Phylogenetic and Pathogenic Characterization of Cytospora Species Causing Apple Canker in Kazakhstan
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Fungal Isolation
2.2. Morphological Identification
2.3. DNA Extraction, PCR Amplification, Sequencing and Phylogenetic Analysis
2.4. Pathogenicity Tests
2.5. Statistical Analysis
3. Results
3.1. Morphological Identification and Description of Cytospora Species
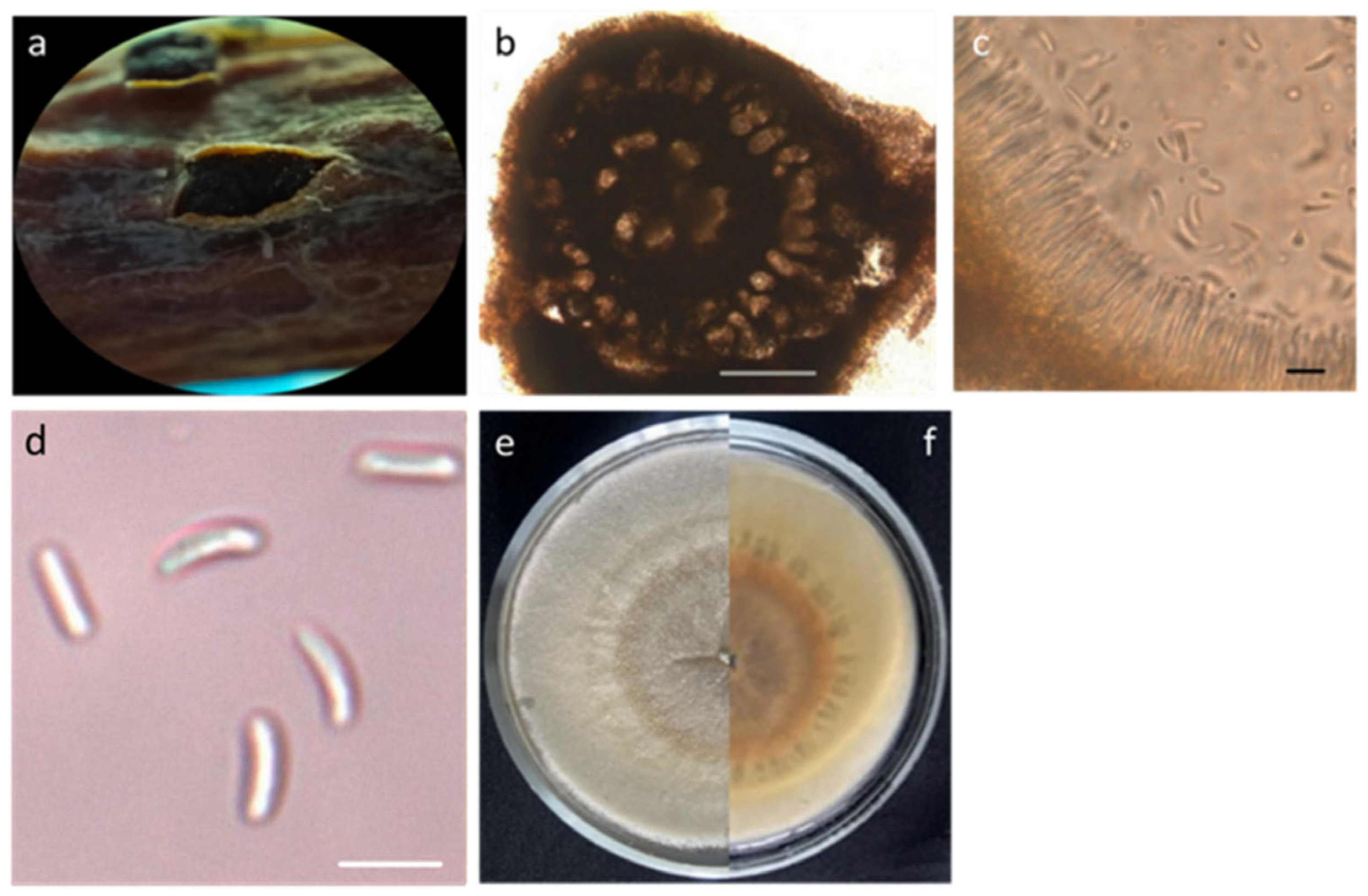
3.1.1. Taxonomic Description of Cytospora leucostoma
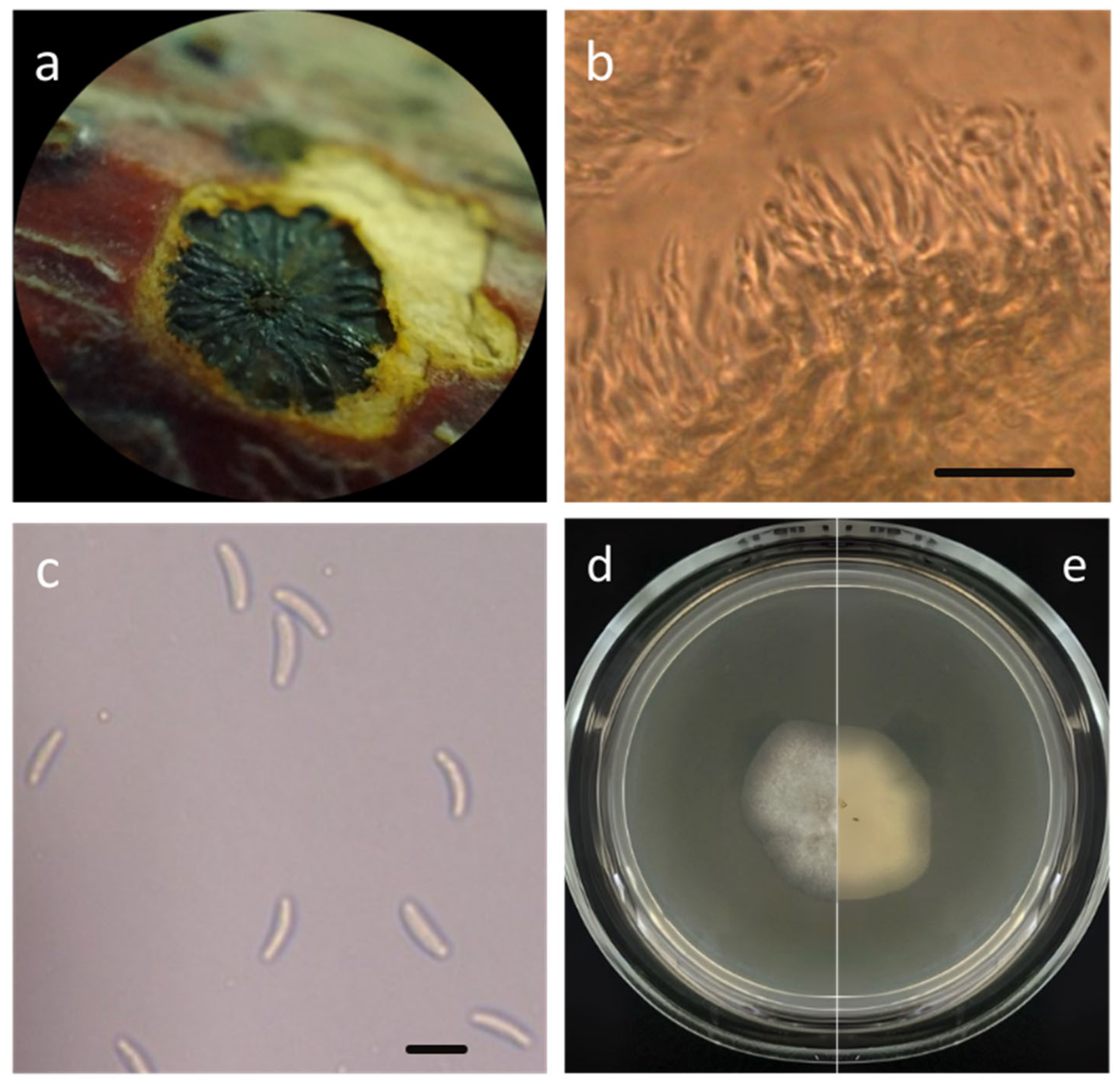
3.1.2. Taxonomic Description of Cytospora sorbicola
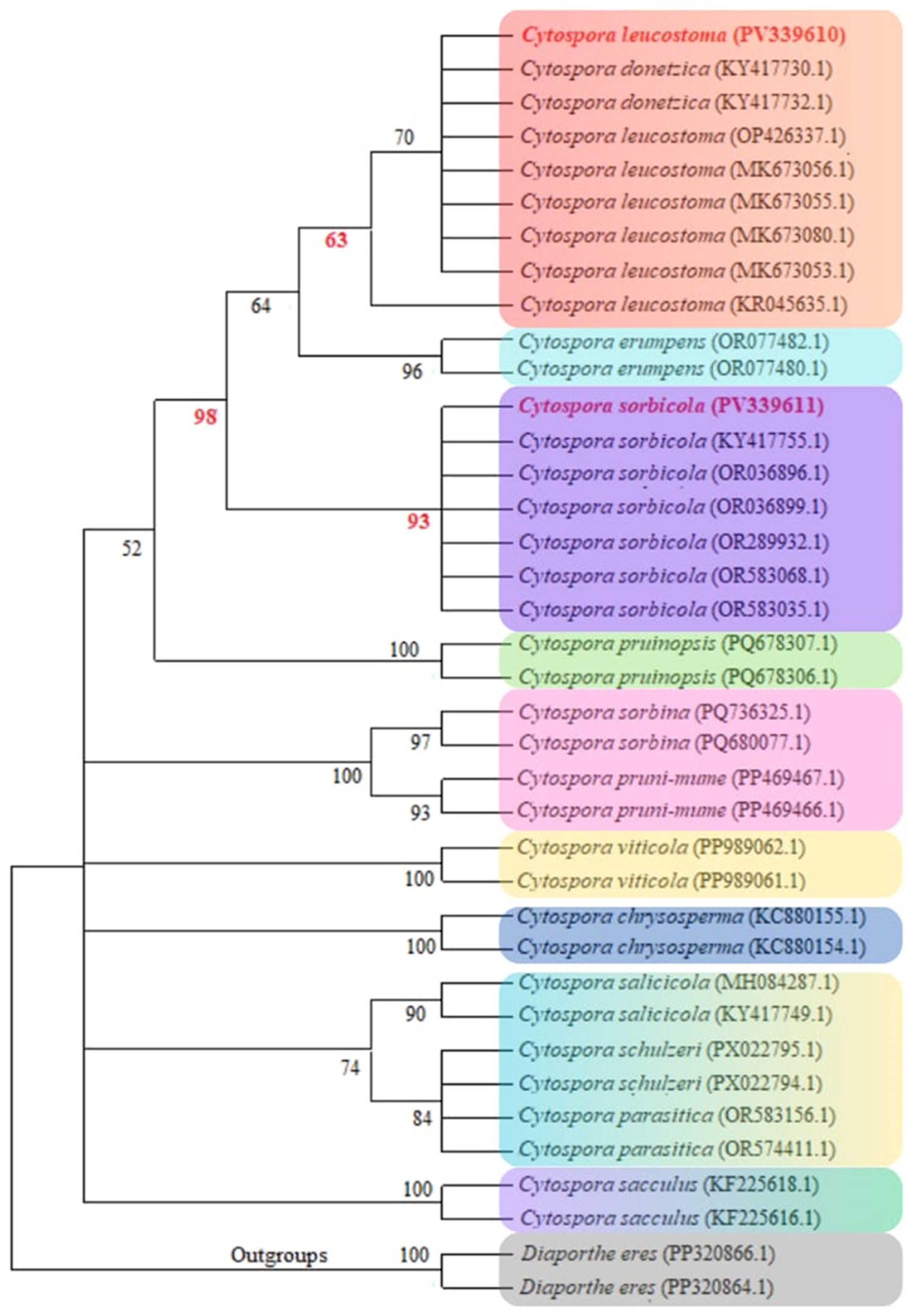
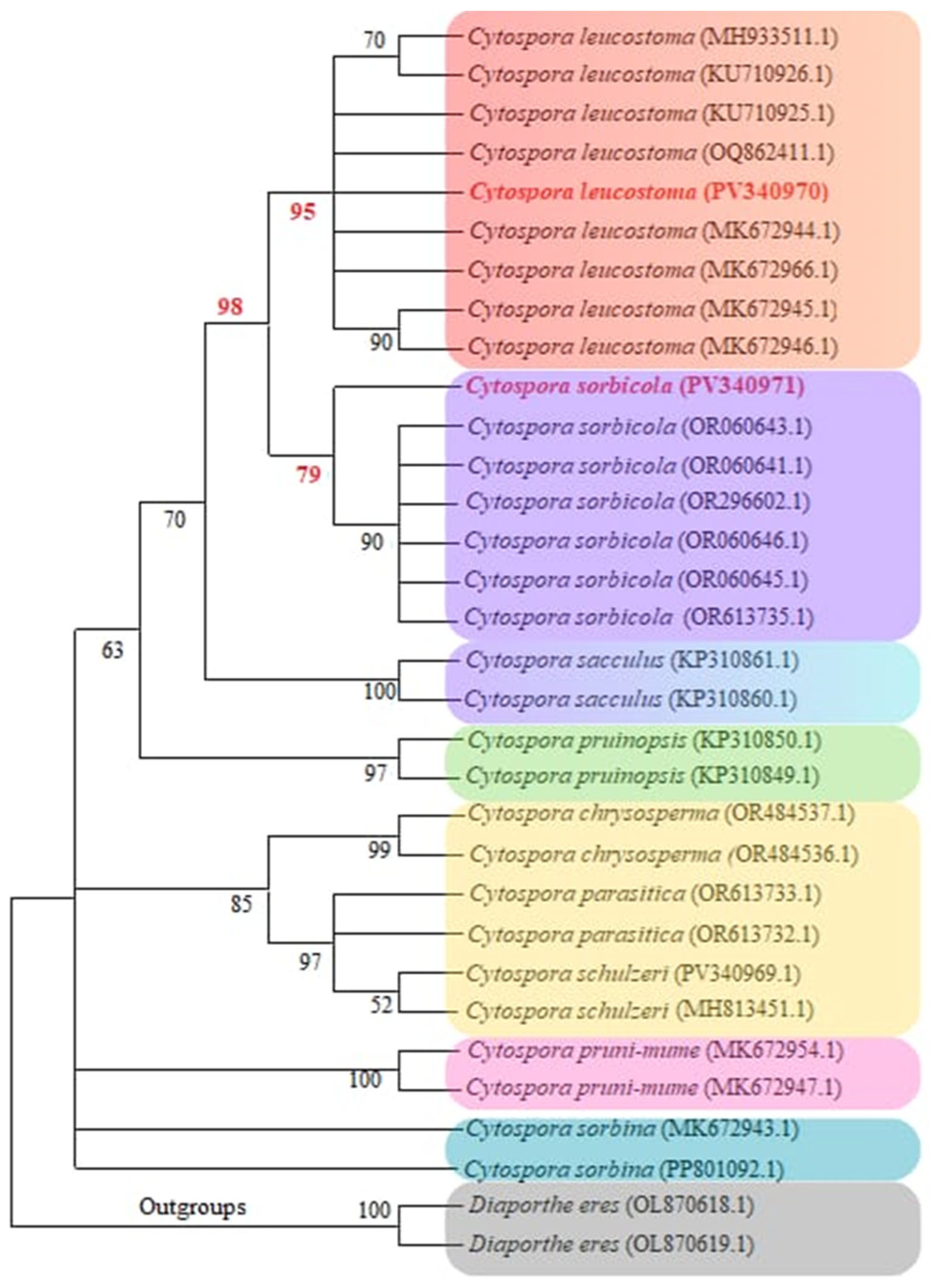
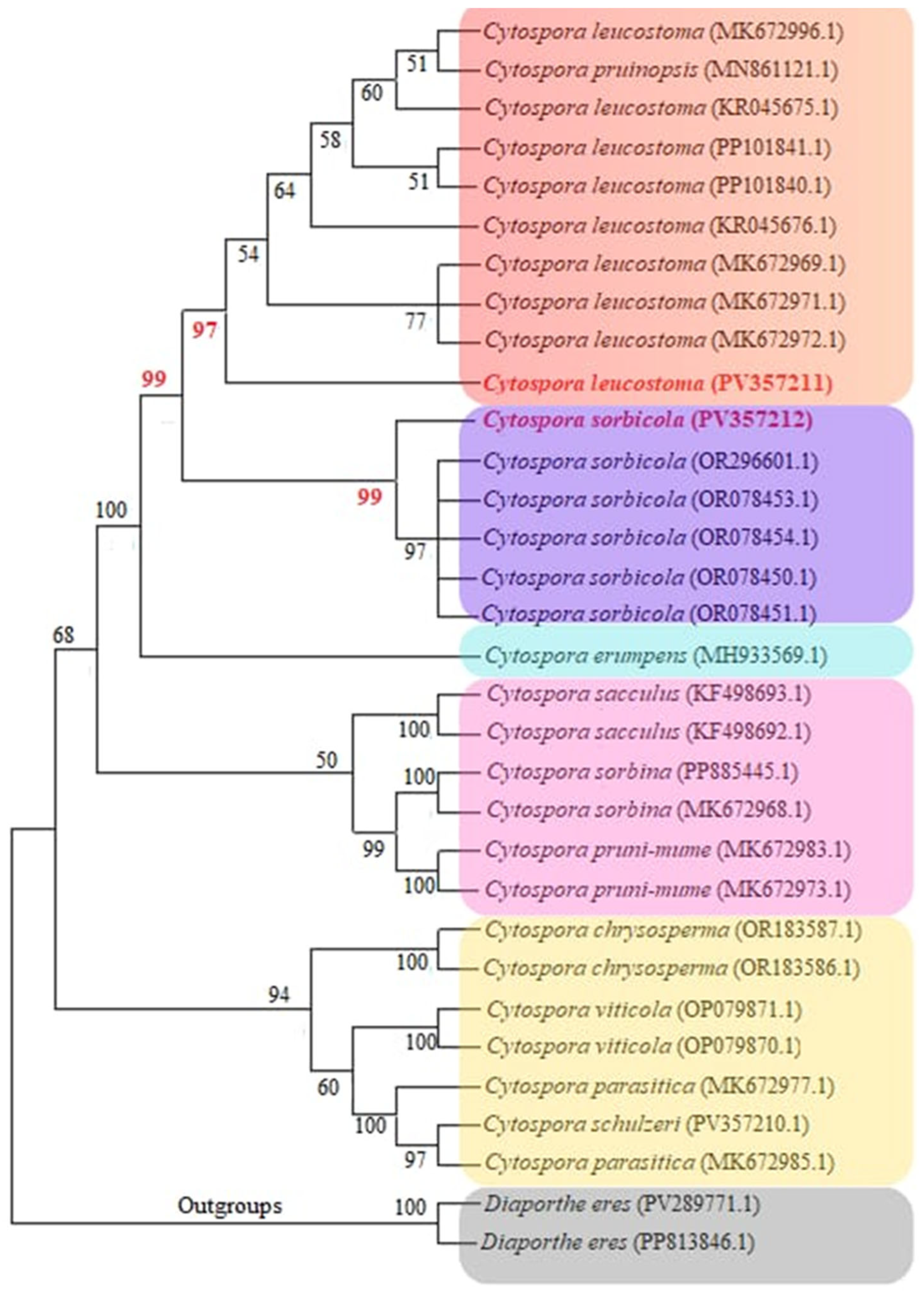
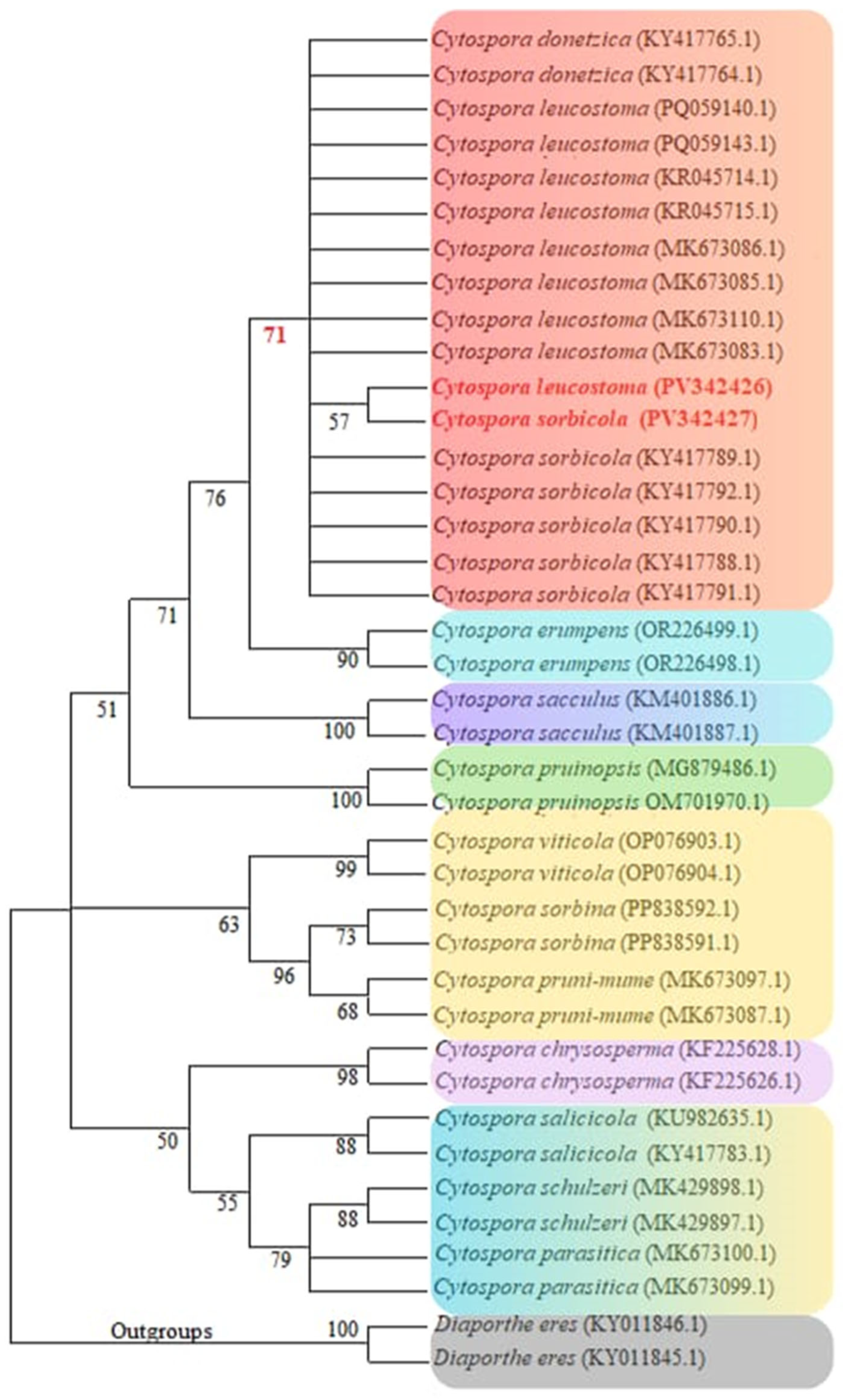
3.2. Cytospora Phylogenetic Analyses
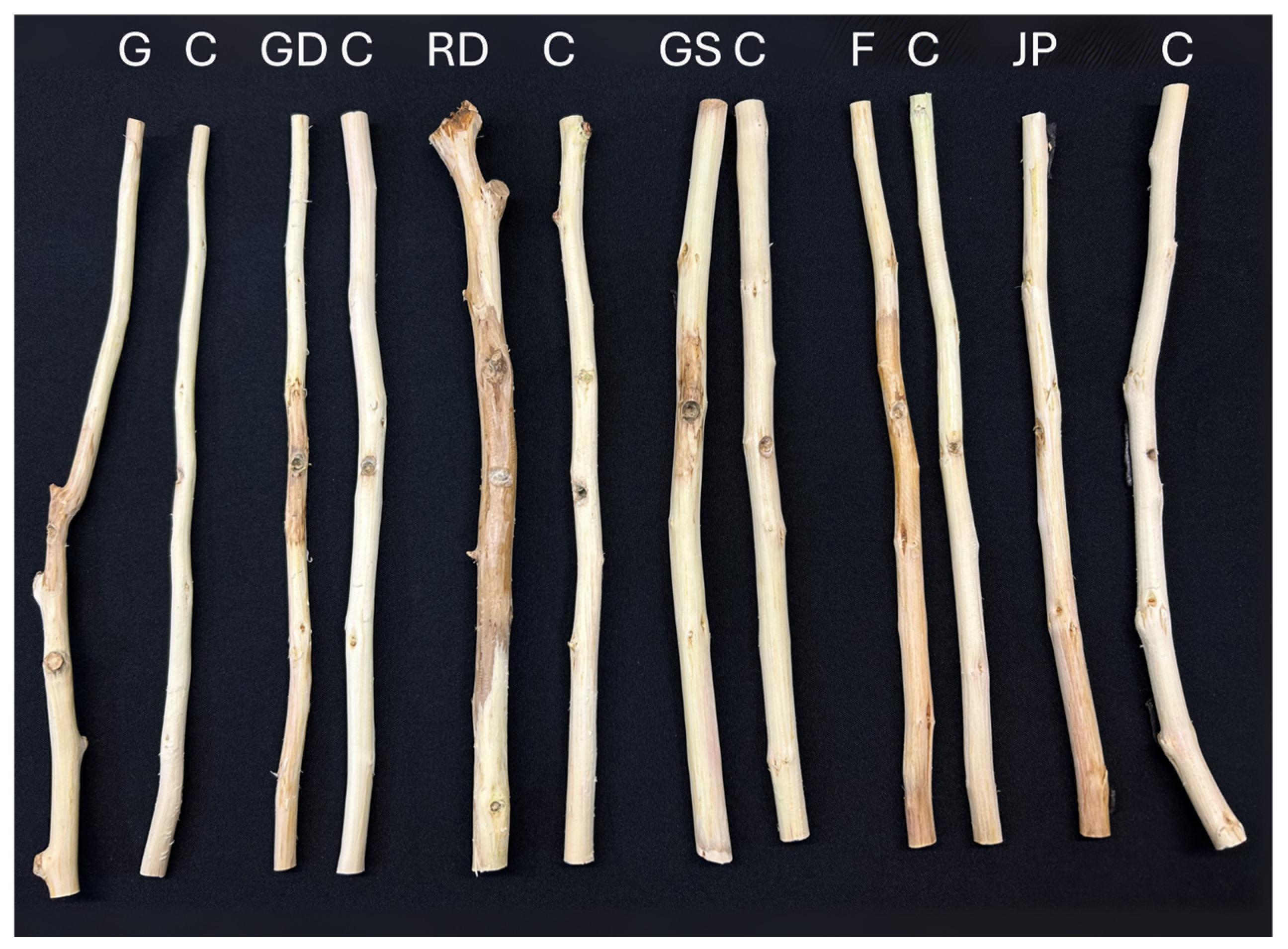
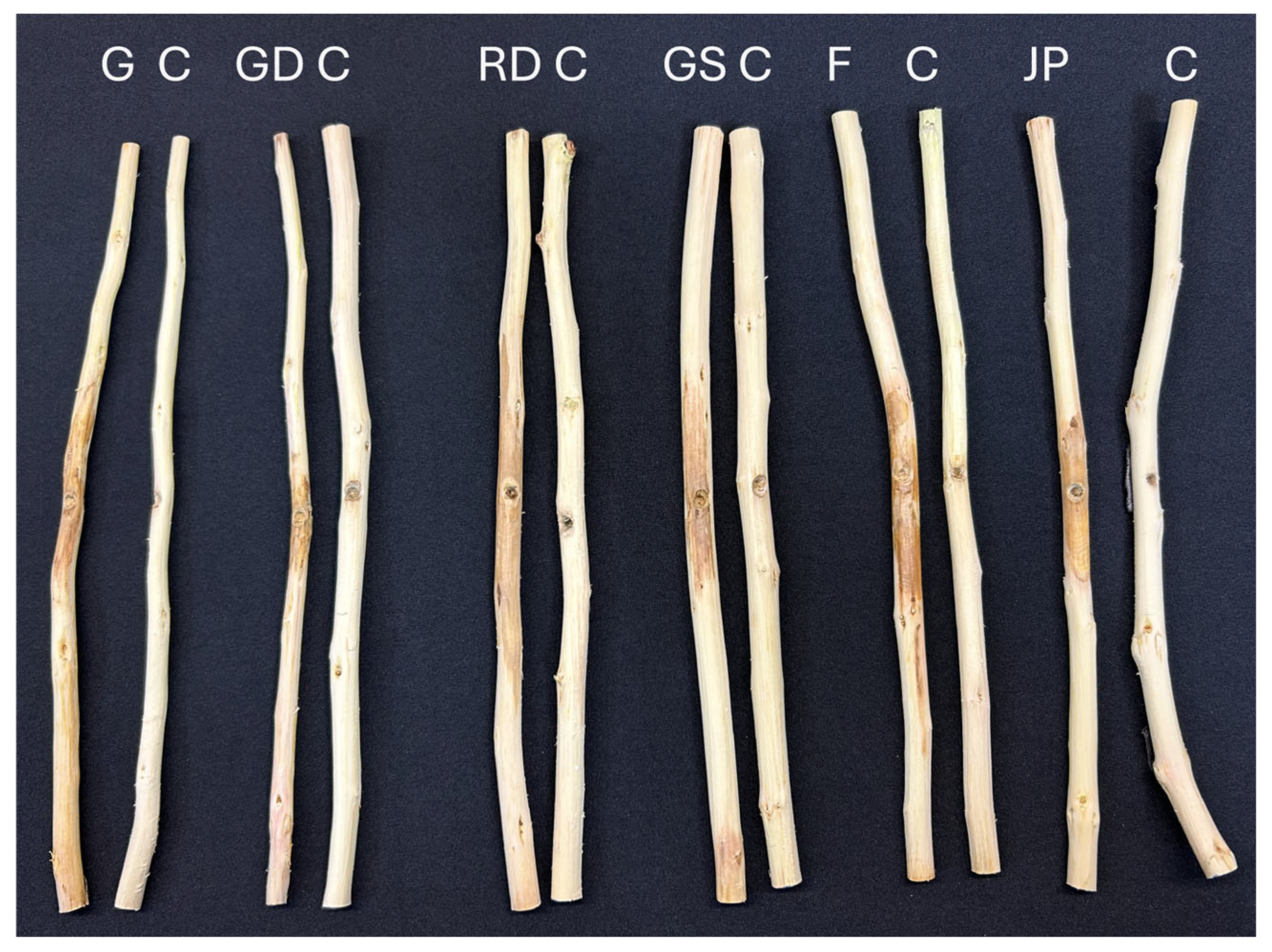
3.3. Pathogenicity Trials
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Robinson, J.P.; Harris, S.A.; Juniper, B.E. Taxonomy of the genus Malus mill. (Rosaceae) with emphasis on the cultivated apple, Malus domestica Borkh. Plant Syst. Evol. 2001, 226, 35–58. [Google Scholar] [CrossRef]
- Tegtmeier, R.; Švara, A.; Gritsenko, D.; Khan, A. Malus sieversii: A historical, genetic, and conservational perspective of the primary progenitor species of domesticated apples. Hortic. Res. 2025, 12, uhae244. [Google Scholar] [CrossRef] [PubMed]
- Gharghani, A.; Zamani, Z.; Talaie, A.; Oraguzie, N.C.; Fatahi, R.; Hajnajari, H.; Wiedow, C.; Gardiner, S.E. Genetic identity and relationships of Iranian apple (Malus × domestica Borkh.) cultivars and landraces, wild Malus species and representative old apple cultivars based on simple sequence repeat (SSR) marker analysis. Genet. Resour. Crop Evol. 2009, 56, 829–842. [Google Scholar] [CrossRef]
- Oyenihi, A.B.; Belay, Z.A.; Mditshwa, A.; Caleb, O.J. “An apple a day keeps the doctor away”: The potentials of apple bioactive constituents for chronic disease prevention. J. Food Sci. 2022, 87, 2291–2309. [Google Scholar] [CrossRef]
- Spengler, R.N. Origins of the apple: The role of megafaunal mutualism in the domestication of Malus and rosaceous trees. Front. Plant Sci. 2019, 10, 617. [Google Scholar] [CrossRef]
- Ha, Y.-H.; Oh, S.-H.; Lee, S.-R. Genetic admixture in the population of wild apple (Malus sieversii) from the Tien Shan Mountains, Kazakhstan. Genes 2021, 12, 104. [Google Scholar] [CrossRef] [PubMed]
- Sutton, T.B.; Aldwinckle, H.S.; Agnello, A.M.; Walgenbach, J.F. Compendium of Apple and Pear Diseases and Pests; APS Press: St. Paul, MN, USA, 2014; pp. 8–116. [Google Scholar] [CrossRef]
- Adams, G.C.; Roux, J.; Wingfield, M.J.; Common, R. Phylogenetic relationships and morphology of Cytospora species and related teleomorphs (Ascomycota, Diaporthales, Valsaceae) from Eucalyptus. Stud. Mycol. 2005, 52, 1–144. [Google Scholar]
- Adams, G.C.; Roux, J.; Wingfield, M.J. Cytospora species (Ascomycota, Diaporthales, Valsaceae), introduced and native pathogens of trees in South Africa. Australas. Plant Pathol. 2006, 35, 521–548. [Google Scholar] [CrossRef]
- Pan, M.; Zhu, H.Y.; Tian, C.M.; Alvarez, L.V.; Fan, X.L. Cytospora piceae sp. nov. associated with canker disease of Picea crassifolia in China. Phytotaxa 2018, 383, 181–196. [Google Scholar] [CrossRef]
- Eken, C.; Sevindik, E. Molecular phylogeny of Cytospora species associated with canker diseases of apple trees in Türkiye. Erwerbs-Obstbau 2023, 65, 2249–2257. [Google Scholar] [CrossRef]
- Azizi, R.; Ghosta, Y.; Ahmadpour, A. Apple crown and collar canker and necrosis caused by Cytospora balanejica sp. nov. in Iran. Sci. Rep. 2024, 14, 6629. [Google Scholar] [CrossRef]
- Zhao, Y.; Cai, G.; Yan, M.; Ma, R.; Zhang, D. Pathogenicity evaluation of Cytospora species in 13 apple (Malus domestica) varieties and wild apple (Malus sieversii) in Xinjiang, China. J. Phytopathol. 2024, 172, e13375. [Google Scholar] [CrossRef]
- Tulegenova, Z.; Amanbayeva, U.; Shalabayeva, A.M.; Yelyubayeva, D.; Zhaxylykov, A.; Uakhit, R.; Smagulova, A.; Kiyan, V.; Dyussembayev, K.; Mukiyanova, G. Identification and pathogenicity of causal agents of apple canker disease in Kazakhstan. Horticulturae 2025, 11, 45. [Google Scholar] [CrossRef]
- Lawrence, D.P.; Holland, L.A.; Nouri, M.T.; Travadon, R.; Abramians, A.; Michailides, T.J.; Trouillas, F.P. Molecular phylogeny of Cytospora species associated with canker diseases of fruit and nut crops in California, with descriptions of ten new species and one new combination. IMA Fungus 2018, 9, 333–370. [Google Scholar] [CrossRef]
- Fan, X.; Bezerra, J.D.P.; Tian, C.-M.; Crous, P.W. Cytospora (Diaporthales) in China. Persoonia 2020, 45, 1–45. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Pan, M.; Bezerra, J.D.P.; Tian, C.; Fan, X. Discovery of Cytospora species associated with canker disease of tree hosts from Mount Dongling of China. MycoKeys 2020, 62, 97–121. [Google Scholar] [CrossRef] [PubMed]
- Chomczynski, P.; Sacchi, N. The single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction: Twenty-something years on. Nat. Protoc. 2006, 1, 581–585. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Academic Press, Inc.: Cambridge, MA, USA, 1990; Volume 18, pp. 315–322. [Google Scholar]
- Alves, A.; Crous, P.W.; Correia, A.; Phillips, A. Morphological and molecular data reveal cryptic speciation in Lasiodiplodia theobromae. Fungal Divers. 2008, 28, 1–13. [Google Scholar]
- O’donnell, K. Fusarium and its near relatives. In The Fungal Holomorph: Mitotic, Meiotic and Pleomorphic Speciation in Fungal Systematics; Reynolds, D.R., Taylor, J.W., Eds.; CABI: Wallingford, UK, 1993. [Google Scholar]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef]
- Hall, T.A. Bioedit: A user-friendly biological sequence alignment editor and analyses program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on the phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Sha, S.S.; Wang, Z.; Yan, C.C.; Hao, H.T.; Wang, L.; Feng, H.Z. Identification of fungal species associated with apple canker in Tarim basin, China. Plant Dis. 2023, 107, 1284–1298. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.H.; Lee, S.W.; Chi, K.H.; Kim, D.A.; Uhm, J.Y. Survey on the occurrence of apple disease in Korea from 1992 to 2000. Plant Pathol. J. 2006, 22, 375–380. [Google Scholar] [CrossRef]
- Meng, X.L.; Qi, X.H.; Han, Z.Y.; Guo, Y.B.; Wang, Y.N.; Hu, T.I.; Wang, L.M.; Cao, K.G.; Wang, S.T. Latent infections of Valsa mali in the seeds, seedlings and twigs of crabapple and apple trees is a potential inoculum source of Valsa canker. Sci. Rep. 2019, 9, 7738. [Google Scholar] [CrossRef]
- Wang, X.; Shi, C.M.; Gleason, M.L.; Huang, L. Fungal species associated with apple Valsa canker in East Asia. Phytopathol. Res. 2020, 2, 35. [Google Scholar] [CrossRef]
- Mehrabi, M.; Mohammadi, G.E.; Fotouhifar, K. Studies on Cytospora canker disease of apple trees in Semirom region of Iran. J. Agric. Technol. 2011, 7, 967–982. [Google Scholar]
- Pan, M.; Zhu, H.Y.; Guido, B.; Tian, C.M.; Fan, X.L. High diversity of Cytospora associated with canker and dieback of Rosaceae in China, with 10 new species described. Front. Plant Sci. 2020, 11, 690. [Google Scholar] [CrossRef]
- Azizi, R.; Ghosta, Y.; Ahmadpour, A. Morphological and molecular characterization of Cytospora species involved in apple decline in Iran. Mycol. Iran. 2020, 7, 205–218. [Google Scholar] [CrossRef]
- Hanifeh, S.; Zafari, D.; Soleimani, M.-J.; Arzanlou, M. Multigene phylogeny, morphology, and pathogenicity trials reveal novel Cytospora species involved in perennial canker disease of apple trees in Iran. Fungal Biol. 2022, 126, 707–726. [Google Scholar] [CrossRef]
- Ilyukhin, E.; Nguyen, H.D.; Castle, A.J.; Ellouze, W. Cytospora paraplurivora sp. nov. isolated from orchards with fruit tree decline syndrome in Ontario, Canada. PLoS ONE 2023, 18, e0279490. [Google Scholar] [CrossRef]
- Rakhimova, E.V.; Sypabekkyzy, G.; Kyzmetova, L.A.; Asylbek, A.M. Genus Cytospora Ehrenb. In the south-east of Kazakhstan. Exp. Biol. 2023, 96, 52–65. [Google Scholar] [CrossRef]
- Norphanphoun, C.; Doilom, M.; Daranagama, D.A.; Phookamsak, R.; Wen, T.C.; Bulgakov, T.S.; Hyde, K.D. Revisiting the genus Cytospora and allied species. Mycosphere 2017, 8, 51–97. [Google Scholar] [CrossRef]
- Norphanphoun, C.; Raspé, O.; Jeewon, R.; Wen, T.C.; Hyde, K.D. Morphological and phylogenetic characterisation of novel Cytospora species associated with mangroves. MycoKeys 2018, 38, 93–120. [Google Scholar] [CrossRef]
- Pan, M.; Zhu, H.Y.; Tian, C.M.; Huang, M.R.; Fan, X. Assessment of Cytospora strains from conifer cankers in China, with the descriptions of four new Cytospora species. Front. Plant Sci. 2021, 12, 636460. [Google Scholar] [CrossRef] [PubMed]
- Travadon, R.; Lawrence, D.P.; Moyer, M.; Fujiyoshi, P.T.; Baumgartner, K. Fungal species associated with grapevine trunk diseases in Washington wine grapes and California table grapes, with novelties in the genera Cadophora, Cytospora, and Sporocadus. Front. Fungal Biol. 2022, 3, 1018140. [Google Scholar] [CrossRef] [PubMed]
- Konta, S.; Tibpromma, S.; Karunarathna, S.C.; Samarakoon, M.C.; Steven, L.S.; Mapook, A.; Boonmee, S.; Senwanna, C.; Balasuriya, A.; Eungwanichayapant, P.D.; et al. Morphology and multigene phylogeny reveal ten novel taxa in Ascomycota from terrestrial palm substrates (Arecaceae) in Thailand. Mycosphere 2023, 14, 107–152. [Google Scholar] [CrossRef]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc. Natl. Acad. Sci. USA 2012, 16, 6241–6246. [Google Scholar] [CrossRef]
- Raja, H.A.; Miller, A.N.; Pearce, C.J.; Oberlies, N.H. Fungal identification using molecular tools: A primer for the natural products research community. J. Nat. Prod. 2017, 80, 756–770. [Google Scholar] [CrossRef]
- Tekpinar, A.; Kalmer, A. Utility of various molecular markers in fungal identification and phylogeny. Nova Hedwig. 2019, 109, 187–224. [Google Scholar] [CrossRef]
- Jiang, N.; Yang, Q.; Fan, X.L.; Tian, C.M. Identification of six Cytospora species on Chinese chestnut in China. MycoKeys 2020, 62, 1–25. [Google Scholar] [CrossRef]
- Fotouhifar, K.B.; Hedjaroude, G.A.; Leuchtmann, A. ITS rDNA phylogeny of Iranian strains of Cytospora and associated teleomorphs. Mycologia 2010, 102, 1369–1382. [Google Scholar] [CrossRef] [PubMed]
- Cai, G.; Zhao, Y.; Zhai, Y.; Yan, M.; Ma, R.; Zhang, D. Two new species of Cytospora (Diaporthales, Cytosporaceae) causing canker disease of Malus domestica and M. sieversii in Xinjiang, China. MycoKeys 2024, 109, 305–318. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.T.; Otto, K.L.; Sterle, D.; Minas, I.S.; Stewart, J.E. Preventive fungicidal control of Cytospora leucostoma in peach orchards in Colorado. Plant Dis. 2019, 103, 1138–1147. [Google Scholar] [CrossRef]
- Agustí-Brisach, C.; Moldero, D.; Raya, M.D.C.; Lorite, I.J.; Orgaz, F.; Trapero, A. Water stress enhances the progression of branch dieback and almond decline under field conditions. Plants 2020, 9, 1213. [Google Scholar] [CrossRef] [PubMed]
- Paap, T.; Brouwers, N.C.; Burgess, T.I.; Hardy, G.E.S.J. Importance of climate, anthropogenic disturbance and pathogens (Quambalaria coyrecup and Phytophthora spp.) on marri (Corymbia calophylla) tree health in southwest Western Australia. Ann. For. Sci. 2017, 74, 62. [Google Scholar] [CrossRef]
- Hossein, M.; Veneklaas, E.J.; Hardy, G.E.S.J.; Poot, P. Tree host-pathogen interactions as influenced by drought timing: Linking physiological performance, biochemical defense and disease severity. Tree Physiol. 2019, 39, 6–18. [Google Scholar] [CrossRef]
- Gomez-Gallego, M.; Galiano, L.; Martínez-Vilalta, J.; Stenlid, J.; Capador-Barreto, H.D.; Elfstrand, M.; Camarero, J.J.; Oliva, J. Interaction of drought- and pathogen-induced mortality in Norway spruce and Scot pine. Plant Cell Environ. 2022, 45, 2292–2305. [Google Scholar] [CrossRef]
- Li, Q.; Cao, S.; Wang, L.; Hou, R.; Sun, W. Impacts of climate change on the potential distribution of three Cytospora species in Xinjiang, China. Forests 2024, 15, 1617. [Google Scholar] [CrossRef]
| Primers Name, 5′-3′ Sequences | References | PCR Components |
|---|---|---|
| ITS1: TCCGTAGGTGAACCTGCGG ITS4: TCCTCCGCTTATTGATATGC | [19] | 1 μL genomic DNA, 12.5 μL master mix, 1 μL ITS 1, 1 Μl ITS4, 9.5 μL ddH2O |
| EF1-688F: CGGTCACTGATCTACAAGTGC EF1-R: CCTCGAACTCACCAGTACCG | [20] | |
| NL1: GCATATCAATAAGCGGAGAAAAG NL4: GGTCCGTGTTTCAAGACGG | [21] | |
| Bt2a: GGTAACCAAATCGGTGCTGCTTTC Bt2b: ACCCTCAGTGTAGTGACCCTTGGC | [22] |
| Apple Cultivars | Cytospora Species | |
|---|---|---|
| C. leucostoma | C. sorbicola | |
| Mean Lesion Length (cm) * | Mean Lesion Length (cm) | |
| Gala | 3.83 ± 0.33 c ** B *** | 6.33 ± 0.33 ab A |
| Golden Delicious | 6.33 ± 0.33 b A | 4.67 ±0.72 bc A |
| Red Delicious | 6.33 ± 0.33 b A | 8.83 ±0.72 a A |
| Granny Smith | 4.50 ± 0.50 bc A | 4.83 ± 0.60 bc A |
| Fuji | 9.00 ± 0.57 a A | 6.33 ± 0.67 ab B |
| Jonaprince | 1.83 ± 0.17 d A | 3.17 ± 0.44 c A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tulegenova, Z.; Nayekova, S.; Zhaxylykov, A.; Spanbayev, A.; Dyussembayev, K.; Mukiyanova, G.; Nariman, T.; Kiyan, V.; Sevindik, E.; Eken, C. Phylogenetic and Pathogenic Characterization of Cytospora Species Causing Apple Canker in Kazakhstan. Agriculture 2025, 15, 2490. https://doi.org/10.3390/agriculture15232490
Tulegenova Z, Nayekova S, Zhaxylykov A, Spanbayev A, Dyussembayev K, Mukiyanova G, Nariman T, Kiyan V, Sevindik E, Eken C. Phylogenetic and Pathogenic Characterization of Cytospora Species Causing Apple Canker in Kazakhstan. Agriculture. 2025; 15(23):2490. https://doi.org/10.3390/agriculture15232490
Chicago/Turabian StyleTulegenova, Zhanar, Saltanat Nayekova, Alikhan Zhaxylykov, Aidar Spanbayev, Kazbek Dyussembayev, Gulzhamal Mukiyanova, Tursunbayev Nariman, Vladimir Kiyan, Emre Sevindik, and Cafer Eken. 2025. "Phylogenetic and Pathogenic Characterization of Cytospora Species Causing Apple Canker in Kazakhstan" Agriculture 15, no. 23: 2490. https://doi.org/10.3390/agriculture15232490
APA StyleTulegenova, Z., Nayekova, S., Zhaxylykov, A., Spanbayev, A., Dyussembayev, K., Mukiyanova, G., Nariman, T., Kiyan, V., Sevindik, E., & Eken, C. (2025). Phylogenetic and Pathogenic Characterization of Cytospora Species Causing Apple Canker in Kazakhstan. Agriculture, 15(23), 2490. https://doi.org/10.3390/agriculture15232490






