High-Density Planting of Panicum virgatum Enhances Soil Carbon Sequestration, Whereas Cultivar Selection and Temporal Dynamics Drive Root and Soil Microbiomes
Abstract
1. Introduction
2. Materials and Methods
2.1. Site Description
2.2. Switchgrass Cultivars
2.3. Soil and Root Sampling
2.4. Soil Chemistry Analyses
2.5. DNA Extraction and Metabarcode Sequencing
2.6. Sequence Data Processing
2.7. Statistical Analysis
2.8. Data Visualization
3. Results
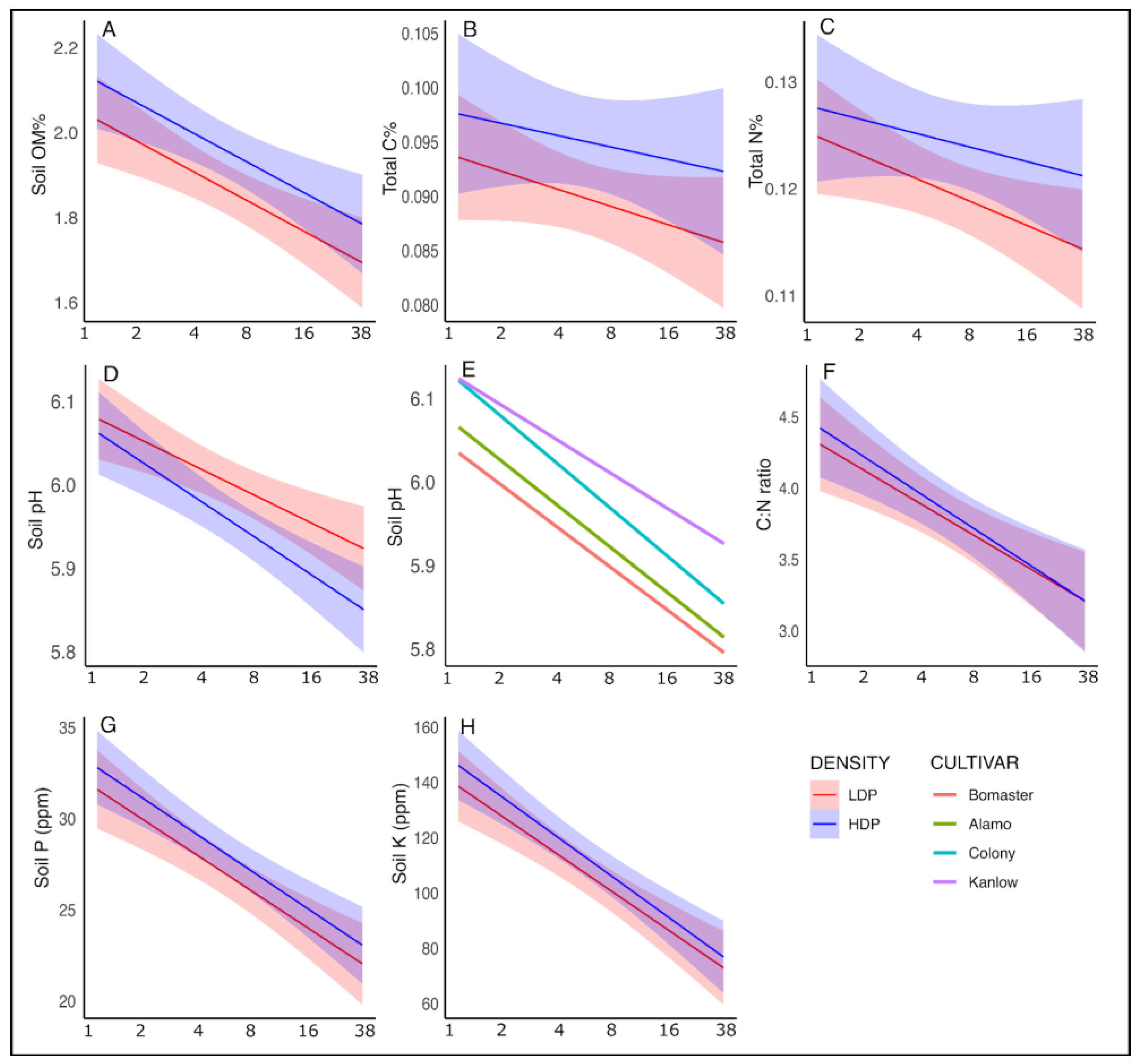
3.1. Effects of Planting Density, Cultivars, and Season on Edaphic Variables
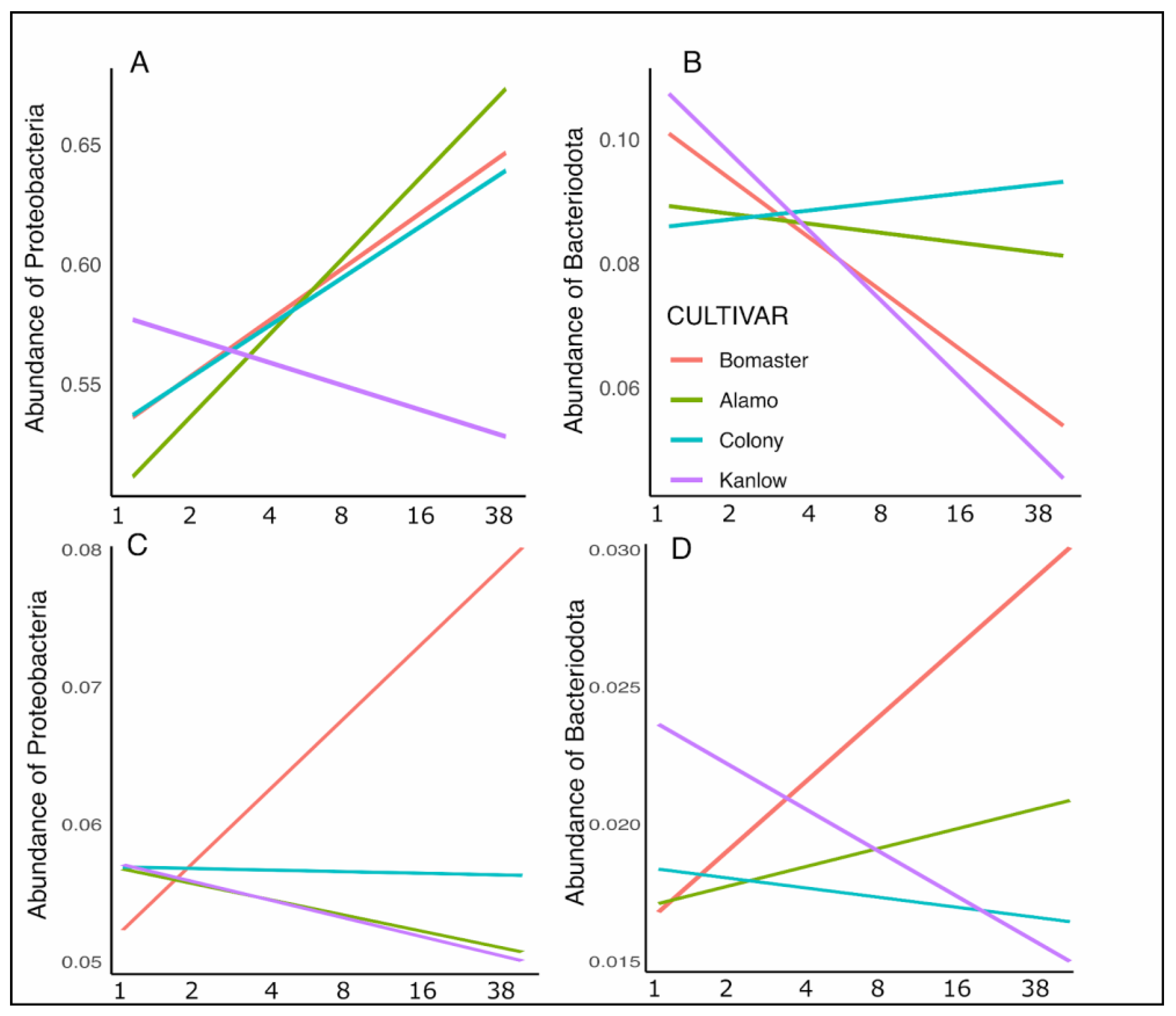
3.2. Relative Taxon Abundances in Soil- and Root-Associated Microbial Communities
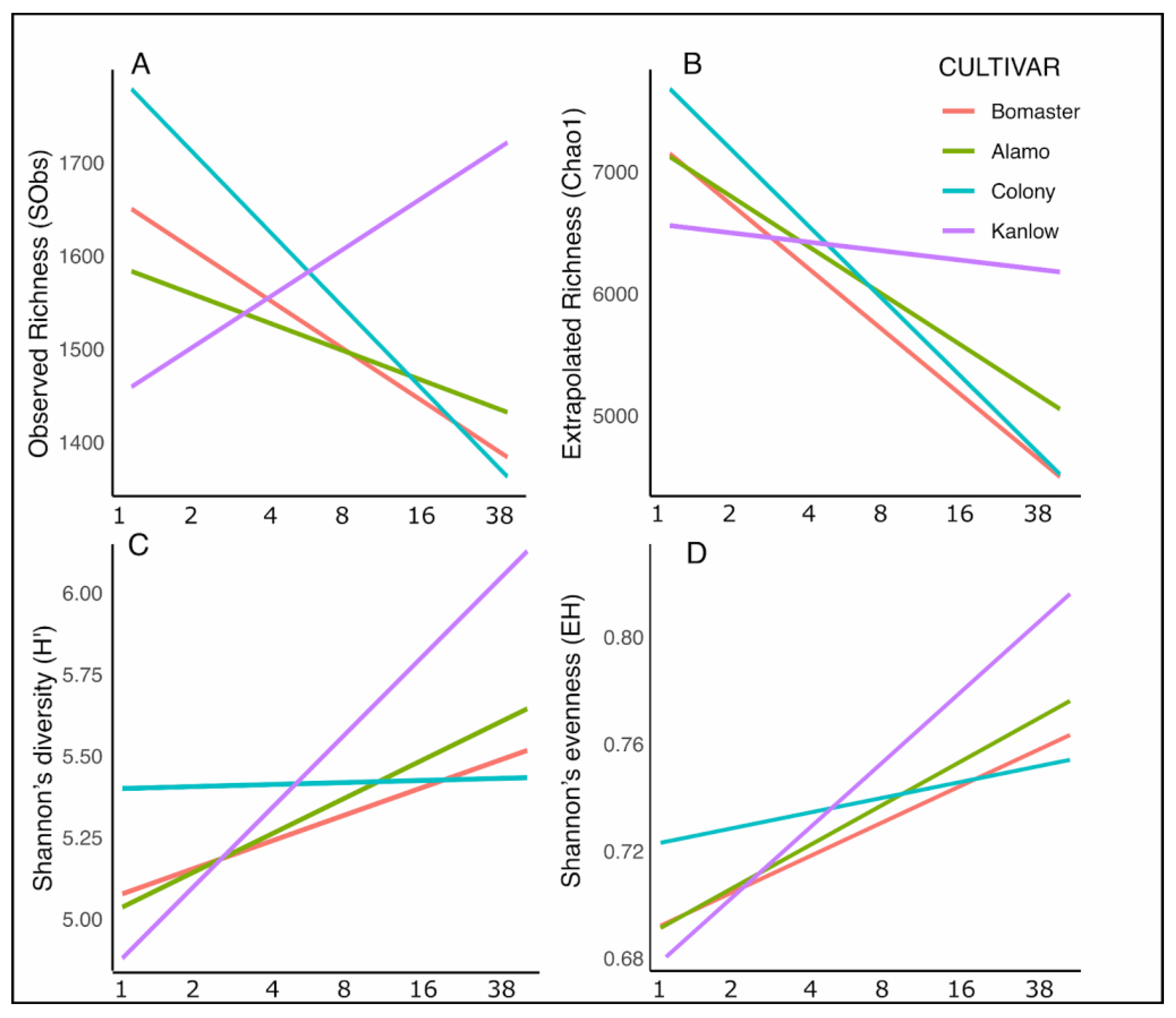
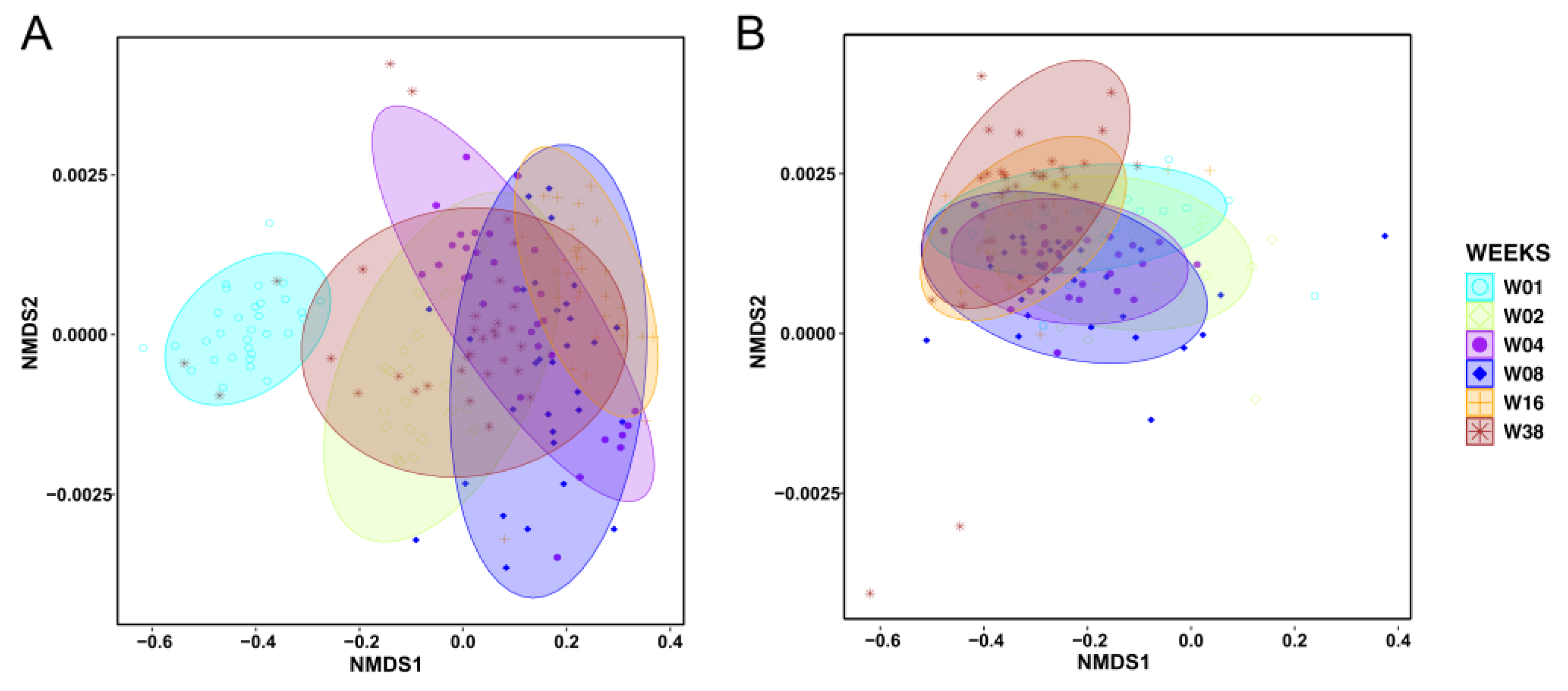
3.3. Effects of Planting Density, Cultivars, and Season on Alpha-Diversity and Community Composition in Soil- and Root-Associated Microbiomes
4. Discussion
4.1. Edaphic Responses
4.2. Soil and Root-Associated Microbiome Diversity and Composition Responses
4.3. Limitations of the Study
4.4. Overall Conclusions and Potential Applications
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Follett, R.F.; Vogel, K.P.; Varvel, G.E.; Mitchell, R.B.; Kimble, J. Soil Carbon Sequestration by Switchgrass and No-Till Maize Grown for Bioenergy. Bioenergy Res. 2012, 5, 866–875. [Google Scholar] [CrossRef]
- Howard Skinner, R.; Zegada-Lizarazu, W.; Schmidt, J.P. Environmental Impacts of Switchgrass Management for Bioenergy Production. In Switchgrass: A Valuable Biomass Crop for Energy; Monti, A., Ed.; Springer: London, UK, 2012; pp. 129–152. ISBN 9781447129035. [Google Scholar]
- Werling, B.P.; Dickson, T.L.; Isaacs, R.; Gaines, H.; Gratton, C.; Gross, K.L.; Liere, H.; Malmstrom, C.M.; Meehan, T.D.; Ruan, L.; et al. Perennial Grasslands Enhance Biodiversity and Multiple Ecosystem Services in Bioenergy Landscapes. Proc. Natl. Acad. Sci. USA 2014, 111, 1652–1657. [Google Scholar] [CrossRef]
- Lowry, D.B.; Behrman, K.D.; Grabowski, P.; Morris, G.P.; Kiniry, J.R.; Juenger, T.E. Adaptations between Ecotypes and along Environmental Gradients in Panicum virgatum. Am. Nat. 2014, 183, 682–692. [Google Scholar] [CrossRef]
- Mitchell, R.; Vogel, K.P.; Sarath, G. Managing and Enhancing Switchgrass as a Bioenergy Feedstock. Biofuels Bioprod. Biorefin. 2008, 2, 530–539. [Google Scholar] [CrossRef]
- Rodrigues, R.R.; Moon, J.; Zhao, B.; Williams, M.A. Microbial Communities and Diazotrophic Activity Differ in the Root-Zone of Alamo and Dacotah Switchgrass Feedstocks. Glob. Chang. Biol. Bioenergy 2017, 9, 1057–1070. [Google Scholar] [CrossRef]
- Sawyer, A.; Staley, C.; Lamb, J.; Sheaffer, C.; Kaiser, T.; Gutknecht, J.; Sadowsky, M.J.; Rosen, C. Cultivar and Phosphorus Effects on Switchgrass Yield and Rhizosphere Microbial Diversity. Appl. Microbiol. Biotechnol. 2019, 103, 1973–1987. [Google Scholar] [CrossRef] [PubMed]
- Zalapa, J.E.; Price, D.L.; Kaeppler, S.M.; Tobias, C.M.; Okada, M.; Casler, M.D. Hierarchical Classification of Switchgrass Genotypes Using SSR and Chloroplast Sequences: Ecotypes, Ploidies, Gene Pools, and Cultivars. Theor. Appl. Genet. 2011, 122, 805–817. [Google Scholar] [CrossRef]
- Zhang, Y.; Zalapa, J.E.; Jakubowski, A.R.; Price, D.L.; Acharya, A.; Wei, Y.; Brummer, E.C.; Kaeppler, S.M.; Casler, M.D. Post-Glacial Evolution of Panicum virgatum: Centers of Diversity and Gene Pools Revealed by SSR Markers and cpDNA Sequences. Genetica 2011, 139, 933–948. [Google Scholar] [CrossRef]
- Beschoren da Costa, P.; Benucci, G.M.N.; Chou, M.-Y.; Van Wyk, J.; Chretien, M.; Bonito, G. Soil Origin and Plant Genotype Modulate Switchgrass Aboveground Productivity and Root Microbiome Assembly. mBio 2022, 13, e0007922. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, J.; Bell, T.; Trexler, R.V.; Carlson, J.E.; Lasky, J.R. Host Genomic Influence on Bacterial Composition in the Switchgrass Rhizosphere. Mol. Ecol. 2022, 31, 3934–3950. [Google Scholar] [CrossRef]
- Bell-Dereske, L.P.; Benucci, G.M.N.; da Costa, P.B.; Bonito, G.; Friesen, M.L.; Tiemann, L.K.; Evans, S.E. Regional Biogeography versus Intra-Annual Dynamics of the Root and Soil Microbiome. Environ. Microbiome 2023, 18, 50. [Google Scholar] [CrossRef]
- Edwards, J.A.; Saran, U.B.; Bonnette, J.; MacQueen, A.; Yin, J.; Nguyen, T.U.; Schmutz, J.; Grimwood, J.; Pennacchio, L.A.; Daum, C.; et al. Genetic Determinants of Switchgrass-Root-Associated Microbiota in Field Sites Spanning Its Natural Range. Curr. Biol. 2023, 33, 1926–1938.e6. [Google Scholar] [CrossRef]
- Stonoha-Arther, C.; Panke-Buisse, K.; Duff, A.J.; Molodchenko, A.; Casler, M.D. Rhizosphere Microbial Community Structure in High-Producing, Low-Input Switchgrass Families. PLoS ONE 2024, 19, e0308753. [Google Scholar] [CrossRef]
- Mosier, S.; Kelly, L.; Ozlu, E.; Robertson, G.P. Switchgrass (Panicum virgatum L.) Cultivars Have Similar Impacts on Soil Carbon and Nitrogen Stocks and Microbial Function. Glob. Chang. Biol. Bioenergy 2024, 16, e13125. [Google Scholar] [CrossRef]
- Sutherland, J.; Bell, T.H.; Bonos, S.; Tkach, C.; Hansen, J.; Crawford, R.; Carlson, J.E.; Lasky, J.R. Bacterial Assembly in the Switchgrass Rhizosphere Is Shaped by Phylogeny, Host Genotype, and Growing Site. Phytobiomes J. 2025, 9, 468–484. [Google Scholar] [CrossRef]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The Rhizosphere Microbiome: Significance of Plant Beneficial, Plant Pathogenic, and Human Pathogenic Microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef] [PubMed]
- Ghimire, S.R.; Craven, K.D. Enhancement of Switchgrass (Panicum virgatum L.) Biomass Production under Drought Conditions by the Ectomycorrhizal Fungus Sebacina Vermifera. Appl. Environ. Microbiol. 2011, 77, 7063–7067. [Google Scholar] [CrossRef]
- Chen, H.; Yang, Z.K.; Yip, D.; Morris, R.H.; Lebreux, S.J.; Cregger, M.A.; Klingeman, D.M.; Hui, D.; Hettich, R.L.; Wilhelm, S.W.; et al. One-Time Nitrogen Fertilization Shifts Switchgrass Soil Microbiomes within a Context of Larger Spatial and Temporal Variation. PLoS ONE 2019, 14, e0211310. [Google Scholar] [CrossRef]
- Grady, K.L.; Sorensen, J.W.; Stopnisek, N.; Guittar, J.; Shade, A. Assembly and Seasonality of Core Phyllosphere Microbiota on Perennial Biofuel Crops. Nat. Commun. 2019, 10, 4135. [Google Scholar] [CrossRef]
- Kim, S.; Lowman, S.; Hou, G.; Nowak, J.; Flinn, B.; Mei, C. Growth Promotion and Colonization of Switchgrass (Panicum virgatum) Cv. Alamo by Bacterial Endophyte Burkholderia Phytofirmans Strain PsJN. Biotechnol. Biofuels 2012, 5, 37. [Google Scholar] [CrossRef] [PubMed]
- Ray, P.; Ishiga, T.; Decker, S.R.; Turner, G.B.; Craven, K.D. A Novel Delivery System for the Root Symbiotic Fungus, Sebacina Vermifera, and Consequent Biomass Enhancement of Low Lignin COMT Switchgrass Lines. Bioenergy Res. 2015, 8, 922–933. [Google Scholar] [CrossRef]
- Wang, B.; Seiler, J.R.; Mei, C. A Microbial Endophyte Enhanced Growth of Switchgrass under Two Drought Cycles Improving Leaf Level Physiology and Leaf Development. Environ. Exp. Bot. 2016, 122, 100–108. [Google Scholar] [CrossRef]
- Sher, Y.; Baker, N.R.; Herman, D.; Fossum, C.; Hale, L.; Zhang, X.; Nuccio, E.; Saha, M.; Zhou, J.; Pett-Ridge, J.; et al. Microbial Extracellular Polysaccharide Production and Aggregate Stability Controlled by Switchgrass (Panicum virgatum) Root Biomass and Soil Water Potential. Soil Biol. Biochem. 2020, 143, 107742. [Google Scholar] [CrossRef]
- Clark, R.B.; Baligar, V.C.; Zobel, R.W. Response of Mycorrhizal Switchgrass to Phosphorus Fractions in Acidic Soil. Commun. Soil Sci. Plant Anal. 2005, 36, 1337–1359. [Google Scholar] [CrossRef]
- Emery, S.M.; Reid, M.L.; Bell-Dereske, L.; Gross, K.L. Soil Mycorrhizal and Nematode Diversity Vary in Response to Bioenergy Crop Identity and Fertilization. Glob. Chang. Biol. Bioenergy 2017, 9, 1644–1656. [Google Scholar] [CrossRef]
- Chamberlain, J.F.; Miller, S.A. Policy Incentives for Switchgrass Production Using Valuation of Non-Market Ecosystem Services. Energy Policy 2012, 48, 526–536. [Google Scholar] [CrossRef]
- Chen, L.; Liu, Y. The Function of Root Exudates in the Root Colonization by Beneficial Soil Rhizobacteria. Biology 2024, 13, 95. [Google Scholar] [CrossRef] [PubMed]
- García de Salamone, I.E.; Funes, J.M.; Di Salvo, L.P.; Escobar-Ortega, J.S.; D’Auria, F.; Ferrando, L.; Fernandez-Scavino, A. Inoculation of Paddy Rice with Azospirillum Brasilense and Pseudomonas Fluorescens: Impact of Plant Genotypes on Rhizosphere Microbial Communities and Field Crop Production. Appl. Soil Ecol. 2012, 61, 196–204. [Google Scholar] [CrossRef]
- Zhao, J.; Bodner, G.; Rewald, B.; Leitner, D.; Nagel, K.A.; Nakhforoosh, A. Root Architecture Simulation Improves the Inference from Seedling Root Phenotyping towards Mature Root Systems. J. Exp. Bot. 2017, 68, 965–982. [Google Scholar] [CrossRef] [PubMed]
- Galindo-Castañeda, T.; Hartmann, M.; Lynch, J.P. Location: Root Architecture Structures Rhizosphere Microbial Associations. J. Exp. Bot. 2024, 75, 594–604. [Google Scholar] [CrossRef]
- Brodsky, O.L.; Shek, K.L.; Dinwiddie, D.; Bruner, S.G.; Gill, A.S.; Hoch, J.M.; Palmer, M.I.; McGuire, K.L. Microbial Communities in Bioswale Soils and Their Relationships to Soil Properties, Plant Species, and Plant Physiology. Front. Microbiol. 2019, 10, 2368. [Google Scholar] [CrossRef]
- Jesus, E.d.C.; Liang, C.; Quensen, J.F.; Susilawati, E.; Jackson, R.D.; Balser, T.C.; Tiedje, J.M. Influence of Corn, Switchgrass, and Prairie Cropping Systems on Soil Microbial Communities in the Upper Midwest of the United States. Glob. Change Biol. Bioenergy 2016, 8, 481–494. [Google Scholar] [CrossRef]
- Whitaker, B.K.; Reynolds, H.L.; Clay, K. Foliar Fungal Endophyte Communities Are Structured by Environment but Not Host Ecotype in Panicum virgatum (switchgrass). Ecology 2018, 99, 2703–2711. [Google Scholar] [CrossRef] [PubMed]
- Hargreaves, S.K.; Williams, R.J.; Hofmockel, K.S. Environmental Filtering of Microbial Communities in Agricultural Soil Shifts with Crop Growth. PLoS ONE 2015, 10, e0134345. [Google Scholar] [CrossRef] [PubMed]
- Ulbrich, T.C.; Friesen, M.L.; Roley, S.S.; Tiemann, L.K.; Evans, S.E. Intraspecific Variability in Root Traits and Edaphic Conditions Influence Soil Microbiomes across 12 Switchgrass Cultivars. Phytobiomes J. 2021, 5, 108–120. [Google Scholar] [CrossRef]
- Elevitch, C.R.; Mazaroli, D.N.; Ragone, D. Agroforestry Standards for Regenerative Agriculture. Sustainability 2018, 10, 3337. [Google Scholar] [CrossRef]
- Gordon, E.; Davila, F.; Riedy, C. Transforming Landscapes and Mindscapes through Regenerative Agriculture. Agric. Human Values 2022, 39, 809–826. [Google Scholar] [CrossRef]
- Hoorman, J.J.; Islam, R.; Sundermeier, A.; Reeder, R. Using Cover Crops to Convert to No-Till. 2009. Available online: https://ohioline.osu.edu/factsheet/SAG-11 (accessed on 25 October 2025).
- Hoorman, J.J.; Sundermeier, A. Using Cover Crops to Improve Soil and Water Quality. Available online: https://ohioline.osu.edu/factsheet/anr-57 (accessed on 25 October 2025).
- Jach-Smith, L.C.; Jackson, R.D. N Addition Undermines N Supplied by Arbuscular Mycorrhizal Fungi to Native Perennial Grasses. Soil Biol. Biochem. 2018, 116, 148–157. [Google Scholar] [CrossRef]
- Jach-Smith, L.C.; Jackson, R.D. Inorganic N Addition Replaces N Supplied to Switchgrass (Panicum virgatum) by Arbuscular Mycorrhizal Fungi. Ecol. Appl. 2020, 30, e02047. [Google Scholar] [CrossRef]
- Oates, L.G.; Duncan, D.S.; Gelfand, I.; Millar, N.; Robertson, G.P.; Jackson, R.D. Nitrous Oxide Emissions during Establishment of Eight Alternative Cellulosic Bioenergy Cropping Systems in the North Central United States. Glob. Chang. Biol. Bioenergy 2016, 8, 539–549. [Google Scholar] [CrossRef]
- Shirzad, M.S.; Bana, R.S.; Bamboriya, S.D. Planting Density and Nitrogen Management Effects on Productivity, Quality and Water-Use-Efficiency of Indian Mustard under Conservation Agriculture Based Pearl Millet–mustard System. J. Agric. Ecol. 2020, 10, 69–75. [Google Scholar] [CrossRef]
- Bai, Y.-F.; Shen, Y.-Y.; Jin, Y.-D.; Hong, Y.; Liu, Y.-Y.; Li, Y.-Q.; Liu, R.; Zhang, Z.-W.; Jiang, C.-Q.; Wang, Y.-J. Selective Thinning and Initial Planting Density Management Promote Biomass and Carbon Storage in a Chronosequence of Evergreen Conifer Plantations in Southeast China. Glob. Ecol. Conserv. 2020, 24, e01216. [Google Scholar] [CrossRef]
- Botare, A.B.; Bondar, U.S.; Patil, N.S.; Ghadage, S.S.; Deshmukh, B.J. Comparative Resource Use Structure and Resource Use Efficiency of Conventional Vs High Density Planting System of Bt Cotton in Yavatmal District of Maharashtra. Int. J. Agric. Food Sci. 2025, 7, 641–645. [Google Scholar] [CrossRef]
- Zaiats, A.; Requena-Mullor, J.M.; Germino, M.J.; Forbey, J.S.; Richardson, B.A.; Caughlin, T.T. Spatial Models Can Improve the Experimental Design of Field-Based Transplant Gardens by Preventing Bias due to Neighborhood Crowding. Ecol. Evol. 2022, 12, e9630. [Google Scholar] [CrossRef]
- Postma, J.A.; Hecht, V.L.; Hikosaka, K.; Nord, E.A.; Pons, T.L.; Poorter, H. Dividing the Pie: A Quantitative Review on Plant Density Responses. Plant Cell Environ. 2021, 44, 1072–1094. [Google Scholar] [CrossRef]
- Qiao, R.; Song, Z.; Chen, Y.; Xu, M.; Yang, Q.; Shen, X.; Yu, D.; Zhang, P.; Ding, C.; Guo, H. Planting Density Effect on Poplar Growth Traits and Soil Nutrient Availability, and Response of Microbial Community, Assembly and Function. BMC Plant Biol. 2024, 24, 1035. [Google Scholar] [CrossRef]
- Cavalieri, A.; Bak, F.; Garcia-Lemos, A.M.; Weiner, J.; Nicolaisen, M.H.; Nybroe, O. Effects of Intra- and Interspecific Plant Density on Rhizosphere Bacterial Communities. Front. Microbiol. 2020, 11, 1045. [Google Scholar] [CrossRef]
- Harker, K.N.; Clayton, G.W.; Blackshaw, R.E.; O’Donovan, J.T.; Stevenson, F.C. Seeding Rate, Herbicide Timing and Competitive Hybrids Contribute to Integrated Weed Management in Canola (Brassica Napus). Can. J. Plant Sci. 2003, 83, 433–440. [Google Scholar] [CrossRef]
- Lay, C.-Y.; Bell, T.H.; Hamel, C.; Harker, K.N.; Mohr, R.; Greer, C.W.; Yergeau, É.; St-Arnaud, M. Canola Root-Associated Microbiomes in the Canadian Prairies. Front. Microbiol. 2018, 9, 1188. [Google Scholar] [CrossRef] [PubMed]
- Ashworth, A.J.; DeBruyn, J.M.; Allen, F.L.; Radosevich, M.; Owens, P.R. Microbial Community Structure Is Affected by Cropping Sequences and Poultry Litter under Long-Term No-Tillage. Soil Biol. Biochem. 2017, 114, 210–219. [Google Scholar] [CrossRef]
- DeBruyn, J.M.; Nixon, L.T.; Fawaz, M.N.; Johnson, A.M.; Radosevich, M. Global Biogeography and Quantitative Seasonal Dynamics of Gemmatimonadetes in Soil. Appl. Environ. Microbiol. 2011, 77, 6295–6300. [Google Scholar] [CrossRef]
- Kramer, S.; Marhan, S.; Haslwimmer, H.; Ruess, L.; Kandeler, E. Temporal Variation in Surface and Subsoil Abundance and Function of the Soil Microbial Community in an Arable Soil. Soil Biol. Biochem. 2013, 61, 76–85. [Google Scholar] [CrossRef]
- Lewandowski, T.E.; Forrester, J.A.; Mladenoff, D.J.; Stoffel, J.L.; Gower, S.T.; D’Amato, A.W.; Balser, T.C. Soil Microbial Community Response and Recovery Following Group Selection Harvest: Temporal Patterns from an Experimental Harvest in a US Northern Hardwood Forest. For. Ecol. Manag. 2015, 340, 82–94. [Google Scholar] [CrossRef]
- Lauber, C.L.; Ramirez, K.S.; Aanderud, Z.; Lennon, J.; Fierer, N. Temporal Variability in Soil Microbial Communities across Land-Use Types. ISME J. 2013, 7, 1641–1650. [Google Scholar] [CrossRef]
- Vance, C.L. Plant Density Effect of Four Varieties of Switchgrass (Panicum virgatum L.) on Biomass Development and Nutrient Build-Up in Heavy Soil. Doctoral Dissertation, Alcorn State University, Lorman, MS, USA, 2015. [Google Scholar]
- Villemereuil, P.; Gaggiotti, O.E. A New FST-based Method to Uncover Local Adaptation Using Environmental Variables. Methods Ecol. Evol. 2015, 6, 1248–1258. [Google Scholar] [CrossRef]
- USDA-NRCS Released Brochure for “Alamo” Switchgrass (Panicum virgatum); Plant Materials Center: Knox City, TX, USA, 2012; Available online: https://www.nrcs.usda.gov/plantmaterials/txpmcrb11189.pdf (accessed on 25 October 2025).
- Burns, J.C.; Godshalk, E.B.; Timothy, D.H. Registration of “BoMaster” Switchgrass. J. Plant Regist. 2008, 2, 31–32. [Google Scholar] [CrossRef]
- Burns, J.C.; Godshalk, E.B.; Timothy, D.H. Registration of “colony” Lowland Switchgrass. J. Plant Regist. 2010, 4, 189–194. [Google Scholar] [CrossRef]
- USDA-NRCS Release Brochure for “Kanlow” Switchgrass (Panicum virgatum L.); USDA-Natural Resources Conservation Service. 2011. Available online: https://www.nrcs.usda.gov/plantmaterials/kspmcrb10373.pdf (accessed on 25 October 2025).
- Mehlich, A. Mehlich 3 Soil Test Extractant: A Modification of Mehlich 2 Extractant. Commun. Soil Sci. Plant Anal. 1984, 15, 1409–1416. [Google Scholar] [CrossRef]
- Combs, S.M.; Nathan, M.V.; Soil Organic Matter. In Recommended Chemical Soil Test Procedures for the North Central Region; North Central Regional Research Publication No. 221; 2015; pp. 57–62. Available online: https://extension.missouri.edu/sites/default/files/legacy_media/wysiwyg/Extensiondata/Pub/pdf/specialb/sb1001.pdf (accessed on 25 October 2025).
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-High-Throughput Microbial Community Analysis on the Illumina HiSeq and MiSeq Platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Sahl, J.W.; Stres, B.; Thallinger, G.G.; Van Horn, D.J.; Weber, C.F.; Ryabin, T.; Hall, J.R.; Hartmann, M.; et al. Introducing Mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a Dual-Index Sequencing Strategy and Curation Pipeline for Analyzing Amplicon Sequence Data on the MiSeq Illumina Sequencing Platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F.O. The SILVA and “All-Species Living Tree Project (LTP)” Taxonomic Frameworks. Nucleic Acids Res. 2014, 42, D643–D648. [Google Scholar] [CrossRef]
- Huse, S.M.; Dethlefsen, L.; Huber, J.A.; Mark Welch, D.; Relman, D.A.; Sogin, M.L. Exploring Microbial Diversity and Taxonomy Using SSU rRNA Hypervariable Tag Sequencing. PLoS Genet. 2008, 4, e1000255. [Google Scholar] [CrossRef]
- Gihring, T.M.; Green, S.J.; Schadt, C.W. Massively Parallel rRNA Gene Sequencing Exacerbates the Potential for Biased Community Diversity Comparisons due to Variable Library Sizes. Environ. Microbiol. 2012, 14, 285–290. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. Available online: https://www.R-project.org/ (accessed on 25 October 2025).
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R. Vegan: Community Ecology Package, R Package Version 2.5-6. 2019. 2020. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 25 October 2025).
- De Cáceres, M.; Legendre, P. Associations between Species and Groups of Sites: Indices and Statistical Inference. Ecology 2009, 90, 3566–3574. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 9783319242774. [Google Scholar]
- Zhang, Y.; Liu, T.; Guo, J.; Tan, Z.; Dong, W.; Wang, H. Changes in the Understory Diversity of Secondary Pinus Tabulaeformis Forests Are the Result of Stand Density and Soil Properties. Glob. Ecol. Conserv. 2021, 28, e01628. [Google Scholar] [CrossRef]
- Ali, A.; Dai, D.; Akhtar, K.; Teng, M.; Yan, Z.; Urbina-Cardona, N.; Mullerova, J.; Zhou, Z. Response of Understory Vegetation, Tree Regeneration, and Soil Quality to Manipulated Stand Density in a Pinus Massoniana Plantation. Glob. Ecol. Conserv. 2019, 20, e00775. [Google Scholar] [CrossRef]
- Na, M.; Sun, X.; Zhang, Y.; Sun, Z.; Rousk, J. Higher Stand Densities Can Promote Soil Carbon Storage after Conversion of Temperate Mixed Natural Forests to Larch Plantations. Eur. J. For. Res. 2021, 140, 373–386. [Google Scholar] [CrossRef]
- Sanderson, M.A.; Reed, R.L. Switchgrass Growth and Development: Water, Nitrogen, and Plant Density Effects. J. Range Manag. 2000, 53, 221–227. [Google Scholar] [CrossRef]
- VanWallendael, A.; Benucci, G.M.N.; da Costa, P.B.; Fraser, L.; Sreedasyam, A.; Fritschi, F.; Juenger, T.E.; Lovell, J.T.; Bonito, G.; Lowry, D.B. Host Genotype Controls Ecological Change in the Leaf Fungal Microbiome. PLoS Biol. 2022, 20, e3001681. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, M.A.; Reed, R.L.; Ocumpaugh, W.R.; Hussey, M.A.; Van Esbroeck, G.; Read, J.C.; Tischler, C.R.; Hons, F.M. Switchgrass Cultivars and Germplasm for Biomass Feedstock Production in Texas. Bioresour. Technol. 1999, 67, 209–219. [Google Scholar] [CrossRef]
- Casler, M.D.; Vogel, K.P.; Taliaferro, C.M.; Wynia, R.L. Latitudinal Adaptation of Switchgrass Populations. Crop Sci. 2004, 44, 293–303. [Google Scholar] [CrossRef]
- Casler, M.D.; Boe, A.R. Cultivar × Environment Interactions in Switchgrass. Crop Sci. 2003, 43, 2226–2233. [Google Scholar] [CrossRef]
- Chen, J.-Z.; Huang, X.-L.; Sun, Q.-W.; Liu, J.-M. Bulk Soil Microbial Reservoir or Plant Recruitment Dominates Rhizosphere Microbial Community Assembly: Evidence from the Rare, Endangered Lauraceae Species Cinmaomum Migao. Ecol. Indic. 2023, 148, 110071. [Google Scholar] [CrossRef]
- Jones, P.; Garcia, B.J.; Furches, A.; Tuskan, G.A.; Jacobson, D. Plant Host-Associated Mechanisms for Microbial Selection. Front. Plant Sci. 2019, 10, 862. [Google Scholar] [CrossRef] [PubMed]
- Revillini, D.; Wilson, G.W.T.; Miller, R.M.; Lancione, R.; Johnson, N.C. Plant Diversity and Fertilizer Management Shape the Belowground Microbiome of Native Grass Bioenergy Feedstocks. Front. Plant Sci. 2019, 10, 1018. [Google Scholar] [CrossRef] [PubMed]
- Singer, E.; Bonnette, J.; Kenaley, S.C.; Woyke, T.; Juenger, T.E. Plant Compartment and Genetic Variation Drive Microbiome Composition in Switchgrass Roots. Environ. Microbiol. Rep. 2019, 11, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Baker, N.R.; Zhalnina, K.; Yuan, M.; Herman, D.; Ceja-Navarro, J.A.; Sasse, J.; Jordan, J.S.; Bowen, B.P.; Wu, L.; Fossum, C.; et al. Nutrient and Moisture Limitations Reveal Keystone Metabolites Linking Rhizosphere Metabolomes and Microbiomes. Proc. Natl. Acad. Sci. USA 2024, 121, e2303439121. [Google Scholar] [CrossRef]
- Hestrin, R.; Lee, M.R.; Whitaker, B.K.; Pett-Ridge, J. The Switchgrass Microbiome: A Review of Structure, Function, and Taxonomic Distribution. Phytobiomes J. 2021, 5, 14–28. [Google Scholar] [CrossRef]
- Mao, Y.; Yannarell, A.C.; Mackie, R.I. Changes in N-Transforming Archaea and Bacteria in Soil during the Establishment of Bioenergy Crops. PLoS ONE 2011, 6, e24750. [Google Scholar] [CrossRef] [PubMed]
- Conradie, T.; Jacobs, K. Seasonal and Agricultural Response of Acidobacteria Present in Two Fynbos Rhizosphere Soils. Diversity 2020, 12, 277. [Google Scholar] [CrossRef]
- de Chaves, M.G.; Silva, G.G.Z.; Rossetto, R.; Edwards, R.A.; Tsai, S.M.; Navarrete, A.A. Acidobacteria Subgroups and Their Metabolic Potential for Carbon Degradation in Sugarcane Soil Amended with Vinasse and Nitrogen Fertilizers. Front. Microbiol. 2019, 10, 1680. [Google Scholar] [CrossRef]
- Hou, S.; Ren, H.; Fan, F.; Zhao, M.; Zhou, W.; Zhou, B.; Li, C. The Effects of Plant Density and Nitrogen Fertilization on Maize Yield and Soil Microbial Communities in the Black Soil Region of Northeast China. Geoderma 2023, 430, 116325. [Google Scholar] [CrossRef]
- Hu, J.; Miller, G.; Shi, W. Abundance, Diversity, and Composition of Root-Associated Microbial Communities Varied with Tall Fescue Cultivars under Water Deficit. Front. Microbiol. 2022, 13, 1078836. [Google Scholar] [CrossRef]
- Wolfgang, A.; Zachow, C.; Müller, H.; Grand, A.; Temme, N.; Tilcher, R.; Berg, G. Understanding the Impact of Cultivar, Seed Origin, and Substrate on Bacterial Diversity of the Sugar Beet Rhizosphere and Suppression of Soil-Borne Pathogens. Front. Plant Sci. 2020, 11, 560869. [Google Scholar] [CrossRef]
- Kazarina, A.; Sarkar, S.; Thapa, S.; Heeren, L.; Kamke, A.; Ward, K.; Hartung, E.; Ran, Q.; Galliart, M.; Jumpponen, A.; et al. Home-Field Advantage Affects the Local Adaptive Interaction between Andropogon Gerardii Ecotypes and Root-Associated Bacterial Communities. Microbiol. Spectr. 2023, 11, e0020823. [Google Scholar] [CrossRef]
- Lavallee, J.M.; Chomel, M.; Alvarez Segura, N.; de Castro, F.; Goodall, T.; Magilton, M.; Rhymes, J.M.; Delgado-Baquerizo, M.; Griffiths, R.I.; Baggs, E.M.; et al. Land Management Shapes Drought Responses of Dominant Soil Microbial Taxa across Grasslands. Nat. Commun. 2024, 15, 29. [Google Scholar] [CrossRef] [PubMed]
- Boeddinghaus, R.S.; Marhan, S.; Berner, D.; Boch, S.; Fischer, M.; Hölzel, N.; Kattge, J.; Klaus, V.H.; Kleinebecker, T.; Oelmann, Y.; et al. Plant Functional Trait Shifts Explain Concurrent Changes in the Structure and Function of Grassland Soil Microbial Communities. J. Ecol. 2019, 107, 2197–2210. [Google Scholar] [CrossRef]
- de Vries, F.T.; Manning, P.; Tallowin, J.R.B.; Mortimer, S.R.; Pilgrim, E.S.; Harrison, K.A.; Hobbs, P.J.; Quirk, H.; Shipley, B.; Cornelissen, J.H.C.; et al. Abiotic Drivers and Plant Traits Explain Landscape-Scale Patterns in Soil Microbial Communities. Ecol. Lett. 2012, 15, 1230–1239. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Reich, P.B.; Trivedi, C.; Eldridge, D.J.; Abades, S.; Alfaro, F.D.; Bastida, F.; Berhe, A.A.; Cutler, N.A.; Gallardo, A.; et al. Multiple Elements of Soil Biodiversity Drive Ecosystem Functions across Biomes. Nat. Ecol. Evol. 2020, 4, 210–220. [Google Scholar] [CrossRef]
- Soliveres, S.; van der Plas, F.; Manning, P.; Prati, D.; Gossner, M.M.; Renner, S.C.; Alt, F.; Arndt, H.; Baumgartner, V.; Binkenstein, J.; et al. Biodiversity at Multiple Trophic Levels Is Needed for Ecosystem Multifunctionality. Nature 2016, 536, 456–459. [Google Scholar] [CrossRef]
- Wu, L.; Chen, H.; Chen, D.; Wang, S.; Wu, Y.; Wang, B.; Liu, S.; Yue, L.; Yu, J.; Bai, Y. Soil Biota Diversity and Plant Diversity Both Contributed to Ecosystem Stability in Grasslands. Ecol. Lett. 2023, 26, 858–868. [Google Scholar] [CrossRef]
- Hu, Z.; Delgado-Baquerizo, M.; Fanin, N.; Chen, X.; Zhou, Y.; Du, G.; Hu, F.; Jiang, L.; Hu, S.; Liu, M. Nutrient-Induced Acidification Modulates Soil Biodiversity-Function Relationships. Nat. Commun. 2024, 15, 2858. [Google Scholar] [CrossRef]
- Xin, Y.; Shi, Y.; He, W.-M. A Shift from Inorganic to Organic Nitrogen-Dominance Shapes Soil Microbiome Composition and Co-Occurrence Networks. Front. Microbiol. 2022, 13, 1074064. [Google Scholar] [CrossRef]
- Kostin, J.E.; Cesarz, S.; Lochner, A.; Schädler, M.; Macdonald, C.A.; Eisenhauer, N. Land-Use Drives the Temporal Stability and Magnitude of Soil Microbial Functions and Modulates Climate Effects. Ecol. Appl. 2021, 31, e02325. [Google Scholar] [CrossRef] [PubMed]
- Staley, C.; Ferrieri, A.P.; Tfaily, M.M.; Cui, Y.; Chu, R.K.; Wang, P.; Shaw, J.B.; Ansong, C.K.; Brewer, H.; Norbeck, A.D.; et al. Diurnal Cycling of Rhizosphere Bacterial Communities Is Associated with Shifts in Carbon Metabolism. Microbiome 2017, 5, 65. [Google Scholar] [CrossRef] [PubMed]
- Malik, A.A.; Martiny, J.B.H.; Brodie, E.L.; Martiny, A.C.; Treseder, K.K.; Allison, S.D. Defining Trait-Based Microbial Strategies with Consequences for Soil Carbon Cycling under Climate Change. ISME J. 2020, 14, 1–9. [Google Scholar] [CrossRef]
- Malik, A.A.; Puissant, J.; Goodall, T.; Allison, S.D.; Griffiths, R.I. Soil Microbial Communities with Greater Investment in Resource Acquisition Have Lower Growth Yield. Soil Biol. Biochem. 2019, 132, 36–39. [Google Scholar] [CrossRef]
- Lee, H.; Bloxham, B.; Gore, J. Resource Competition Can Explain Simplicity in Microbial Community Assembly. Proc. Natl. Acad. Sci. USA 2023, 120, e2212113120. [Google Scholar] [CrossRef]
- Tilman, D. Resource Competition and Community Structure. Monogr. Popul. Biol. 1982, 17, 1–296. [Google Scholar]
- White, E.P.; Adler, P.B.; Lauenroth, W.K.; Gill, R.A.; Greenberg, D.; Kaufman, D.M.; Rassweiler, A.; Rusak, J.A.; Smith, M.D.; Steinbeck, J.R.; et al. A Comparison of the Species–time Relationship across Ecosystems and Taxonomic Groups. Oikos 2006, 112, 185–195. [Google Scholar] [CrossRef]
- Pascale, A.; Proietti, S.; Pantelides, I.S.; Stringlis, I.A. Modulation of the Root Microbiome by Plant Molecules: The Basis for Targeted Disease Suppression and Plant Growth Promotion. Front. Plant Sci. 2019, 10, 1741. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.A.; Ke, P.-J.; Adler, P.B. Mechanistic Approaches to Investigate Soil Microbe-mediated Plant Competition. J. Ecol. 2023, 111, 1590–1597. [Google Scholar] [CrossRef]
- Hannula, S.E.; Kielak, A.M.; Steinauer, K.; Huberty, M.; Jongen, R.; De Long, J.R.; Heinen, R.; Bezemer, T.M. Time after Time: Temporal Variation in the Effects of Grass and Forb Species on Soil Bacterial and Fungal Communities. mBio 2019, 10, e02635-19. [Google Scholar] [CrossRef]
- Wei, G.; Li, M.; Zhang, G.; Chen, Z.; Wei, F.; Jiao, S.; Qian, J.; Wang, Y.; Wei, J.; Wang, Y.; et al. Temporal Dynamics of Rhizosphere Communities across the Life Cycle of Panax Notoginseng. Front. Microbiol. 2022, 13, 853077. [Google Scholar] [CrossRef]
- Zhalnina, K.; Louie, K.B.; Hao, Z.; Mansoori, N.; da Rocha, U.N.; Shi, S.; Cho, H.; Karaoz, U.; Loqué, D.; Bowen, B.P.; et al. Dynamic Root Exudate Chemistry and Microbial Substrate Preferences Drive Patterns in Rhizosphere Microbial Community Assembly. Nat. Microbiol. 2018, 3, 470–480. [Google Scholar] [CrossRef]
- Avolio, M.L.; Forrestel, E.J.; Chang, C.C.; La Pierre, K.J.; Burghardt, K.T.; Smith, M.D. Demystifying Dominant Species. New Phytol. 2019, 223, 1106–1126. [Google Scholar] [CrossRef]
- Cleland, E.E.; Collins, S.L.; Dickson, T.L.; Farrer, E.C.; Gross, K.L.; Gherardi, L.A.; Hallett, L.M.; Hobbs, R.J.; Hsu, J.S.; Turnbull, L.; et al. Sensitivity of Grassland Plant Community Composition to Spatial vs. Temporal Variation in Precipitation. Ecology 2013, 94, 1687–1696. [Google Scholar] [CrossRef] [PubMed]
- Collins, S.L.; Suding, K.N.; Cleland, E.E.; Batty, M.; Pennings, S.C.; Gross, K.L.; Grace, J.B.; Gough, L.; Fargione, J.E.; Clark, C.M. Rank Clocks and Plant Community Dynamics. Ecology 2008, 89, 3534–3541. [Google Scholar] [CrossRef] [PubMed]
- Kazarina, A.; Sarkar, S.; Adams, B.; Rodela, L.; Pogranichny, S.; Hartung, E.; Johnson, L.; Jumpponen, A.M.; Lee, S.T.M. Interaction of Plant-Derived Metabolites and Rhizobiome Functions Enhances Drought Stress Tolerance. Genome Biol. 2024, 26, 310. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kazarina, A.; Mandyam, K.; Panicker, G.; Tyler, H.L.; Jumpponen, A. High-Density Planting of Panicum virgatum Enhances Soil Carbon Sequestration, Whereas Cultivar Selection and Temporal Dynamics Drive Root and Soil Microbiomes. Agriculture 2025, 15, 2274. https://doi.org/10.3390/agriculture15212274
Kazarina A, Mandyam K, Panicker G, Tyler HL, Jumpponen A. High-Density Planting of Panicum virgatum Enhances Soil Carbon Sequestration, Whereas Cultivar Selection and Temporal Dynamics Drive Root and Soil Microbiomes. Agriculture. 2025; 15(21):2274. https://doi.org/10.3390/agriculture15212274
Chicago/Turabian StyleKazarina, Anna, Keerthi Mandyam, Girish Panicker, Heather L. Tyler, and Ari Jumpponen. 2025. "High-Density Planting of Panicum virgatum Enhances Soil Carbon Sequestration, Whereas Cultivar Selection and Temporal Dynamics Drive Root and Soil Microbiomes" Agriculture 15, no. 21: 2274. https://doi.org/10.3390/agriculture15212274
APA StyleKazarina, A., Mandyam, K., Panicker, G., Tyler, H. L., & Jumpponen, A. (2025). High-Density Planting of Panicum virgatum Enhances Soil Carbon Sequestration, Whereas Cultivar Selection and Temporal Dynamics Drive Root and Soil Microbiomes. Agriculture, 15(21), 2274. https://doi.org/10.3390/agriculture15212274





