Multi-Omics Annotation and Residual Split Strategy-Based Deep Learning Model for Efficient and Robust Genomic Prediction in Pigs
Abstract
1. Introduction
2. Materials and Methods
2.1. Pig Dataset
2.2. Multi-Omics Functional Annotation Information
2.3. Methods and Models
2.3.1. The Models Used for Comparisons
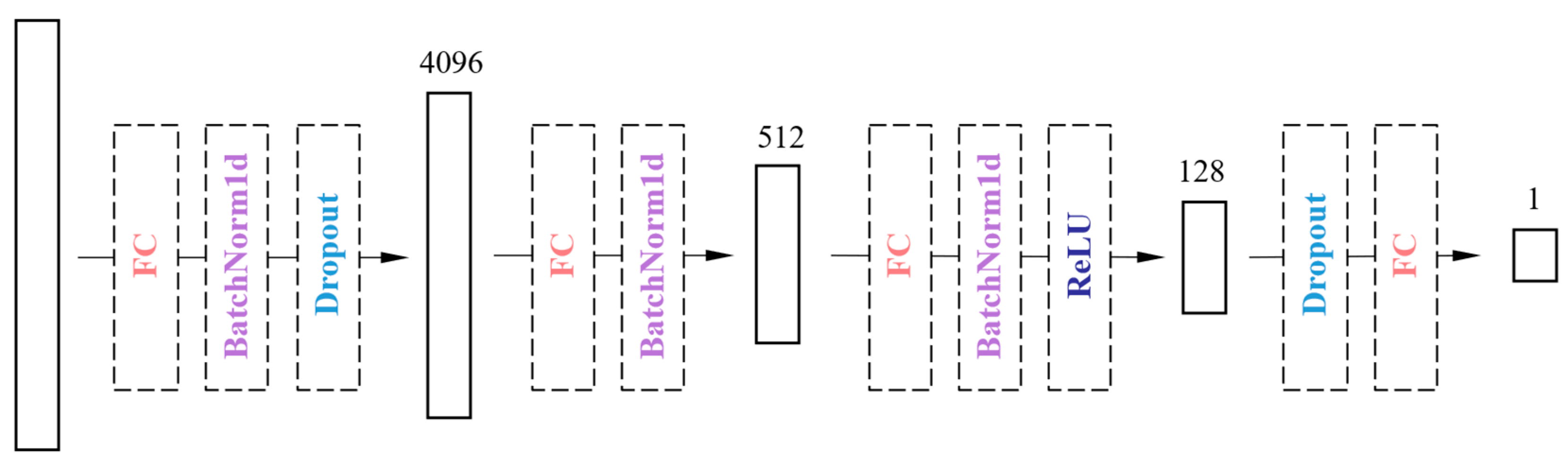
2.3.2. The Proposed Model ‘MARS’
2.3.3. Evaluation Methods and Indicators
3. Results
3.1. Data Analysis
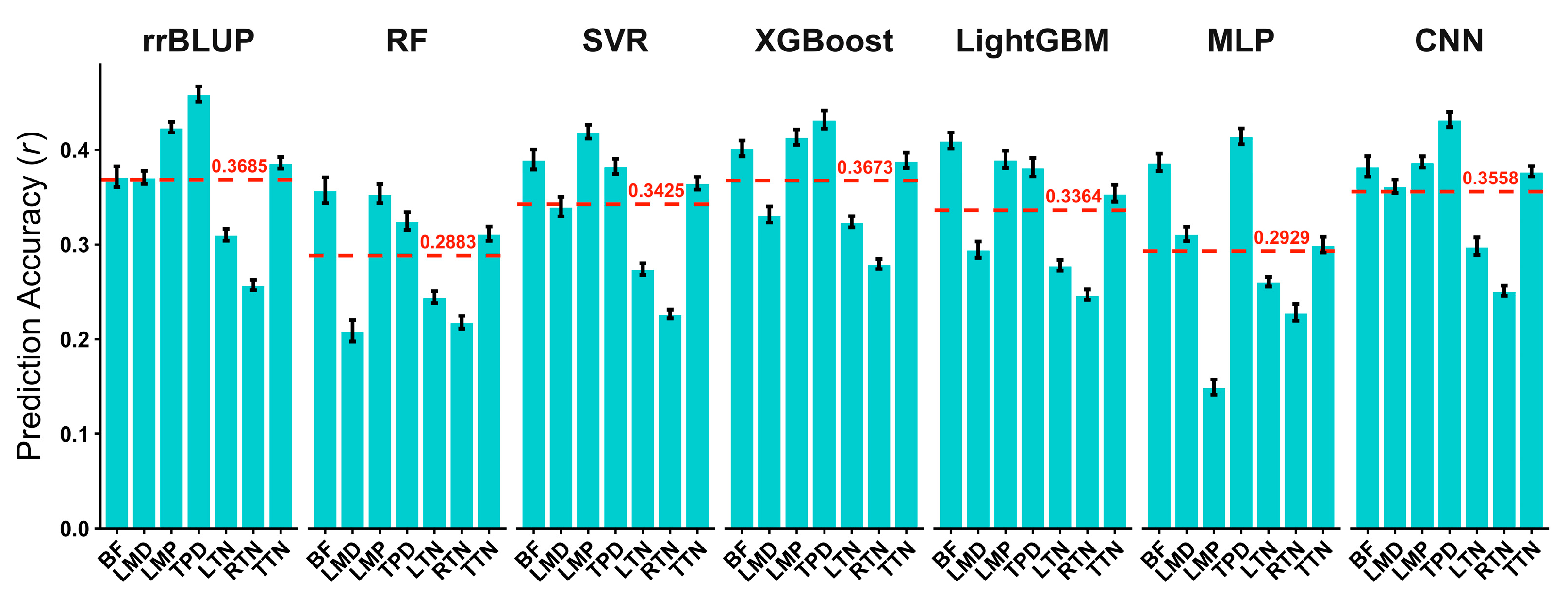
3.2. Comparison of Prediction Performances of Different Models
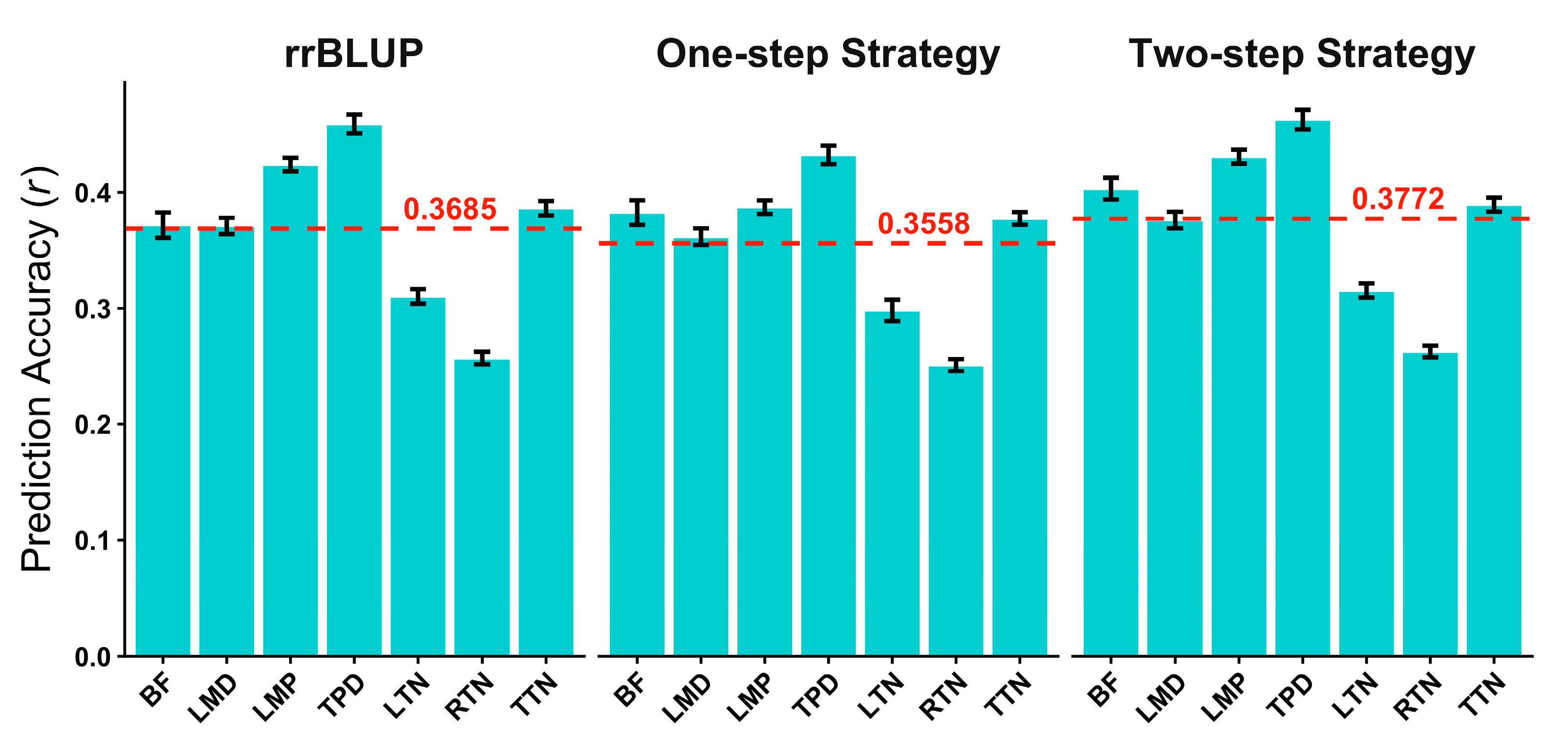
3.3. Effectiveness of Residual Split Strategy on Deep Learning Model
3.4. Multi-Omics Annotation-Based Hierarchical Feature Selection
4. Discussion
5. Conclusions
6. Availability of Used Software Tools and Packages
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Meuwissen, T.H.; Hayes, B.J.; Goddard, M.E. Prediction of total genetic value using genome-wide dense marker maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef]
- Georges, M.; Charlier, C.; Hayes, B. Harnessing genomic information for livestock improvement. Nat. Rev. Genet. 2019, 20, 135–156. [Google Scholar]
- Chatterjee, N.; Shi, J.; García-Closas, M. Developing and evaluating polygenic risk prediction models for stratified disease prevention. Nat. Rev. Genet. 2016, 17, 392–406. [Google Scholar] [CrossRef]
- Xu, Y.; Ma, K.; Zhao, Y.; Wang, X.; Zhou, K.; Yu, G.; Li, C.; Li, P.; Yang, Z.; Xu, C. Genomic selection: A breakthrough technology in rice breeding. Crop J. 2021, 9, 669–677. [Google Scholar]
- Schaeffer, L. Strategy for applying genome-wide selection in dairy cattle. J. Anim. Breed. Genet. Genet. 2006, 123, 218–223. [Google Scholar]
- Lilin, Y.; Yunlong, M.; Tao, X.; Mengjin, Z.; Mei, Y.; Xinyun, L.; Xiaolei, L.; Shuhong, Z. The Progress and Prospect of Genomic Selection Models. Acta Vet. Zootech. Sin. 2019, 50, 233–242. [Google Scholar]
- Zuk, O.; Hechter, E.; Sunyaev, S.R.; Lander, E.S. The mystery of missing heritability: Genetic interactions create phantom heritability. Proc. Natl. Acad. Sci. USA 2012, 109, 1193–1198. [Google Scholar] [CrossRef]
- Lappalainen, T.; Li, Y.I.; Ramachandran, S.; Gusev, A. Genetic and molecular architecture of complex traits. Cell 2024, 187, 1059–1075. [Google Scholar] [CrossRef]
- Avasthi, P.; Celebi, F.M.; Hochstrasser, M.L.; Mets, D.G.; York, R. Harnessing genotype-phenotype nonlinearity to accelerate biological prediction. Arcadia Sci. 2023. [Google Scholar] [CrossRef]
- Costa-Neto, G.; Fritsche-Neto, R.; Crossa, J. Nonlinear kernels, dominance, and envirotyping data increase the accuracy of genome-based prediction in multi-environment trials. Heredity 2021, 126, 92–106. [Google Scholar] [CrossRef]
- Hay, E.H. Machine Learning for the Genomic Prediction of Growth Traits in a Composite Beef Cattle Population. Animals 2024, 14, 3014. [Google Scholar] [CrossRef]
- Zhu, W.; Li, W.; Zhang, H.; Li, L. Big data and artificial intelligence-aided crop breeding: Progress and prospects. J. Integr. Plant Biol. 2024, 67, 722–739. [Google Scholar] [CrossRef]
- Montesinos-López, O.A.; Montesinos-López, A.; Pérez-Rodríguez, P.; Barrón-López, J.A.; Martini, J.W.; Fajardo-Flores, S.B.; Gaytan-Lugo, L.S.; Santana-Mancilla, P.C.; Crossa, J. A review of deep learning applications for genomic selection. BMC Genom. 2021, 22, 19. [Google Scholar] [CrossRef]
- Chafai, N.; Hayah, I.; Houaga, I.; Badaoui, B. A review of machine learning models applied to genomic prediction in animal breeding. Front. Genet. 2023, 14, 1150596. [Google Scholar] [CrossRef]
- Srivastava, S.; Lopez, B.I.; Kumar, H.; Jang, M.; Chai, H.-H.; Park, W.; Park, J.-E.; Lim, D. Prediction of Hanwoo Cattle Phenotypes from Genotypes Using Machine Learning Methods. Animals 2021, 11, 2066. [Google Scholar] [CrossRef]
- Shokor, F.; Croiseau, P.; Gangloff, H.; Saintilan, R.; Tribout, T.; Mary-Huard, T.; Cuyabano, B.C.D. Deep Learning and GBLUP Integration: An Approach that Identifies Nonlinear Genetic Relationships Between Traits. bioRxiv 2024. [Google Scholar] [CrossRef]
- An, B.; Liang, M.; Chang, T.; Duan, X.; Du, L.; Xu, L.; Zhang, L.; Gao, X.; Li, J.; Gao, H. KCRR: A nonlinear machine learning with a modified genomic similarity matrix improved the genomic prediction efficiency. Brief Bioinform 2021, 22, 132. [Google Scholar] [CrossRef]
- Pérez-Enciso, M.; Zingaretti, L.M. A guide on deep learning for complex trait genomic prediction. Genes 2019, 10, 553. [Google Scholar] [CrossRef]
- Habier, D.; Fernando, R.L.; Kizilkaya, K.; Garrick, D.J. Extension of the bayesian alphabet for genomic selection. BMC Bioinform. 2011, 12, 186. [Google Scholar] [CrossRef]
- Yin, L.; Zhang, H.; Zhou, X.; Yuan, X.; Zhao, S.; Li, X.; Liu, X. KAML: Improving genomic prediction accuracy of complex traits using machine learning determined parameters. Genome Biol. 2020, 21, 146. [Google Scholar] [CrossRef]
- Ma, W.; Qiu, Z.; Song, J.; Cheng, Q.; Ma, C. DeepGS: Predicting phenotypes from genotypes using Deep Learning. bioRxiv 2017. bioRxiv:241414. [Google Scholar]
- Wang, K.; Abid, M.A.; Rasheed, A.; Crossa, J.; Hearne, S.; Li, H. DNNGP, a deep neural network-based method for genomic prediction using multi-omics data in plants. Mol. Plant 2023, 16, 279–293. [Google Scholar] [CrossRef]
- e Silva, F.F.; Zambrano, M.F.B.; Varona, L.; Glória, L.S.; Lopes, P.S.; Silva, M.V.G.B.; Arbex, W.; Lázaro, S.F.; Resende, M.D.V.d.; Guimarães, S.E.F. Genome association study through nonlinear mixed models revealed new candidate genes for pig growth curves. Sci. Agric. 2017, 74, 1–7. [Google Scholar] [CrossRef]
- Xu, Y.; Liu, X.; Fu, J.; Wang, H.; Wang, J.; Huang, C.; Prasanna, B.M.; Olsen, M.S.; Wang, G.; Zhang, A. Enhancing genetic gain through genomic selection: From livestock to plants. Plant Commun. 2020, 1, 100005. [Google Scholar] [CrossRef] [PubMed]
- Romero, A.; Carrier, P.L.; Erraqabi, A.; Sylvain, T.; Auvolat, A.; Dejoie, E.; Legault, M.-A.; Dubé, M.-P.; Hussin, J.G.; Bengio, Y. Diet Networks: Thin Parameters for Fat Genomic. arXiv 2016, arXiv:1611.09340. [Google Scholar]
- Nani, J.P.; Rezende, F.M.; Peñagaricano, F. Predicting male fertility in dairy cattle using markers with large effect and functional annotation data. BMC Genom. 2019, 20, 258. [Google Scholar] [CrossRef]
- Edwards, S.M.; Sørensen, I.F.; Sarup, P.; Mackay, T.F.C.; Sørensen, P. Genomic Prediction for Quantitative Traits Is Improved by Mapping Variants to Gene Ontology Categories in Drosophila melanogaster. Genetics 2016, 203, 1871–1883. [Google Scholar] [CrossRef] [PubMed]
- Gao, N.; Martini, J.W.R.; Zhang, Z.; Yuan, X.; Zhang, H.; Simianer, H.; Li, J. Incorporating Gene Annotation into Genomic Prediction of Complex Phenotypes. Genetics 2017, 207, 489–501. [Google Scholar] [CrossRef] [PubMed]
- Speed, D.; Balding, D.J. MultiBLUP: Improved SNP-based prediction for complex traits. Genome Res. 2014, 24, 1550–1557. [Google Scholar] [CrossRef]
- Dunham, I.; Kundaje, A.; Aldred, S.F.; Collins, P.J.; Davis, C.A.; Doyle, F.; Epstein, C.B.; Frietze, S.; Harrow, J.; Kaul, R.; et al. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- Yang, R.; Guo, X.; Zhu, D.; Tan, C.; Bian, C.; Ren, J.; Huang, Z.; Zhao, Y.; Cai, G.; Liu, D.; et al. Accelerated deciphering of the genetic architecture of agricultural economic traits in pigs using a low-coverage whole-genome sequencing strategy. GigaScience 2021, 10, 048. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Ayres, D.L.; Darling, A.; Zwickl, D.J.; Beerli, P.; Holder, M.T.; Lewis, P.O.; Huelsenbeck, J.P.; Ronquist, F.; Swofford, D.L.; Cummings, M.P.; et al. BEAGLE: An Application Programming Interface and High-Performance Computing Library for Statistical Phylogenetics. Syst. Biol. 2012, 61, 170–173. [Google Scholar] [CrossRef]
- Ma, R.; Kuang, R.; Zhang, J.; Sun, J.; Xu, Y.; Zhou, X.; Han, Z.; Hu, M.; Wang, D.; Fu, Y.; et al. Annotation and assessment of functional variants in regulatory regions using epigenomic data in farm animals. bioRxiv 2024, 6, 578787. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Smola, A.J.; Schölkopf, B.J.S. A tutorial on support vector regression. Stat. Comput. 2004, 14, 199–222. [Google Scholar] [CrossRef]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.-Y. Lightgbm: A highly efficient gradient boosting decision tree. Adv. Neural Inf. Process. Syst. 2017, 30, 3149–3157. [Google Scholar]
- Gilmour, A.R.; Thompson, R.; Cullis, B.R. Average information REML: An efficient algorithm for variance parameter estimation in linear mixed models. Biometrics 1995, 51, 1440–1450. [Google Scholar] [CrossRef]
- Yin, L.; Zhang, H.; Tang, Z.; Yin, D.; Fu, Y.; Yuan, X.; Li, X.; Liu, X.; Zhao, S. HIBLUP: An integration of statistical models on the BLUP framework for efficient genetic evaluation using big genomic data. Nucleic Acids Res. 2023, 51, 3501–3512. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Adam, K.D.B.J. A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar] [CrossRef]
- Yin, L.; Zhang, H.; Tang, Z.; Xu, J.; Yin, D.; Zhang, Z.; Yuan, X.; Zhu, M.; Zhao, S.; Li, X.; et al. rMVP: A Memory-efficient, Visualization-enhanced, and Parallel-accelerated Tool for Genome-wide Association Study. Genom. Proteom. Bioinform. 2021, 19, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Montesinos-López, A.; Montesinos-López, O.A.; Gianola, D.; Crossa, J.; Hernández-Suárez, C.M. Multi-environment genomic prediction of plant traits using deep learners with dense architecture. G3 Genes Genomes Genet. 2018, 8, 3813–3828. [Google Scholar] [CrossRef]
- Wang, J.; Tiezzi, F.; Huang, Y.; Maltecca, C.; Jiang, J. Benchmarking of feed-forward neural network models for genomic prediction of quantitative traits in pigs. Front. Genet. 2025, 16, 1618891. [Google Scholar] [CrossRef]
- Vabalas, A.; Gowen, E.; Poliakoff, E.; Casson, A.J. Machine learning algorithm validation with a limited sample size. PLoS ONE 2019, 14, e0224365. [Google Scholar] [CrossRef]
- VanRaden, P.M. Efficient methods to compute genomic predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef] [PubMed]
- Goddard, M. Genomic selection: Prediction of accuracy and maximisation of long term response. Genetica 2009, 136, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Habier, D.; Fernando, R.L.; Dekkers, J.C.M. The Impact of Genetic Relationship Information on Genome-Assisted Breeding Values. Genetics 2007, 177, 2389–2397. [Google Scholar] [CrossRef]
- Töpner, K.; Rosa, G.J.M.; Gianola, D.; Schön, C.-C. Bayesian Networks Illustrate Genomic and Residual Trait Connections in Maize (Zea mays L.). G3 Genes|Genomes|Genet. 2017, 7, 2779–2789. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Xie, L.; Wang, F. An improved deep convolutional neural network to predict airfoil lift coefficient. In Proceedings of the International Conference on Aerospace System Science and Engineering, Toronto, ON, Canada, 30 July–1 August 2019; pp. 275–286. [Google Scholar]
- Xiang, T.; Li, T.; Li, J.; Li, X.; Wang, J. Using machine learning to realize genetic site screening and genomic prediction of productive traits in pigs. FASEB J. 2023, 37, 22961. [Google Scholar] [CrossRef]


| Category | Supporting Data |
|---|---|
| 1a | QTL + OCR + NFR + Motif + Footprint |
| 1b | OCR + NFR + Motif + Footprint |
| 1c | QTL + OCR + NFR + Footprint |
| 1d | OCR + NFR + Footprint |
| 2a | QTL + OCR + Motif + Footprint |
| 2b | OCR + Motif + Footprint |
| 2c | QTL + OCR + Footprint |
| 2d | OCR + Footprint |
| 3a | OCR + NFR |
| 3b | OCR/NFR + Motif |
| 4 | OCR/NFR/Footprint + TF binding site |
| 5 | H3K27ac peak |
| 6 | rest |
| Traits | Number | Max | Min | Median | Mean | SD | CV |
|---|---|---|---|---|---|---|---|
| BF | 2796 | 38.27 | 6.1 | 10.74 | 10.99 | 2.26 | 0.21 |
| LMD | 2796 | 59.4 | 5.2 | 46 | 46.15 | 3.93 | 0.09 |
| LMP | 2795 | 59.89 | 47.15 | 54.01 | 54.02 | 1.58 | 0.03 |
| TPD | 2604 | 93.09 | 10 | 62.49 | 62.96 | 9.99 | 0.16 |
| LTN | 2796 | 9 | 4 | 5 | 5.35 | 0.66 | 0.12 |
| RTN | 2796 | 8 | 4 | 5 | 5.37 | 0.64 | 0.12 |
| TTN | 2796 | 15 | 8 | 11 | 10.73 | 1.07 | 0.10 |
| Annotation Category | Number of Classified SNPs |
|---|---|
| QTL | 13,594 |
| OCR | 19,122 |
| NFR | 5493 |
| Footprint | 1154 |
| Matched Motif | 56 |
| Intersect Motif | 167 |
| Any motif | 6641 |
| Enhancer | 39,750 |
| Enhancer Narrow Peak | 62,506 |
| Active promoter | 9262 |
| Active promoter narrow peak | 19,852 |
| Category | Number of Classified SNPs |
|---|---|
| 1 | 45 |
| 2 | 68 |
| 3 | 643 |
| 4 | 34,436 |
| 5 | 54,169 |
| rest | 276,597 |
| Total | 365,958 |
| Traits | Feature Selection | |
|---|---|---|
| No | Yes | |
| BF | 0.4032 (0.0094) | 0.3988 (0.0099) |
| LMD | 0.3760 (0.0072) | 0.3752 (0.0068) |
| LMP | 0.4307 (0.0060) | 0.4293 (0.0059) |
| TPD | 0.4626 (0.0084) | 0.4659 (0.0079) |
| LTN | 0.3153 (0.0061) | 0.3123 (0.0065) |
| RTN | 0.2629 (0.0050) | 0.2640 (0.0055) |
| TTN | 0.3895 (0.0061) | 0.3915 (0.0058) |
| Mean | 0.3772 | 0.3767 |
| Traits/ Time (h) | CNN_Without_HFS 1 | CNN_with_HFS 2 | ||
|---|---|---|---|---|
| Training Time 3 | HFS Time 4 | Total Time 5 | ||
| BF | 7.74 | 1.49 | 0.29 | 1.78 |
| LMD | 0.66 | 0.20 | 0.30 | 0.5 |
| LMP | 1.66 | 0.13 | 0.34 | 0.47 |
| TPD | 0.27 | 0.05 | 0.31 | 0.36 |
| LTN | 0.62 | 0.16 | 0.35 | 0.51 |
| RTN | 0.34 | 0.07 | 0.31 | 0.38 |
| TTN | 0.27 | 0.04 | 0.32 | 0.36 |
| Mean | 1.65 | 0.31 | 0.32 | 0.62 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, J.; Tang, Z.; Zhang, H.; Liu, Y.; Xiong, X.; Liu, X.; Yin, L.; Lei, M. Multi-Omics Annotation and Residual Split Strategy-Based Deep Learning Model for Efficient and Robust Genomic Prediction in Pigs. Agriculture 2025, 15, 2354. https://doi.org/10.3390/agriculture15222354
Ma J, Tang Z, Zhang H, Liu Y, Xiong X, Liu X, Yin L, Lei M. Multi-Omics Annotation and Residual Split Strategy-Based Deep Learning Model for Efficient and Robust Genomic Prediction in Pigs. Agriculture. 2025; 15(22):2354. https://doi.org/10.3390/agriculture15222354
Chicago/Turabian StyleMa, Jingnan, Zhenshuang Tang, Haohao Zhang, Yangfan Liu, Xiong Xiong, Xiaolei Liu, Lilin Yin, and Minggang Lei. 2025. "Multi-Omics Annotation and Residual Split Strategy-Based Deep Learning Model for Efficient and Robust Genomic Prediction in Pigs" Agriculture 15, no. 22: 2354. https://doi.org/10.3390/agriculture15222354
APA StyleMa, J., Tang, Z., Zhang, H., Liu, Y., Xiong, X., Liu, X., Yin, L., & Lei, M. (2025). Multi-Omics Annotation and Residual Split Strategy-Based Deep Learning Model for Efficient and Robust Genomic Prediction in Pigs. Agriculture, 15(22), 2354. https://doi.org/10.3390/agriculture15222354






