Molecular Regulatory Networks Underlying Root Growth and Development in Crested Wheatgrass (Agropyron cristatum L.)
Abstract
1. Introduction
2. Material and Methods
2.1. Plant Materials and Growth Conditions
2.2. Root Morphology Measurements
2.3. RNA Extraction and RNA-Seq Library Construction
2.4. Transcriptome Sequencing and Quality Assessment
2.5. Differentially Expressed Gene Analysis and Functional Annotation
2.6. Quantitative Real-Time PCR Analysis
2.7. Data Analysis and Statistical Methods
3. Results
3.1. Plant Root Growth Index Determination
3.2. Differential Expression Genes in Roots of A. cristatum s at Different Sampling Days
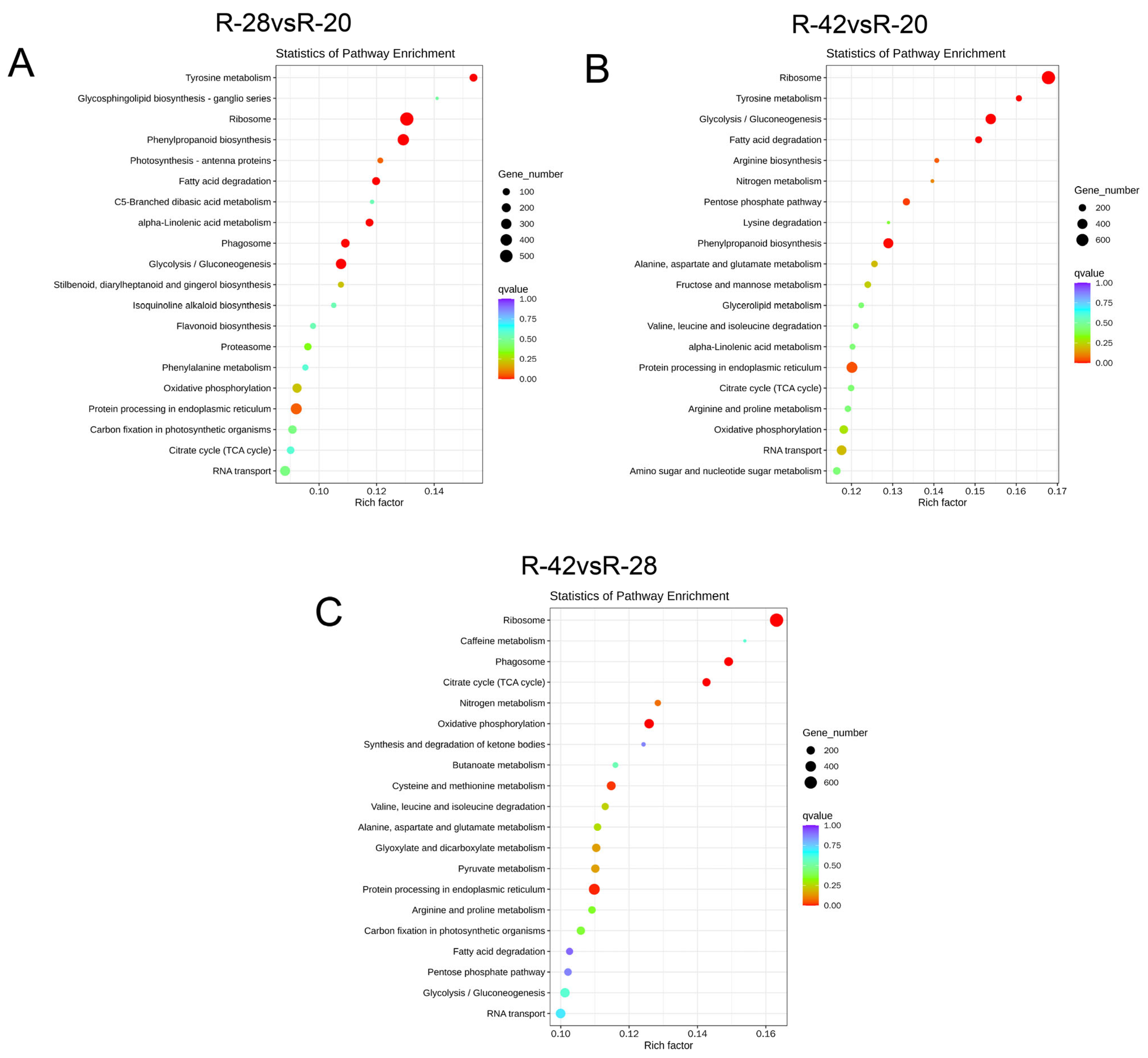
3.3. KEGG Classification and Enrichment Analysis of Differentially Expressed Genes
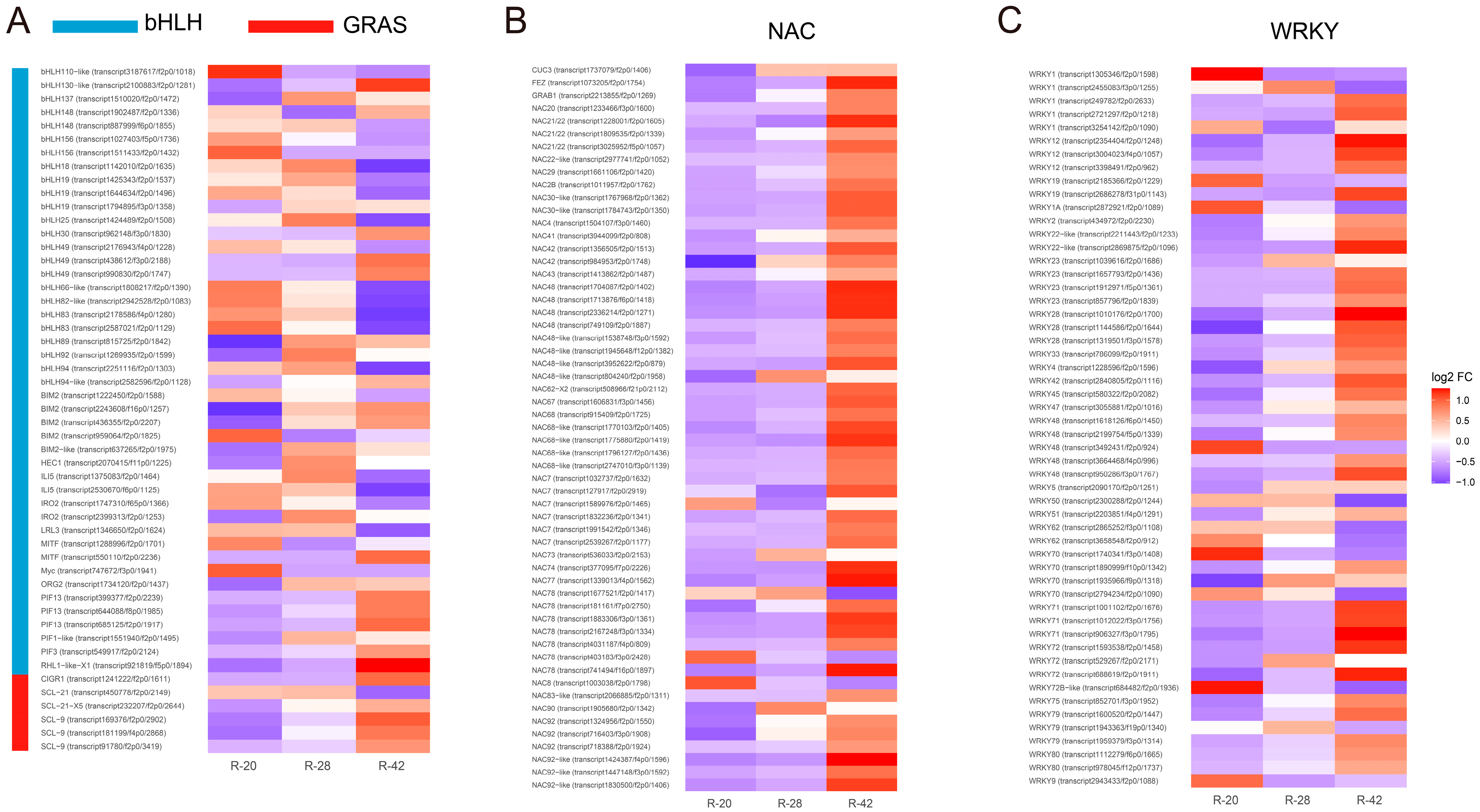
3.4. Transcription Factor Analysis During Root Development
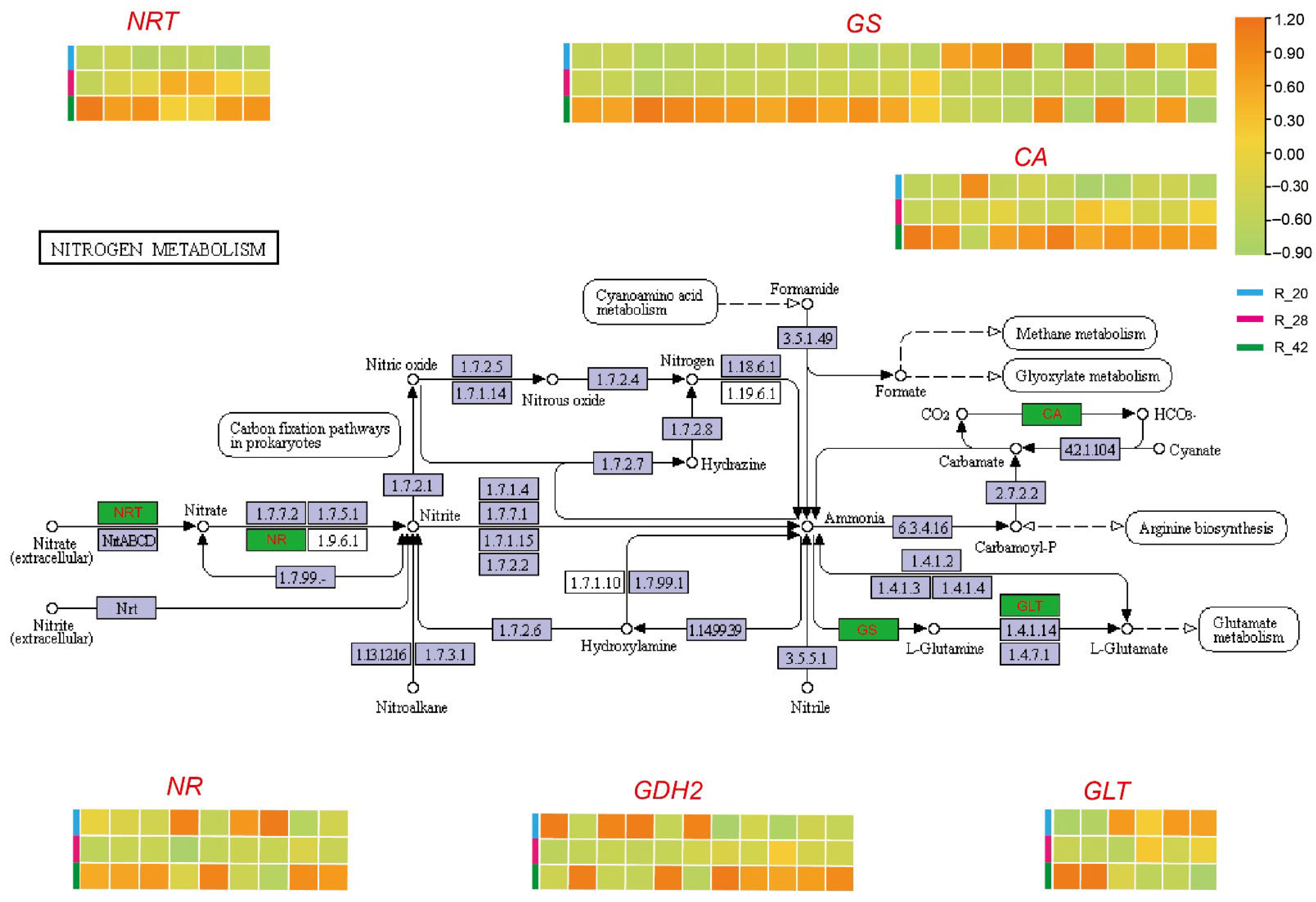
3.5. Nitrogen Metabolic Network Dynamics During Root Development
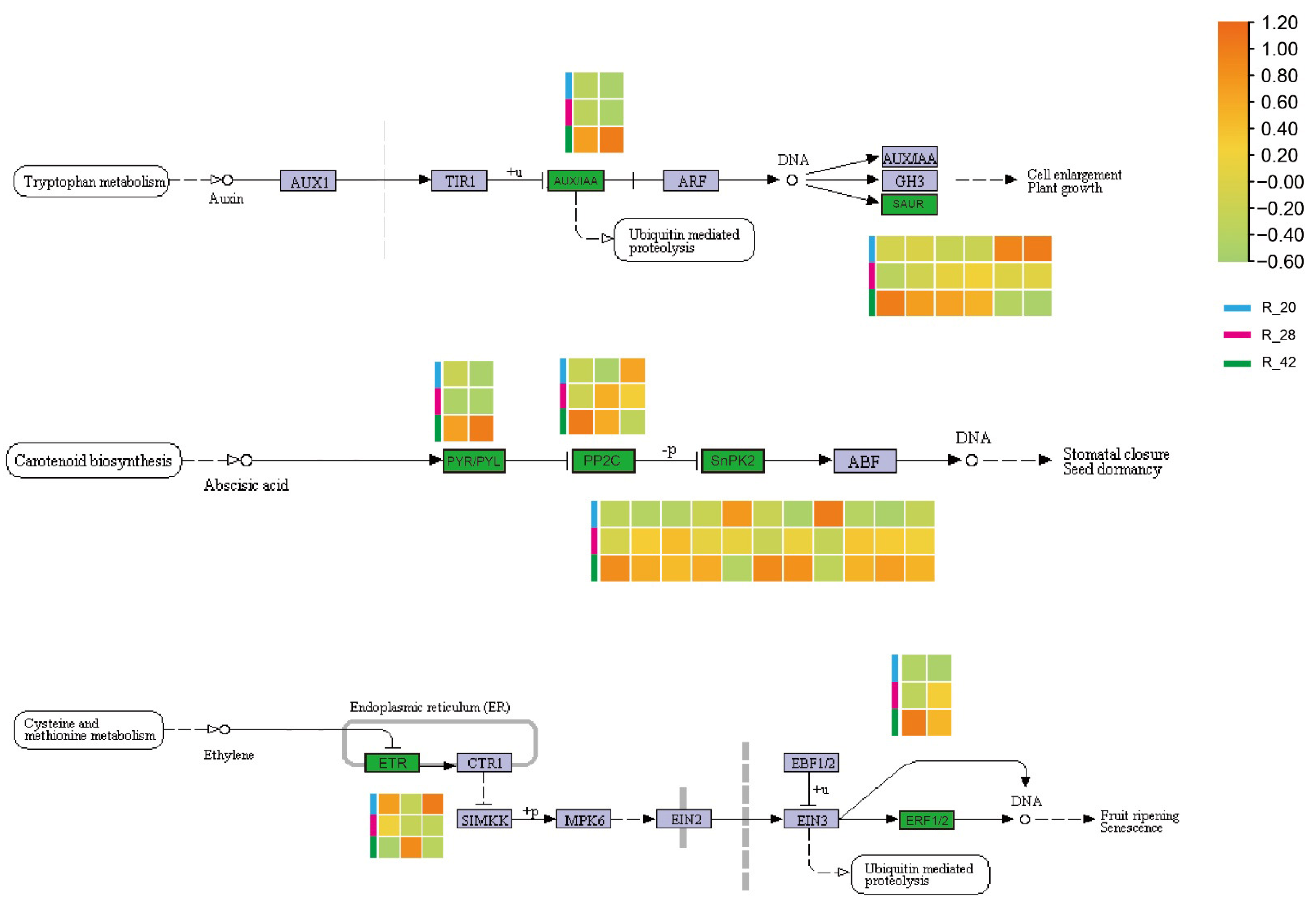
3.6. Phytohormone Signaling Networks in Root Development
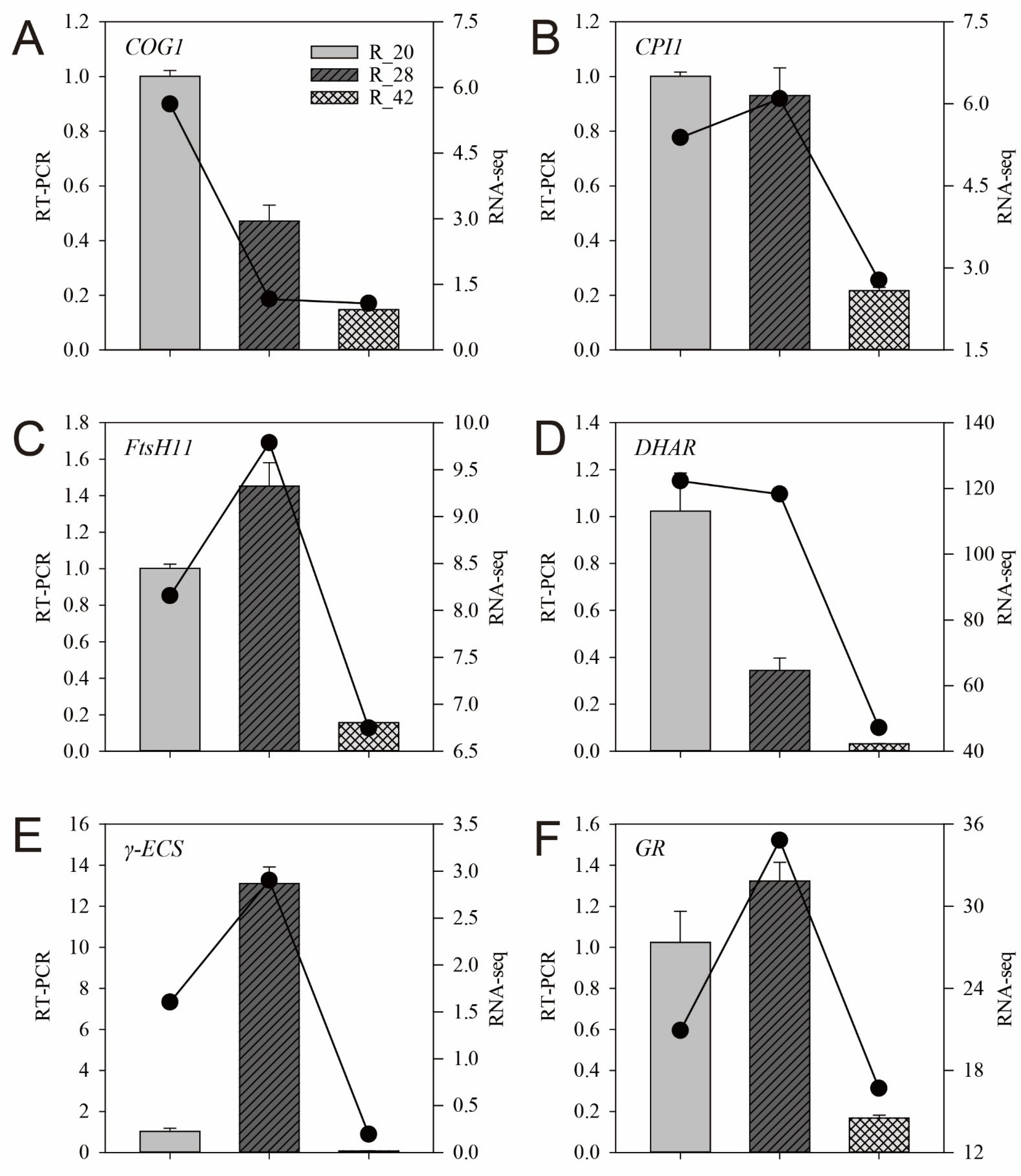
3.7. RT-qPCR Validation of RNA-Seq Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Robins, J.; Jensen, K. Breeding of the Crested Wheatgrass Complex (Agropyron spp.) for North American Temperate Rangeland Agriculture and Conservation. Agronomy 2020, 10, 1134. [Google Scholar] [CrossRef]
- Koryakina, V.; Kochegina, A. Results of studying wheatgrass (Agropyron Gaertn.) accessions from the VIR global genetic resources collection in Yakutia. Proc. Appl. Bot. Genet. Breed. 2021, 182, 59–71. [Google Scholar] [CrossRef]
- Asay, K.H.; Johnson, D.A. Genetic Variances for Forage Yield in Crested Wheatgrass at Six Levels of Irrigation. Crop Sci. 1990, 30, 79–82. [Google Scholar] [CrossRef]
- Wei, X.; Yu, W.; Qinghua, Y. Effect of NaCl on Growth and Activities of Protective Enzymes in Seedlings of White Clovers (Trifolium repens L.). Acta Agrestia Sinica. 2011, 19, 492–496. Available online: https://manu40.magtech.com.cn/Jweb_cdxb/CN/Y2011/V19/I3/492 (accessed on 16 November 2025).
- Abdelrady, W.A.; Ma, Z.; Elshawy, E.E.; Wang, L.; Askri, S.M.H.; Ibrahim, Z.; Dennis, E.; Kanwal, F.; Zeng, F.; Shamsi, I.H. Physiological and biochemical mechanisms of salt tolerance in barley under salinity stress. Plant Stress. 2024, 11, 100403. [Google Scholar] [CrossRef]
- Shi, Y.; Wan, L.; Liu, J.; Wang, Y.; Guo, R.; Wu, X.; Li, X. Analysis of the Principal Components and the Subordinate Function of Lolium perenne Drought Resistance. Acta Agrestia Sin. 2010, 18, 669–672. Available online: https://manu40.magtech.com.cn/Jweb_cdxb/EN/Y2010/V18/I5/669 (accessed on 16 November 2025).
- Pierret, A.; Moran, C.J. Plant Roots and Soil Structure. In Encyclopedia of Agrophysics; Encyclopedia of Earth Sciences Series; Springer: Dordrecht, The Netherlands, 2011; pp. 628–632. [Google Scholar] [CrossRef]
- Robins, J.G.; Waldron, B.L.; Jensen, K.B. Productivity, stability, and resilience of cool-season perennial grasses used for rangeland revegetation. Agrosystems Geosci. Environ. 2020, 3, e20002. [Google Scholar] [CrossRef]
- Xu, C.; Mi, F.; Wang, Y.; Li, X.; Zhao, H. Factors influencing plant regeneration systems in mature embryo culture of Agropyron. Acta Prataculturae Sin. 2009, 18, 80–85. Available online: http://cyxb.magtech.com.cn/EN/Y2009/V18/I1/80 (accessed on 16 November 2025).
- Ruiz Herrera, L.F.; Shane, M.W.; López-Bucio, J. Nutritional regulation of root development. Wiley Interdiscip. Rev. Dev. Biol. 2015, 4, 431–443. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Singh, D.; Saksena, H.B.; Sharma, M.; Tiwari, A.; Awasthi, P.; Botta, H.K.; Shukla, B.N.; Laxmi, A. Understanding the Intricate Web of Phytohormone Signalling in Modulating Root System Architecture. Int. J. Mol. Sci. 2021, 22, 5508. [Google Scholar] [CrossRef]
- Dewey, D.R. The Genomic System of Classification as a Guide to Intergeneric Hybridization with the Perennial Triticeae. BMC Plant Biol. 2021, 21, 9.209–279. [Google Scholar]
- Asay, K.H.; Jensen, K.B.; Hsiao, C.; Dewey, D.R. Probable origin of standard crested wheatgrass, Agropyron desertorum Fisch. ex Link, Schultes. Can. J. Plant Sci. 1992, 72, 763–772. [Google Scholar] [CrossRef]
- Fan, L.; Tang, J.; Ma, L.; Zhang, Z.; Jiang, Q.; Li, E.; Ma, Q.; Wang, X.; Zhao, W. Screening Genes Related to Drought Resistance in Agropyron mongolicum Keng Based on Transcriptome. Acta Agrestia Sin. 2024, 32, 3344–3357. [Google Scholar] [CrossRef]
- Satam, H.; Joshi, K.; Mangrolia, U.; Waghoo, S.; Zaidi, G.; Rawool, S.; Thakare, R.P.; Banday, S.; Mishra, A.K.; Das, G.; et al. Next-Generation Sequencing Technology: Current Trends and Advancements. Biology 2023, 12, 997. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Hu, L.; Li, H.; Chen, L.; Lou, Y.; Amombo, E.; Fu, J. RNA-seq for gene identification and transcript profiling in relation to root growth of bermudagrass (Cynodon dactylon) under salinity stress. BMC Genom. 2015, 16, 575. [Google Scholar] [CrossRef]
- Hu, T.; Sun, X.; Zhang, X.; Nevo, E.; Fu, J. An RNA sequencing transcriptome analysis of the high-temperature stressed tall fescue reveals novel insights into plant thermotolerance. BMC Genom. 2014, 15, 1147. [Google Scholar] [CrossRef]
- Du, J.; Li, X.; Li, T.; Yu, D.; Han, B. Genome-wide transcriptome profiling provides overwintering mechanism of Agropyron mongolicum. BMC Plant Biol. 2017, 17, 138. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, W.; Han, H.; Song, L.; Bai, L.; Gao, Z.; Zhang, Y.; Yang, X.; Li, X.; Gao, A.; et al. De novo transcriptome sequencing of Agropyron cristatum to identify available gene resources for the enhancement of wheat. Genomics 2015, 106, 129–136. [Google Scholar] [CrossRef]
- Zhou, S.; Zhang, J.; Han, H.; Zhang, J.; Ma, H.; Zhang, Z.; Lu, Y.; Liu, W.; Yang, X.; Li, X.; et al. Full-length transcriptome sequences of Agropyron cristatum facilitate the prediction of putative genes for thousand-grain weight in a wheat-A. cristatum translocation line. BMC Genom. 2019, 20, 1025. [Google Scholar] [CrossRef]
- Wang, S.; Ren, X.; Huang, B.; Wang, G.; Zhou, P.; An, Y. Aluminium-induced reduction of plant growth in alfalfa (Medicago sativa) is mediated by interrupting auxin transport and accumulation in roots. Sci. Rep. 2016, 6, 30079. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Wray, G.A. Transcriptional regulation and the evolution of development. Int. J. Dev. Biol. 2003, 47, 675–684. Available online: https://pubmed.ncbi.nlm.nih.gov/14756343/ (accessed on 2 July 2025).
- Makkena, S.; Lamb, R.S. The bHLH transcription factor SPATULA regulates root growth by controlling the size of the root meristem. BMC Plant Biol. 2013, 13, 1. [Google Scholar] [CrossRef]
- Zhao, Q.; Fan, Z.; Qiu, L.; Che, Q.; Wang, T.; Li, Y.; Wang, Y. MdbHLH130, an Apple bHLH Transcription Factor, Confers Water Stress Resistance by Regulating Stomatal Closure and ROS Homeostasis in Transgenic Tobacco. Front. Plant Sci. 2020, 11, 543696. [Google Scholar] [CrossRef]
- Ding, W.; Yu, Z.; Tong, Y.; Huang, W.; Chen, H.; Wu, P. A transcription factor with a bHLH domain regulates root hair development in rice. Cell Res. 2009, 19, 1309–1311. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Yang, Q.; Wang, X.; Chen, Y.; He, R.; Li, X.; Pan, H.; Zhuo, R.; Qu, T.; Qiu, W. Transcription Factors Involved in Plant Stress and Growth and Development: NAC. Agronomy 2025, 15, 949. [Google Scholar] [CrossRef]
- Vidal, E.; Álvarez, J.; Gutiérrez, R.A. Nitrate regulation of AFB3 and NAC4 gene expression in Arabidopsis roots depends on NRT1.1 nitrate transport function. Plant Signal Behav. 2014, 9, e28501. [Google Scholar] [CrossRef]
- Han, S.; Wang, Y.; Li, Y.; Zhu, R.; Gu, Y.; Li, J.; Guo, H.; Ye, W.; Nabi, H.; Yang, T.; et al. The OsNAC41-RoLe1-OsAGAP module promotes root development and drought resistance in upland rice. Mol. Plant 2024, 7, 1573–1593. [Google Scholar] [CrossRef] [PubMed]
- Grunewald, W.; Karimi, M.; Wieczorek, K.; Van de Cappelle, E.; Wischnitzki, E.; Grundler, F.; Inzé, D.; Beeckman, T.; Gheysen, G. A role for AtWRKY23 in feeding site establishment of plant-parasitic nematodes. Plant Physiol. 2008, 148, 358–368. [Google Scholar] [CrossRef]
- Shtin, M.; Dello Ioio, R.; Del Bianco, M. It’s Time for a Change: The Role of Gibberellin in Root Meristem Development. Front. Plant Sci. 2022, 13, 882517. [Google Scholar] [CrossRef]
- Neves, C.; Ribeiro, B.; Amaro, R.; Expósito, J.; Grimplet, J.; Fortes, A.M. Network of GRAS transcription factors in plant development, fruit ripening and stress responses. Hortic. Res. 2023, 10, uhad220. [Google Scholar] [CrossRef]
- Gaudinier, A.; Rodriguez-Medina, J.; Zhang, L.; Olson, A.; Liseron-Monfils, C.; Bågman, A.M.; Foret, J.; Abbitt, S.; Tang, M.; Li, B.; et al. Transcriptional regulation of nitrogen-associated metabolism and growth. Nature 2018, 563, 259–264. [Google Scholar] [CrossRef]
- Orosa-Puente, B.; Leftley, N.; Wangenheim, D.; Banda, J.; Srivastava, A.K.; Hill, K.; Truskina, J.; Bhosale, R.; Morris, E.; Srivastava, M.; et al. Root branching toward water involves posttranslational modification of transcription factor ARF7. Science 2018, 362, 1407–1410. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Gu, Y.; Liu, Y.; Liu, Z.; Wang, P. Nitrogen-Driven Orchestration of Lateral Root Development: Molecular Mechanisms and Systemic Integration. Biology 2025, 14, 1099. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Boisson-Dernier, A.; Israelsson-Nordström, M.; Böhmer, M.; Xue, S.; Ries, A.; Godoski, J.; Kuhn, J.M.; Schroeder, J.I. Carbonic anhydrases are upstream regulators of CO2-controlled stomatal movements in guard cells. Nat. Cell Biol. 2010, 12, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Cheng, Y.H.; Chen, K.E.; Tsay, Y.F. Nitrate Transport, Signaling, and Use Efficiency. Annu. Rev. Plant Biol. 2018, 69, 85–122. [Google Scholar] [CrossRef]
- Naz, M.; Luo, B.; Guo, X.; Li, B.; Chen, J.; Fan, X. Overexpression of Nitrate Transporter OsNRT2.1 Enhances Nitrate-Dependent Root Elongation. Genes 2019, 10, 290. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, Y.; Nie, G.; Liu, H. Leaf and Root Growth, Carbon and Nitrogen Contents, and Gene Expression of Perennial Ryegrass to Different Nitrogen Supplies. J. Am. Soc. Hortic. Sci. 2016, 141, 555–562. [Google Scholar] [CrossRef]
- Remans, T.; Nacry, P.; Pervent, M.; Filleur, S.; Diatloff, E.; Mounier, E.; Tillard, P.; Forde, B.G.; Gojon, A. The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc. Natl. Acad. Sci. USA 2006, 103, 19206–19211. [Google Scholar] [CrossRef]
- Pozo, M.J.; López-Ráez, J.A.; Azcón-Aguilar, C.; García-Garrido, J.M. Phytohormones as integrators of environmental signals in the regulation of mycorrhizal symbioses. New Phytol. 2015, 205, 1431–1436. [Google Scholar] [CrossRef]
- Yoshida, T.; Fernie, A.R.; Shinozaki, K.; Takahashi, F. Long-distance stress and developmental signals associated with abscisic acid signaling in environmental responses. Plant J. 2021, 105, 477–488. [Google Scholar] [CrossRef]
- Spartz, A.K.; Lee, S.H.; Wenger, J.P.; Gonzalez, N.; Itoh, H.; Inzé, D.; Peer, W.A.; Murphy, A.S.; Overvoorde, P.J.; Gray, W.M. The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J. 2012, 70, 978–990. [Google Scholar] [CrossRef]
- Qiu, T.; Chen, Y.; Li, M.; Kong, Y.; Zhu, Y.; Han, N.; Bia, H.; Zhu, M.; Wang, J. The tissue-specific and developmentally regulated expression patterns of the SAUR41 subfamily of small auxin up RNA genes: Potential implications. Plant Signal Behav. 2013, 8, e25283. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Gray, W.M. SAUR Proteins as Effectors of Hormonal and Environmental Signals in Plant Growth. Mol. Plant 2015, 8, 1153–1164. [Google Scholar] [CrossRef] [PubMed]
- Umezawa, T.; Nakashima, K.; Miyakawa, T.; Kuromori, T.; Tanokura, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular basis of the core regulatory network in ABA responses: Sensing, signaling and transport. Plant Cell Physiol. 2010, 51, 1821–1839. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, Z.; Gao, J.; Wang, P.; Hu, T.; Wang, Z.; Hou, Y.J.; Wan, Y.; Liu, W.; Xie, S.; et al. Arabidopsis Duodecuple Mutant of PYL ABA Receptors Reveals PYL Repression of ABA-Independent SnRK2 Activity. Cell Rep. 2018, 23, 3340–3351.e5. [Google Scholar] [CrossRef]
- Kieber, J.J.; Rothenberg, M.; Roman, G.; Feldmann, K.A.; Ecker, J.R. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 1993, 72, 427–441. [Google Scholar] [CrossRef]
- Yoo, S.D.; Cho, Y.; Sheen, J. Emerging connections in the ethylene signaling network. Trends Plant Sci. 2009, 14, 270–279. [Google Scholar] [CrossRef]
- Meng, X.; Xu, J.; He, Y.; Yang, K.Y.; Mordorski, B.; Liu, Y.; Zhang, S. Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell 2013, 25, 1126–1142. [Google Scholar] [CrossRef]
- Contreras-Cornejo, H.; López-Bucio, J.; Méndez-Bravo, A.; Macías-Rodríguez, L.; Ramos-Vega, M.; Guevara-García, Á.A.; López-Bucio, J. Mitogen-Activated Protein Kinase 6 and Ethylene and Auxin Signaling Pathways Are Involved in Arabidopsis Root-System Architecture Alterations by Trichoderma atroviride. Mol. Plant Microbe Interact. 2015, 28, 701–710. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, H.; Li, X.; Xu, Y.; Zhang, X.; Long, R.; Ding, W.; Li, R.; Zhao, Y.; Wang, X.; Li, M. Molecular Regulatory Networks Underlying Root Growth and Development in Crested Wheatgrass (Agropyron cristatum L.). Agriculture 2025, 15, 2392. https://doi.org/10.3390/agriculture15222392
Zhu H, Li X, Xu Y, Zhang X, Long R, Ding W, Li R, Zhao Y, Wang X, Li M. Molecular Regulatory Networks Underlying Root Growth and Development in Crested Wheatgrass (Agropyron cristatum L.). Agriculture. 2025; 15(22):2392. https://doi.org/10.3390/agriculture15222392
Chicago/Turabian StyleZhu, He, Xinyu Li, Yanran Xu, Xiaxiang Zhang, Ruicai Long, Wang Ding, Ruyue Li, Yan Zhao, Xuemin Wang, and Mingna Li. 2025. "Molecular Regulatory Networks Underlying Root Growth and Development in Crested Wheatgrass (Agropyron cristatum L.)" Agriculture 15, no. 22: 2392. https://doi.org/10.3390/agriculture15222392
APA StyleZhu, H., Li, X., Xu, Y., Zhang, X., Long, R., Ding, W., Li, R., Zhao, Y., Wang, X., & Li, M. (2025). Molecular Regulatory Networks Underlying Root Growth and Development in Crested Wheatgrass (Agropyron cristatum L.). Agriculture, 15(22), 2392. https://doi.org/10.3390/agriculture15222392





