Comparative Analysis of Chloroplast Genomes in Sansevieria Reveals Phylogenetic Relationships and High Variability Molecular Markers
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and DNA Extraction
2.2. Chloroplast Genome Sequencing, Assembly, and Annotation
2.3. Phylogenetic Analysis
2.4. Nucleotide Diversity Analysis and IR Analysis
2.5. Molecular Marker Development
3. Results
3.1. Assembly and Annotation of Chloroplast Genomes
3.2. Codon Usage Bias
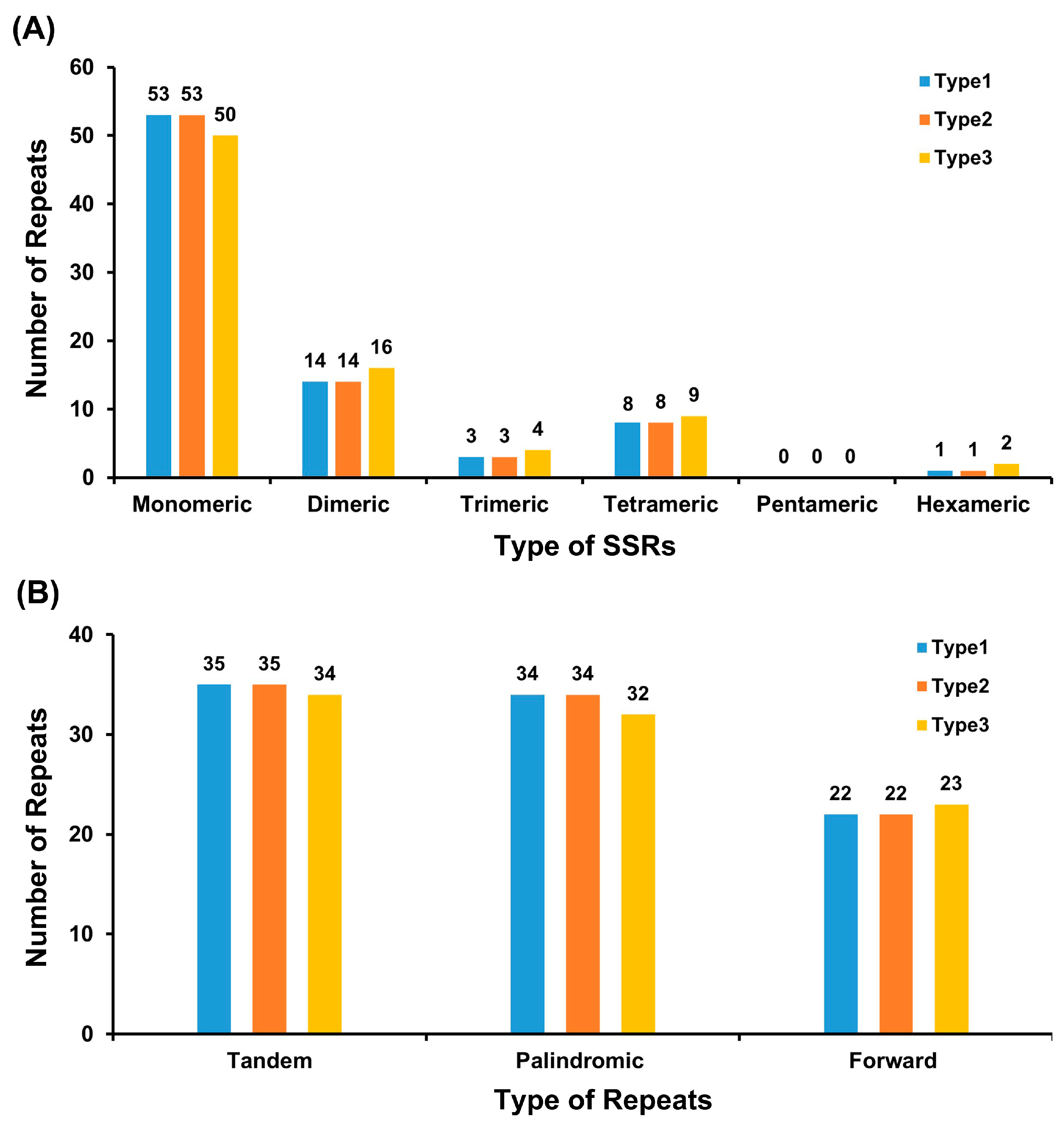
3.3. Repeat Sequence Analysis
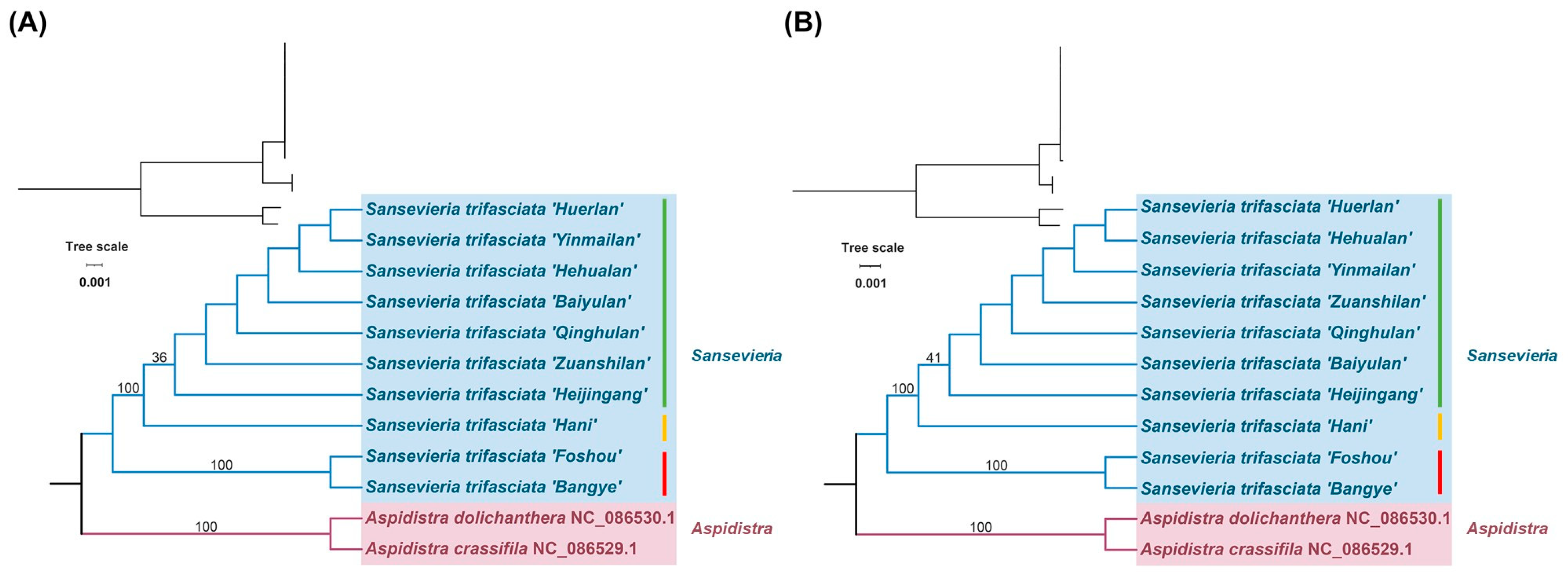
3.4. Phylogenetic Analysis
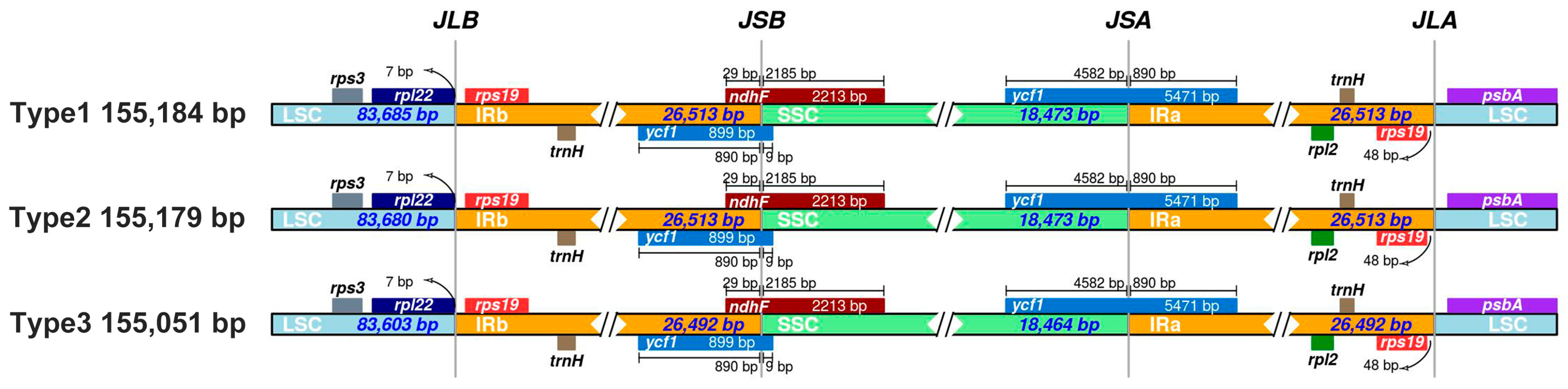
3.5. IR Boundary Expansion/Contraction Analysis
3.6. Nucleotide Diversity (Pi) Analysis
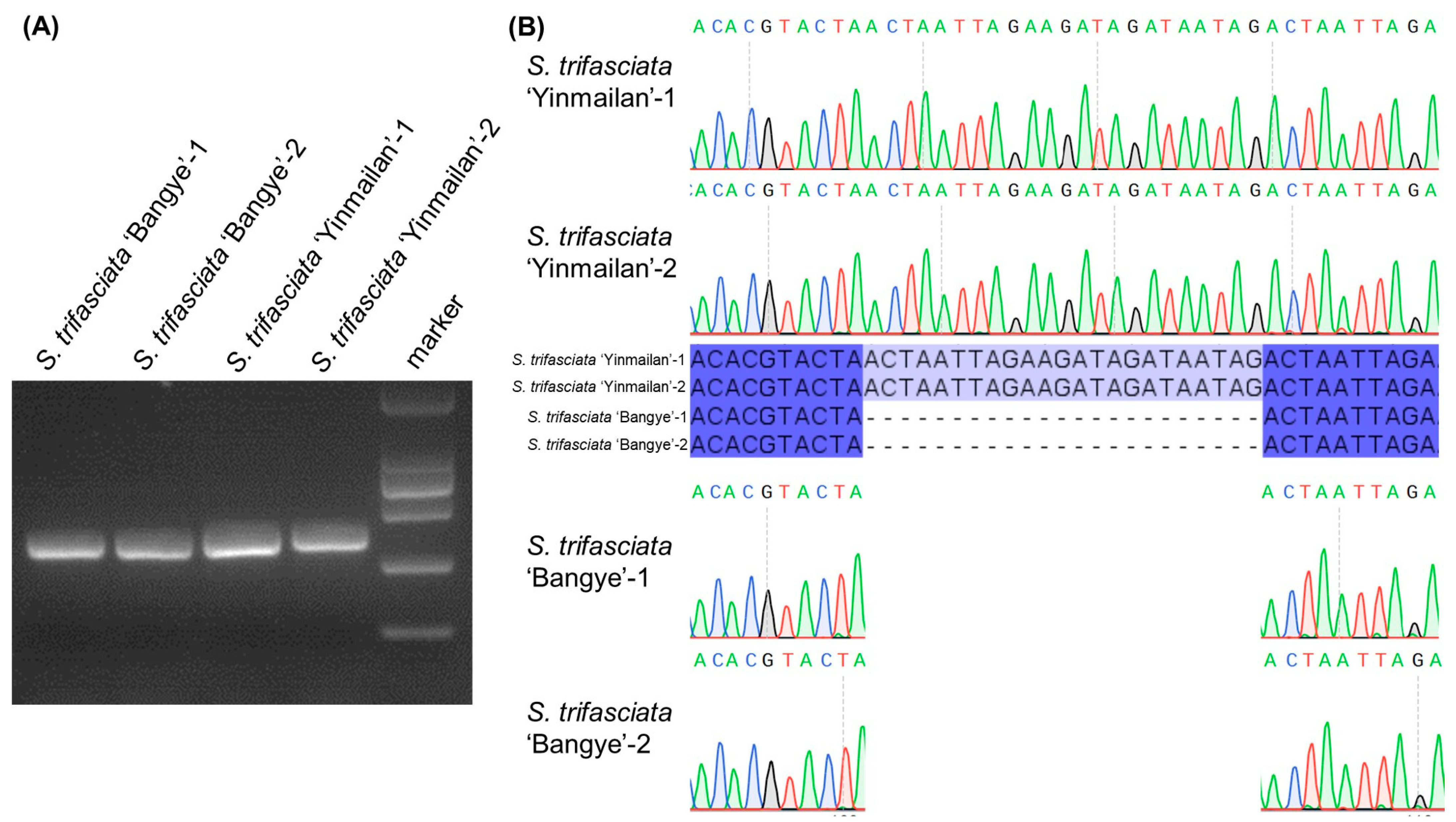
3.7. Molecular Marker Development
4. Discussion
4.1. Structural Conservation
4.2. Codon Bias
4.3. Hypervariable IRs
4.4. Phylogeny and Molecular Marker Development
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PCR | Polymerase chain reaction |
| SSR | Simple sequence repeat |
| ML | Maximum-likelihood |
| NCBI | National Center for Biotechnology Information |
| BLAST | Basic Local Alignment Search Tool |
| PCGs | Protein-coding gene sequences |
References
- Kasmawati, H.; Mustarichie, R.; Halimah, E.; Ruslin, R.; Arfan, A.; Sida, N.A. Unrevealing the Potential of Sansevieria trifasciata Prain Fraction for the Treatment of Androgenetic Alopecia by Inhibiting Androgen Receptors Based on LC-MS/MS Analysis, and In-Silico Studies. Molecules 2022, 27, 4358. [Google Scholar] [CrossRef]
- Van Kleinwee, I.; Larridon, I.; Shah, T.; Bauters, K.; Asselman, P.; Goetghebeur, P.; Leliaert, F.; Veltjen, E. Plastid phylogenomics of the Sansevieria Clade of Dracaena (Asparagaceae) resolves a recent radiation. Mol. Phylogenet. Evol. 2022, 169, 107404. [Google Scholar] [CrossRef]
- Garcia-Hernandez, E.; Loera-Quezada, M.M.; Moran-Velazquez, D.C.; Lopez, M.G.; Chable-Vega, M.A.; Santillan-Fernandez, A.; Zavaleta-Mancera, H.A.; Tang, J.Z.; Azadi, P.; Ibarra-Laclette, E.; et al. Indirect organogenesis for high frequency shoot regeneration of two cultivars of Sansevieria trifasciata Prain differing in fiber production. Sci. Rep. 2022, 12, 8507. [Google Scholar] [CrossRef]
- Dewatisari, W.F.; Nugroho, L.H.; Retnaningrum, E.; Purwestri, Y.A. Antibacterial and Anti-biofilm-Forming Activity of Secondary Metabolites from Sansevieria trifasciata Leaves Against Pseudomonas aeruginosa. Indones. J. Pharm. 2022, 33, 100–109. [Google Scholar] [CrossRef]
- Hoornenborg, C.W.; Qomariyah, N.; Gonzalez-Ponce, H.A.; Van Beek, A.P.; Moshage, H.; Van Dijk, G. Dracaena trifasciata (Prain) Mabb leaf extract protects MIN6 pancreas-derived beta cells against the diabetic toxin streptozotocin: Role of the NF-kappaB pathway. Front. Pharmacol. 2025, 16, 1485952. [Google Scholar] [CrossRef]
- Tallei, T.E.; Rembet, R.E.; Pelealu, J.J.; Kolondam, B.J. Sequence Variation and Phylogenetic Analysis of Sansevieria trifasciata (Asparagaceae). Biosci. Res. 2016, 13, 1–7. [Google Scholar]
- Khalumba, M.L.; Mbugua, P.K.; Kung’u, J.B. Uses and conservation of some highland species of the genus Sansevieria Thunb in Kenya. In African Crop Science Conference Proceedings; African Crop Science Society (ACSS): Kampala, Uganda, 2005; pp. 527–532. [Google Scholar]
- Kee, Y.J.; Zakaria, L.; Mohd, M.H. Identification, pathogenicity and histopathology of Colletotrichum sansevieriae causing anthracnose of Sansevieria trifasciata in Malaysia. J. Appl. Microbiol. 2020, 129, 626–636. [Google Scholar] [CrossRef]
- Guo, W.; Li, P.; Lei, K.; Ji, L. Characterization of the complete chloroplast genome of Sansevieria trifasciata var. Laurentii. Mitochondrial DNA B 2021, 6, 198–199. [Google Scholar] [CrossRef]
- Kasmawati, H.; Ruslin, R.; Arfan, A.; Sida, N.A.; Saputra, D.I.; Halimah, E.; Mustarichie, R. Antibacterial Potency of an Active Compound from Sansevieria trifasciata Prain: An Integrated In Vitro and In Silico Study. Molecules 2023, 28, 6096. [Google Scholar] [CrossRef]
- Lin, L.; Hao, Z.; Zhou, L.; Liu, W.; Liu, N.; Wang, K.; Jia, R. Elucidating phylogenetic relationships within the genus Curcuma through the comprehensive analysis of the chloroplast genome of Curcuma viridiflora Roxb. 1810 (Zingiberaceae). Mitochondrial DNA Part B 2024, 9, 371–375. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Qian, Z.; Gichira, A.W.; Chen, J.; Li, Z. Assembly and comparative analysis of the first complete mitochondrial genome of the invasive water hyacinth, Eichhornia crassipes. Gene 2024, 914, 148416. [Google Scholar] [CrossRef]
- Hao, Z.; Zhang, Z.; Jiang, J.; Pan, L.; Zhang, J.; Cui, X.; Li, Y.; Li, J.; Luo, L. Complete mitochondrial genome of Melia azedarach L., reveals two conformations generated by the repeat sequence mediated recombination. BMC Plant Biol. 2024, 24, 645. [Google Scholar] [CrossRef]
- Hao, Z.; Zhang, Z.; Zhang, J.; Cui, X.; Li, J.; Luo, L.; Li, Y. The complete mitochondrial genome of Aglaia odorata, insights into its genomic structure and RNA editing sites. Front. Plant Sci. 2024, 15, 1362045. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Duan, B.; Zhou, Z.; Fang, H.; Yang, M.; Xia, C.; Zhou, Y.; Wang, J. Comparative analysis of medicinal plants Scutellaria baicalensis and common adulterants based on chloroplast genome sequencing. BMC Genom. 2024, 25, 39. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.; Liu, C.; Yang, X.; Li, M.; Liu, L.; Jia, K.; Li, W. Comparative and Phylogenetic Analysis of the Chloroplast Genomes of Four Wild Species of the Genus Prunus. Genes 2025, 16, 239. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Shen, F.; Wang, L.; Dou, J.; Dong, T.; Li, M.; Wang, Q.; Dong, S.; Zhang, G.; Zhao, J.; et al. Accelerating Moss Identification Through the Development of Specific DNA Barcodes Based on the Whole Chloroplast Genome. Mol. Ecol. Resour. 2025, e70004. [Google Scholar] [CrossRef]
- Huang, Y.; Jin, X.J.; Zhang, C.Y.; Li, P.; Meng, H.H.; Zhang, Y.H. Plastome evolution of Engelhardia facilitates phylogeny of Juglandaceae. BMC Plant. Biol. 2024, 24, 634. [Google Scholar] [CrossRef]
- Yue, M.; Chen, H.; Xuan, L.; Yang, Y.; Chong, X.; Li, M.; Yu, C.; Lu, X.; Zhang, F. Novel molecular markers for Taxodium breeding from the chloroplast genomes of four artificial Taxodium hybrids. Front. Genet. 2023, 14, 1193023. [Google Scholar] [CrossRef]
- Verstraeten, L.; Fokias, K.; Saerens, G.; Bekaert, B. Cost-effective optimisation and validation of the VISAGE enhanced tool assay on the Illumina NovaSeq 6000 platform. Forensic Sci. Int. Genet. 2025, 78, 103299. [Google Scholar] [CrossRef]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; DePamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Shi, L.; Chen, H.; Jiang, M.; Wang, L.; Wu, X.; Huang, L.; Liu, C. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 2019, 47, W65–W73. [Google Scholar] [CrossRef] [PubMed]
- Lewis, S.E.; Searle, S.M.; Harris, N.; Gibson, M.; Lyer, V.; Richter, J.; Wiel, C.; Bayraktaroglu, L.; Birney, E.; Crosby, M.A.; et al. Apollo: A sequence annotation editor. Genome Biol. 2002, 3, H82. [Google Scholar] [CrossRef]
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. tRNAscan-SE 2.0: Improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 2021, 49, 9077–9096. [Google Scholar] [CrossRef]
- Chen, Y.; Ye, W.; Zhang, Y.; Xu, Y. High speed BLASTN: An accelerated MegaBLAST search tool. Nucleic Acids Res. 2015, 43, 7762–7768. [Google Scholar] [CrossRef] [PubMed]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlic, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sanchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sanchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, W273–W279. [Google Scholar] [CrossRef]
- Amiryousefi, A.; Hyvonen, J.; Poczai, P. IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics 2018, 34, 3030–3031. [Google Scholar] [CrossRef]
- Darling, A.C.; Mau, B.; Blattner, F.R.; Perna, N.T. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004, 14, 1394–1403. [Google Scholar] [CrossRef]
- Steane, D.A.; De Kok, R.P.; Olmstead, R.G. Phylogenetic relationships between Clerodendrum (Lamiaceae) and other Ajugoid genera inferred from nuclear and chloroplast DNA sequence data. Mol. Phylogenet. Evol. 2004, 32, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.M.; Agarwal, A.; Dev, B.; Kumar, K.; Prakash, O.; Arya, M.C.; Nasim, M. Assessment of photosynthetic potential of indoor plants under cold stress. Photosynthetica 2016, 54, 138–142. [Google Scholar] [CrossRef]
- Kittipornkul, P.; Treesubsuntorn, C.; Kobthong, S.; Yingchutrakul, Y.; Julpanwattana, P.; Thiravetyan, P. The potential of proline as a key metabolite to design real-time plant water deficit and low-light stress detector in ornamental plants. Environ. Sci. Pollut. R 2024, 31, 36152–36162. [Google Scholar] [CrossRef]
- Chen, Y.; Du, X.; Pan, L.; Huang, Q.; Hao, Z. Comparative analysis of chloroplast genomes in Carica species reveals evolutionary relationships of papaya and the development of efficient molecular markers. Front. Plant Sci. 2025, 16, 1686914. [Google Scholar] [CrossRef]
- Shi, N.; Yuan, Y.; Huang, R.; Wen, G. Analysis of codon usage patterns in complete plastomes of four medicinal Polygonatum species (Asparagaceae). Front. Genet. 2024, 15, 1401013. [Google Scholar] [CrossRef]
- Lu, Q.X.; Chang, X.; Gao, J.; Wu, X.; Wu, J.; Qi, Z.C.; Wang, R.H.; Yan, X.L.; Li, P. Evolutionary Comparison of the Complete Chloroplast Genomes in Convallaria Species and Phylogenetic Study of Asparagaceae. Genes 2022, 13, 1724. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Li, A.; Rochaix, J.D.; Liu, Z. Architecture of chloroplast TOC-TIC translocon supercomplex. Nature 2023, 615, 349–357. [Google Scholar] [CrossRef]
- Liu, J.; Gong, P.; Lu, R.; Lozano-Duran, R.; Zhou, X.; Li, F. Chloroplast immunity: A cornerstone of plant defense. Mol. Plant 2024, 17, 686–688. [Google Scholar] [CrossRef] [PubMed]
- Xing, J.; Pan, J.; Yang, W. Chloroplast protein translocation complexes and their regulation. J. Integr. Plant Biol. 2025, 67, 912–925. [Google Scholar] [CrossRef] [PubMed]
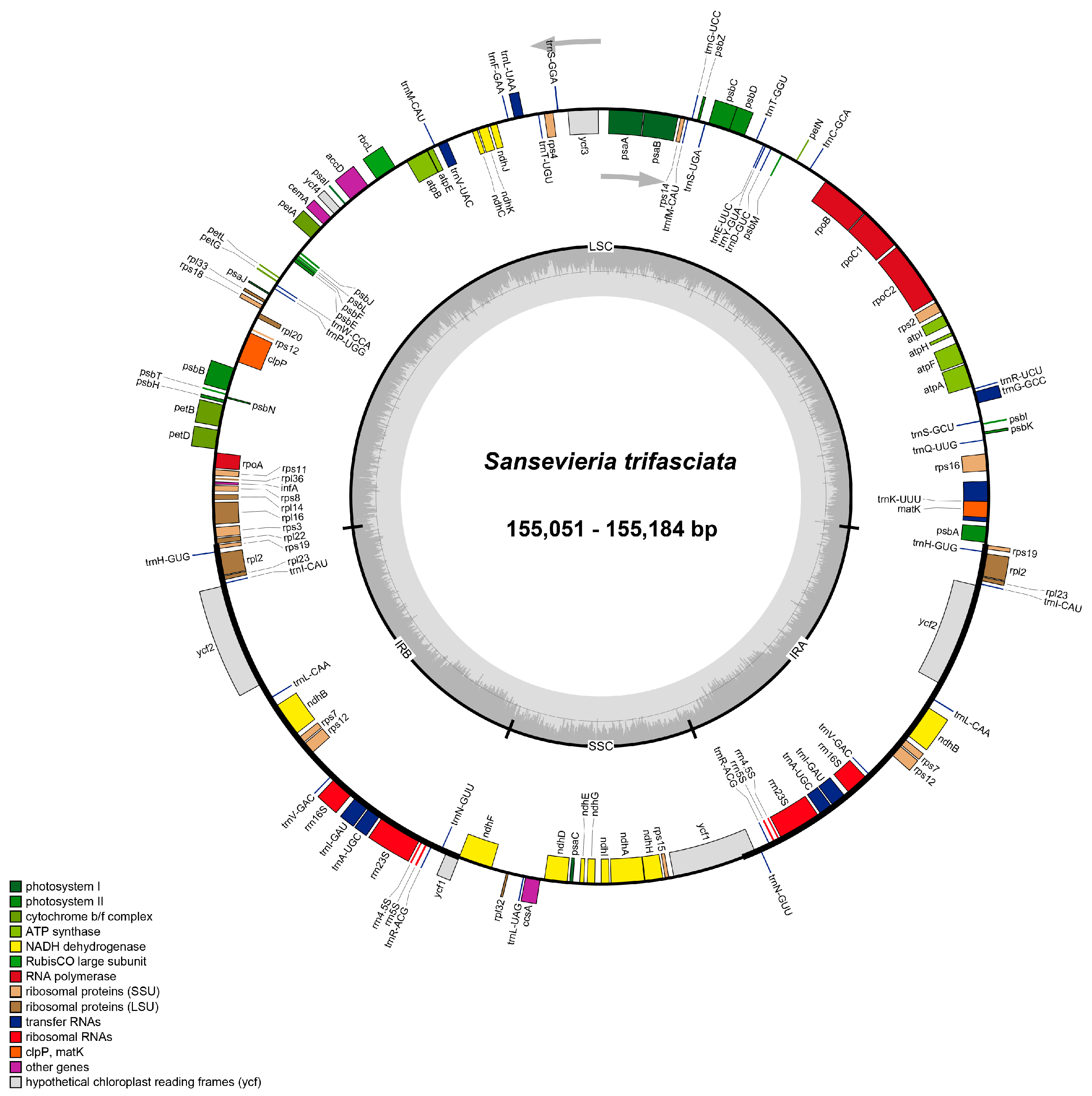


| Chloroplast Genome | Name of S. trifasciata Cultivars | Total Length (bp) | GC Content (%) |
|---|---|---|---|
| hpl-1 | S. trifasciata ‘Hani’. | 155,184 | 37.46 |
| hpl-2 | S. trifasciata ‘Heijingang’ | 155,179 | 37.46 |
| hpl-5 | S. trifasciata ‘Hehualan’ | 155,179 | 37.46 |
| hpl-6 | S. trifasciata ‘Qinghulan’ | 155,179 | 37.46 |
| hpl-7 | S. trifasciata ‘Zuanshilan’ | 155,179 | 37.46 |
| hpl-8 | S. trifasciata ‘Yinmailan’ | 155,179 | 37.46 |
| hpl-9 | S. trifasciata ‘Baiyulan’ | 155,179 | 37.46 |
| hpl-10 | S. trifasciata ‘Huerlan’ | 155,179 | 37.46 |
| hpl-11 | S. trifasciata ‘Foshou’ | 155,051 | 37.44 |
| hpl-12 | S. trifasciata ‘Bangye’ | 155,051 | 37.44 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, Z.; Kong, H.; Lv, X.; Du, X.; Zhao, H.; Lu, J. Comparative Analysis of Chloroplast Genomes in Sansevieria Reveals Phylogenetic Relationships and High Variability Molecular Markers. Agriculture 2025, 15, 2606. https://doi.org/10.3390/agriculture15242606
Hao Z, Kong H, Lv X, Du X, Zhao H, Lu J. Comparative Analysis of Chloroplast Genomes in Sansevieria Reveals Phylogenetic Relationships and High Variability Molecular Markers. Agriculture. 2025; 15(24):2606. https://doi.org/10.3390/agriculture15242606
Chicago/Turabian StyleHao, Zhigang, Hua Kong, Xiaojun Lv, Xiaoxi Du, Hui Zhao, and Jinghua Lu. 2025. "Comparative Analysis of Chloroplast Genomes in Sansevieria Reveals Phylogenetic Relationships and High Variability Molecular Markers" Agriculture 15, no. 24: 2606. https://doi.org/10.3390/agriculture15242606
APA StyleHao, Z., Kong, H., Lv, X., Du, X., Zhao, H., & Lu, J. (2025). Comparative Analysis of Chloroplast Genomes in Sansevieria Reveals Phylogenetic Relationships and High Variability Molecular Markers. Agriculture, 15(24), 2606. https://doi.org/10.3390/agriculture15242606





