020, 10, 11791. [Google Scholar] [CrossRef]
- Tian, S.; Liang, S.; Qiao, K.; Wang, F.; Zhang, Y.; Chai, T. Co-expression of multiple heavy metal transporters changes the translocation, accumulation, and potential oxidative stress of Cd and Zn in rice (Oryza sativa). J. Hazard. Mater. 2019, 380, 120853. [Google Scholar] [CrossRef]
- Yamaji, N.; Xia, J.; Mitani-Ueno, N.; Yokosho, K.; Feng Ma, J. Preferential delivery of zinc to developing tissues in rice is mediated by P-type heavy metal ATPase OsHMA2. Plant Physiol. 2013, 162, 927–939. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.U.; Rono, J.K.; Liu, X.S.; Feng, S.J.; Yang, Z.M. Functional characterization of a new metallochaperone for reducing cadmium concentration in rice crop. J. Clean. Prod. 2020, 272, 123152. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, Y.; Kong, X.; Li, J.; Liu, C.; Wang, S.; Xie, S.; Wang, J.; Liu, H.; Lei, L.; Zheng, H.; et al. The m6A Methylation Profile Identified That OsHMT9.1 Deregulates Chromium Toxicity in Rice (Oryza sativa L.) Through Negative Regulatory Functions. Agriculture 2025, 15, 519. https://doi.org/10.3390/agriculture15050519
Hou Y, Kong X, Li J, Liu C, Wang S, Xie S, Wang J, Liu H, Lei L, Zheng H, et al. The m6A Methylation Profile Identified That OsHMT9.1 Deregulates Chromium Toxicity in Rice (Oryza sativa L.) Through Negative Regulatory Functions. Agriculture. 2025; 15(5):519. https://doi.org/10.3390/agriculture15050519
Chicago/Turabian StyleHou, Yushan, Xuejiao Kong, Jingwen Li, Changsheng Liu, Shuo Wang, Shupeng Xie, Jingguo Wang, Hualong Liu, Lei Lei, Hongliang Zheng, and et al. 2025. "The m6A Methylation Profile Identified That OsHMT9.1 Deregulates Chromium Toxicity in Rice (Oryza sativa L.) Through Negative Regulatory Functions" Agriculture 15, no. 5: 519. https://doi.org/10.3390/agriculture15050519
APA StyleHou, Y., Kong, X., Li, J., Liu, C., Wang, S., Xie, S., Wang, J., Liu, H., Lei, L., Zheng, H., Xin, W., Zou, D., Wei, Z., & Yang, L. (2025). The m6A Methylation Profile Identified That OsHMT9.1 Deregulates Chromium Toxicity in Rice (Oryza sativa L.) Through Negative Regulatory Functions. Agriculture, 15(5), 519. https://doi.org/10.3390/agriculture15050519





