Multi-Environment Evaluation of Soybean Variety Heike 88: Transgressive Segregation and Regional Adaptation in Northern China
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design and Statistical Framework
2.2. Plant Materials and Breeding Strategy
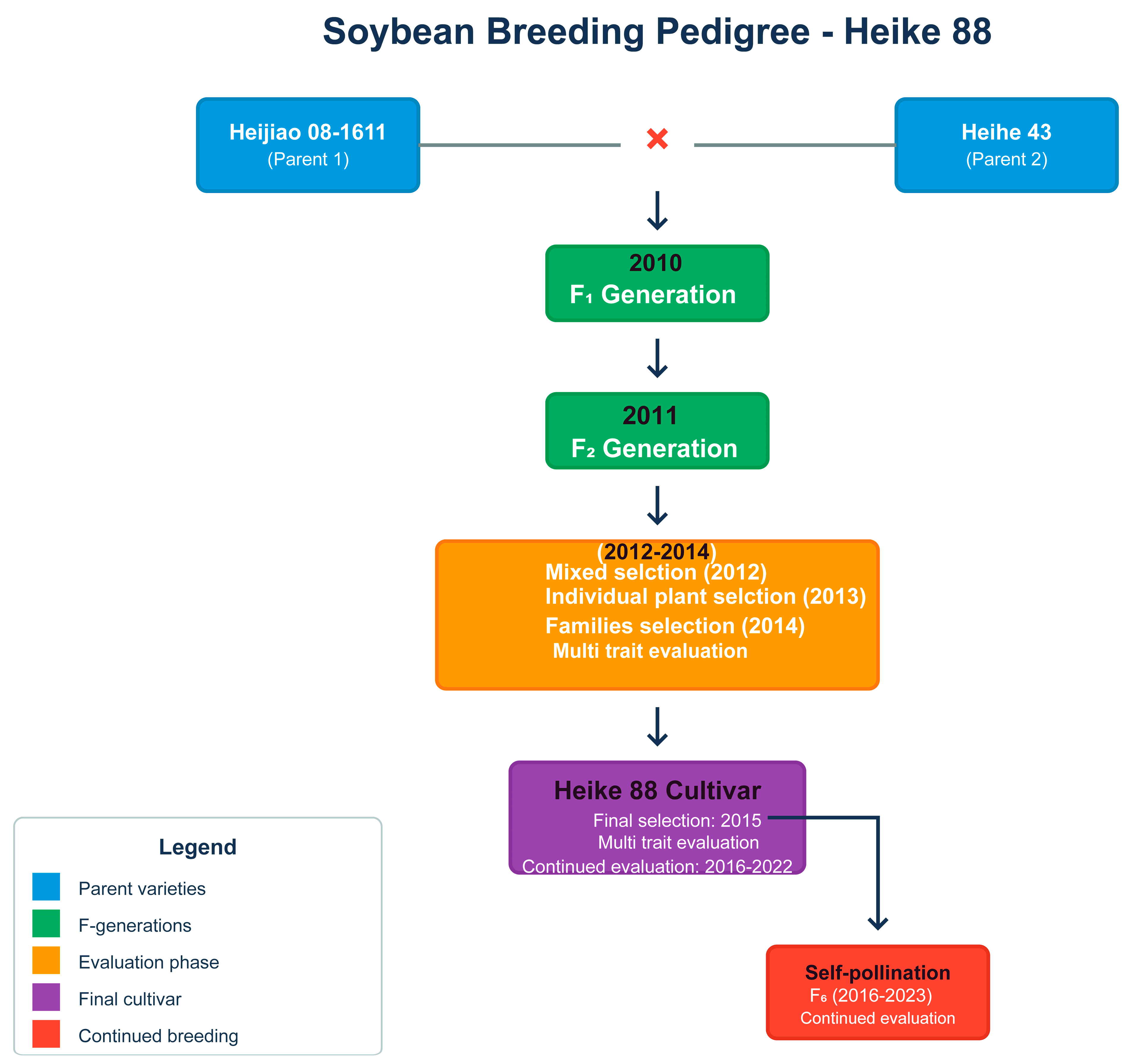
2.2.1. Parent Line Selection and Characterization
2.2.2. Hybridization and Population Development
2.2.3. Performance Evaluation Design and Parent Performance Assessment
2.3. Multi-Phase Evaluation Program
2.4. Field Management and Cultural Practices
2.5. Agronomic Measurements and Data Collection
2.5.1. Growth and Development Characteristics
2.5.2. Yield and Yield Components
2.5.3. Quality Analysis Protocols
2.6. Disease Resistance Evaluation
2.7. Environmental Characterization
2.8. Statistical Analysis
3. Results
3.1. Analysis of Variance for Agronomic and Quality Traits
3.2. Multi-Location Performance Evaluation of Soybean Variety Heike 88
3.2.1. Growth and Development Characteristics
3.2.2. Morphological Characteristics and Plant Architecture
3.2.3. Yield Performance and Components
3.2.4. Seed Quality Components
3.2.5. Disease Resistance and Seed Quality
3.3. Comprehensive Performance Analysis and Multi-Dimensional Performance Characterization
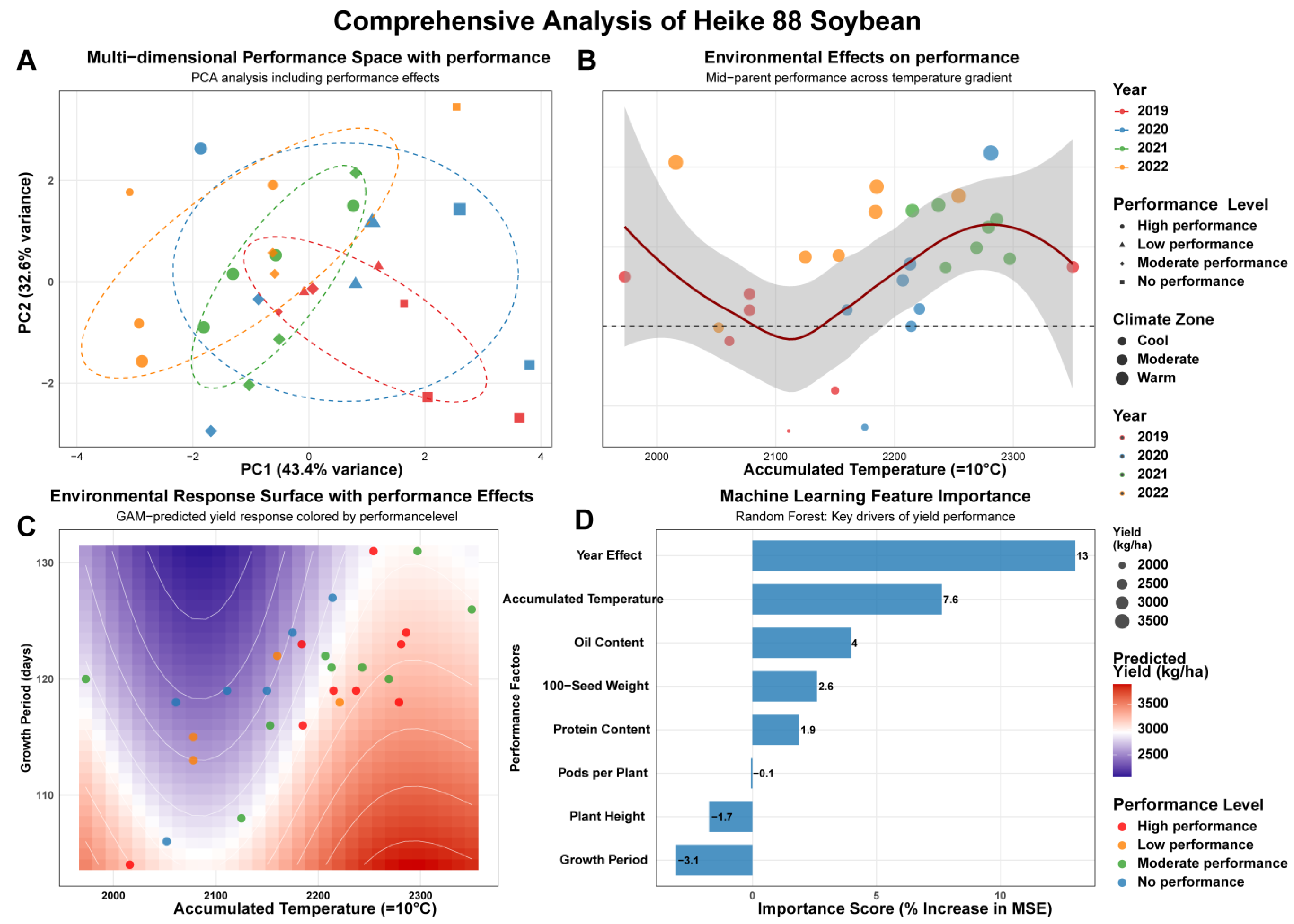
3.3.1. Principal Component Analysis and Performance Space Mapping
3.3.2. Environmental Modulation of Performance Expression
3.3.3. Environmental Response Surface Analysis
3.3.4. Machine Learning Feature Importance Analysis
3.4. Temporal Performance Dynamics and Trait Relationships
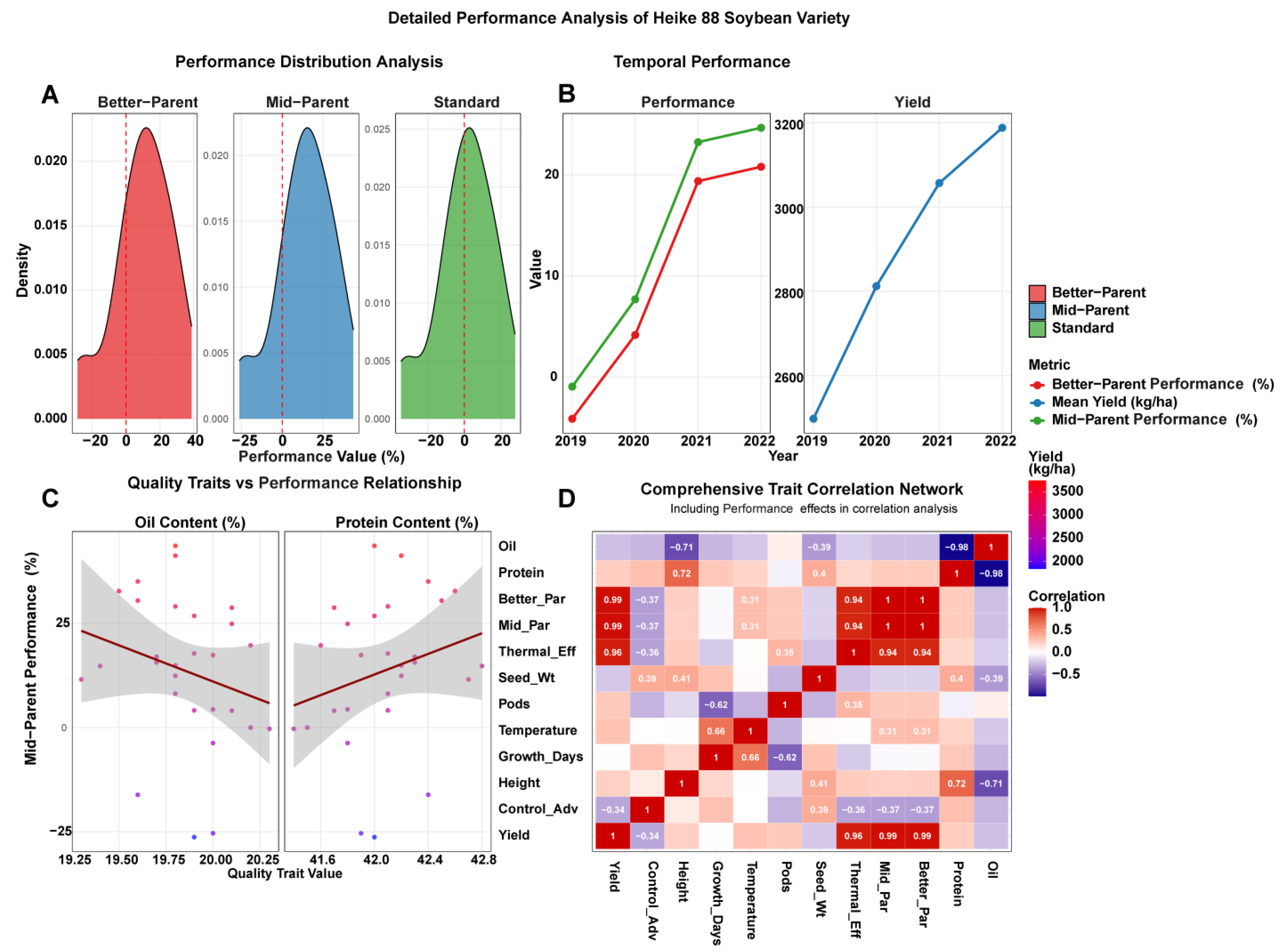
3.4.1. Performance Distribution Patterns
3.4.2. Temporal Performance and Performance Trends
3.4.3. Quality Trait-Performance Relationships
3.4.4. Comprehensive Trait Correlation Network
4. Discussion
4.1. Transgressive Segregation and Genetic Improvement Through Pedigree Breeding
4.2. Temperature-Dependent Performance and Adaptive Breeding
4.3. Disease Resistance and Durability
4.4. Methodological Implications
4.5. Quality Trait Independence
4.6. Study Limitations and Methodological Considerations
4.7. Future Research Directions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, F.; Hong, H.; Liu, X.; Wang, X.; Zhang, C.; Zhao, K.; Yuan, R.; Abdelghany, A.M.; Zhang, B.; Lamlom, S.F. Large-scale evaluation of soybean germplasm reveals geographic patterns in shade tolerance and identifies elite genotypes for intercropping systems. BMC Plant Biol. 2025, 25, 1092. [Google Scholar] [CrossRef] [PubMed]
- Abdelghany, A.M.; Zhang, S.; Azam, M.; Shaibu, A.S.; Feng, Y.; Qi, J.; Li, J.; Li, Y.; Tian, Y.; Hong, H. Exploring the phenotypic stability of soybean seed compositions using multi-trait stability index approach. Agronomy 2021, 11, 2200. [Google Scholar] [CrossRef]
- Beckman, J.; Johnson, M.E.; Ajewole, K.; Kaufman, J.; Sabala, E. The Growing Demand for Animal Products and Feed in India: Future Prospects for Production, Trade, and Technology Innovation. Res. Agric. Appl. Econ. 2025, 54. [Google Scholar] [CrossRef]
- Musah, S. Determinants of Agricultural Productivity in the Leading Producing Countries Worldwide. Master’s Thesis, Southern Illinois University at Carbondale, Carbondale, IL, USA, 2025. [Google Scholar]
- Zhu, W.; Li, J.; Xie, T. Impact of climate change on soybean production: Research progress and response strategies. Adv. Resour. Res. 2024, 4, 474–496. [Google Scholar]
- Shea, Z.; Singer, W.M.; Zhang, B. Soybean production, versatility, and improvement. Legume Crops Prospect. Prod. Uses 2020, 10, 29–71. [Google Scholar]
- Anderson, E.J.; Ali, M.L.; Beavis, W.D.; Chen, P.; Clemente, T.E.; Diers, B.W.; Graef, G.L.; Grassini, P.; Hyten, D.L.; McHale, L.K. Soybean [Glycine max (L.) Merr.] breeding: History, improvement, production and future opportunities. In Advances in Plant Breeding Strategies: Legumes; Springer: Berlin/Heidelberg, Germany, 2019; Volume 7, pp. 431–516. [Google Scholar]
- Miedaner, T. Breeding strategies for improving plant resistance to diseases. In Advances in Plant Breeding Strategies: Agronomic, Abiotic and Biotic Stress Traits; Springer: Berlin/Heidelberg, Germany, 2016; pp. 561–599. [Google Scholar]
- Ahmar, S.; Gill, R.A.; Jung, K.-H.; Faheem, A.; Qasim, M.U.; Mubeen, M.; Zhou, W. Conventional and molecular techniques from simple breeding to speed breeding in crop plants: Recent advances and future outlook. Int. J. Mol. Sci. 2020, 21, 2590. [Google Scholar] [CrossRef]
- Buch, K.; Kaushik, A.; Mishra, U.; Beese, S.; Samanta, S.; Singh, R. Unravelling the complexity of plant breeding through modern genetic techniques and tools: A review. Int. J. Plant Soil Sci. 2023, 35, 97–105. [Google Scholar] [CrossRef]
- Das, A.K.; Choudhary, M.; Kumar, P.; Karjagi, C.G.; KR, Y.; Kumar, R.; Singh, A.; Kumar, S.; Rakshit, S. Heterosis in genomic era: Advances in the molecular understanding and techniques for rapid exploitation. Crit. Rev. Plant Sci. 2021, 40, 218–242. [Google Scholar] [CrossRef]
- Xu, J. Exploration du Polymorphisme Moléculaire et Protéique de la Tomate Pour l’Identification de QTL de Qualité du Fruit. Ph.D. Thesis, Université d’Avignon, Avignon, France, 2012. [Google Scholar]
- Wu, X.; Liu, Y.; Zhang, Y.; Gu, R. Advances in research on the mechanism of heterosis in plants. Front. Plant Sci. 2021, 12, 745726. [Google Scholar] [CrossRef] [PubMed]
- Snodgrass, S.J. The Consequences of Hybridization in the Short-and Long-Term Among the Tripsacinae Subtribe of Grasses. Ph.D. Thesis, Iowa State University, Ames, IA, USA, 2024. [Google Scholar]
- Ren, X.; Chen, L.; Deng, L.; Zhao, Q.; Yao, D.; Li, X.; Cong, W.; Zang, Z.; Zhao, D.; Zhang, M. Comparative transcriptomic analysis reveals the molecular mechanism underlying seedling heterosis and its relationship with hybrid contemporary seeds DNA methylation in soybean. Front. Plant Sci. 2024, 15, 1364284. [Google Scholar] [CrossRef]
- Shahzad, K.; Zhang, X.; Guo, L.; Qi, T.; Bao, L.; Zhang, M.; Zhang, B.; Wang, H.; Tang, H.; Qiao, X. Comparative transcriptome analysis between inbred and hybrids reveals molecular insights into yield heterosis of upland cotton. BMC Plant Biol. 2020, 20, 239. [Google Scholar] [CrossRef]
- Liu, J.; Li, M.; Zhang, Q.; Wei, X.; Huang, X. Exploring the molecular basis of heterosis for plant breeding. J. Integr. Plant Biol. 2020, 62, 287–298. [Google Scholar] [CrossRef]
- Ouyang, Y.; Li, X.; Zhang, Q. Understanding the genetic and molecular constitutions of heterosis for developing hybrid rice. J. Genet. Genom. 2022, 49, 385–393. [Google Scholar] [CrossRef]
- Belcapo, S.; Réthoré, E.; Nguema-Ona, E.; Ezquer, I. Unraveling Novel Mechanisms Controlling Heterosis in seeds: Advances and Biotechnological Applications in crops. J. Exp. Bot. 2025, eraf400. [Google Scholar] [CrossRef]
- Ruff, L.A. Soybean Heterosis and Response to Water: Yield, Yield Components, and Morphology; The University of Nebraska-Lincoln: Lincoln, NE, USA, 2016. [Google Scholar]
- Hochholdinger, F.; Yu, P. Molecular concepts to explain heterosis in crops. Trends Plant Sci. 2025, 30, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Charles, D.R. Optimizing Multi-Trait Selection Index in Maize Breeding with Advanced Machine Learning and Robust Uncertainty Analysis. Ph.D. Thesis, Université Côte d’Azur, Nice, France, 2024. [Google Scholar]
- Hairong, Y.; Yiyuan, C.; Bun, K.H. China’s soybean crisis: The logic of modernization and its discontents. In Soy, Globalization, and Environmental Politics in South America; Routledge: Oxfordshire, UK, 2017; pp. 123–145. [Google Scholar]
- Zhao, J.; Wang, Y.; Zhao, M.; Wang, K.; Li, S.; Gao, Z.; Shi, X.; Chu, Q. Prospects for soybean production increase by closing yield gaps in the Northeast Farming Region, China. Field Crops Res. 2023, 293, 108843. [Google Scholar] [CrossRef]
- Xin, M.; Zhang, Z.; Han, Y.; Feng, L.; Lei, Y.; Li, X.; Wu, F.; Wang, J.; Wang, Z.; Li, Y. Soybean phenological changes in response to climate warming in three northeastern provinces of China. Field Crops Res. 2023, 302, 109082. [Google Scholar] [CrossRef]
- Zhao, J.; Wang, C.; Shi, X.; Bo, X.; Li, S.; Shang, M.; Chen, F.; Chu, Q. Modeling climatically suitable areas for soybean and their shifts across China. Agric. Syst. 2021, 192, 103205. [Google Scholar] [CrossRef]
- He, H.; Chen, M.; Li, M.; Qu, K.; Dang, H.; Li, Q.; Hu, Z.; Zhang, Q. Impact of climate change on the potential allocation of resources of rice cultivation in Yangtze-Huai Rivers region: A case study of Anhui Province, China. Theor. Appl. Climatol. 2024, 155, 6697–6708. [Google Scholar] [CrossRef]
- Gao, J.; Liu, Y. Climate warming and land use change in Heilongjiang Province, Northeast China. Appl. Geogr. 2011, 31, 476–482. [Google Scholar] [CrossRef]
- Wu, M.; Pei, W.; Wedegaertner, T.; Zhang, J.; Yu, J. Genetics, breeding and genetic engineering to improve cottonseed oil and protein: A review. Front. Plant Sci. 2022, 13, 864850. [Google Scholar] [CrossRef]
- Pandarinathan, S.; Adhimoolam, P.K.; Gurav, N.P.; Shamkuwar, S.; Panigrahi, C.K.; Huded, S.; Kumar, M. A Review on Genetic Mechanisms of Plant-pathogen Resistance in Crop Breeding. Plant Cell Biotechnol. Mol. Biol. 2024, 25, 221–234. [Google Scholar] [CrossRef]
- Mural, R.V. Genotype-By-Environment Interaction and Stability: Concepts, Analysis, And Applications. Des. Exp. Biom. Anal. 2025, 2, 127–144. [Google Scholar]
- Allard, R.W. Principles of Plant Breeding; John Wiley & Sons: Hoboken, NJ, USA, 1999. [Google Scholar]
- Hallauer, A.R.; Carena, M.J.; Miranda Filho, J.D. Quantitative Genetics in Maize Breeding; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2010; Volume 6, pp. 1–650. [Google Scholar]
- Wilcox, J.; Sediyama, T. Interrelationships among height, lodging and yield in determinate and indeterminate soybeans. Euphytica 1981, 30, 323–326. [Google Scholar] [CrossRef]
- Paez, V.; Barrett, W.B.; Deng, X.; Diaz-Amigo, C.; Fiedler, K.; Fuerer, C.; Hostetler, G.L.; Johnson, P.; Joseph, G.; Konings, E.J. AOAC SMPR® 2016.002. J. AOAC Int. 2016, 99, 1122–1124. [Google Scholar] [CrossRef]
- Dohlman, E.; Hansen, J.; Boussios, D. USDA Agricultural Projections to 2029; USDA: Washington, DC, USA, 2020.
- Nyvall, R.F. Diseases of Soybeans: Glycine max (L.) Merr. In Field Crop Diseases Handbook; Springer: Berlin/Heidelberg, Germany, 1989; pp. 503–559. [Google Scholar]
- Niblack, T.; Arelli, P.; Noel, G.; Opperman, C.; Orf, J.; Schmitt, D.; Shannon, J.; Tylka, G. A revised classification scheme for genetically diverse populations of Heterodera glycines. J. Nematol. 2002, 34, 279. [Google Scholar]
- McMaster, G.S.; Wilhelm, W. Growing degree-days: One equation, two interpretations. Agric. For. Meteorol. 1997, 87, 291–300. [Google Scholar] [CrossRef]
- Wickham, H.; Bryan, J. R Packages; O’Reilly Media, Inc.: Sebastopol, CA, USA, 2023; pp. 1–341. [Google Scholar]
- de Mendiburu, F. Agricolae Tutorial, Version 1.3–5; Universidad Nacional Agraria: La Molina, Peru, 2021. [Google Scholar]
- Wickham, H. Programming with ggplot2. In Ggplot2: Elegant Graphics for Data Analysis; Springer: Berlin/Heidelberg, Germany, 2016; pp. 241–253. [Google Scholar]
- Finlay, K.; Wilkinson, G. The analysis of adaptation in a plant-breeding programme. Aust. J. Agric. Res. 1963, 14, 742–754. [Google Scholar] [CrossRef]
- Dumble, S.; Frutos Bernal, E.; Galindo, V. Package GGEBiplots; Version 0.1; 2022; Volume 3, Available online: https://CRAN.R-project.org/package=GGEBiplots (accessed on 1 August 2025).
- Yan, W.; Hunt, L.A.; Sheng, Q.; Szlavnics, Z. Cultivar evaluation and mega-environment investigation based on the GGE biplot. Crop Sci. 2000, 40, 597–605. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Rieseberg, L.H. Hybrid origins of plant species. Annu. Rev. Ecol. Syst. 1997, 28, 359–389. [Google Scholar] [CrossRef]
- DeVicente, M.; Tanksley, S. QTL analysis of transgressive segregation in an interspecific tomato cross. Genetics 1993, 134, 585–596. [Google Scholar] [CrossRef]
- Rai, N.; Rai, M. Heterosis Breeding in Vegetable Crops; New India Publishing: New Delhi, India, 2006; pp. 1–258. [Google Scholar]
- Nyadanu, D.; Lowor, S.; Tchokponhoue, D.; Pobee, P.; Nunekpeku, W.; Brako-Marfo, M.; Okyere, D.; Owusu-Ansah, F.; Ofori, A. Combining ability and gene action for sexual compatibility and pattern of nut colour segregation among ten elite clones of kola (Cola nitida (Vent) Schott and Endl.). Euphytica 2021, 217, 62. [Google Scholar] [CrossRef]
- Yan, W.; Tinker, N.A. Biplot analysis of multi-environment trial data: Principles and applications. Can. J. Plant Sci. 2006, 86, 623–645. [Google Scholar] [CrossRef]
- Krause, M.D.; Piepho, H.-P.; Dias, K.O.; Singh, A.K.; Beavis, W.D. Models to estimate genetic gain of soybean seed yield from annual multi-environment field trials. Theor. Appl. Genet. 2023, 136, 252. [Google Scholar] [CrossRef]
- Hochholdinger, F.; Baldauf, J.A. Heterosis in plants. Curr. Biol. 2018, 28, R1089–R1092. [Google Scholar] [CrossRef]
- Li, Y.; Chang, J.; Gao, X.; Zhang, L.; Wang, L.; Ren, C. A case study on the impacts of future climate change on soybean yield and countermeasures in Fujin city of Heilongjiang province, China. Front. Agron. 2024, 6, 1257830. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, C.; Lamlom, S.F.; Zhao, K.; Abdelghany, A.M.; Wang, X.; Zhang, F.; Yuan, R.; Han, D.; Zha, B. Genetic adaptations of soybean to cold stress reveal key insights through transcriptomic analysis. Biology 2024, 13, 856. [Google Scholar] [CrossRef]
- Zha, B.; Zhang, C.; Yuan, R.; Zhao, K.; Sun, J.; Liu, X.; Wang, X.; Zhang, F.; Zhang, B.; Lamlom, S.F. Integrative QTL mapping and candidate gene analysis for main stem node number in soybean. BMC Plant Biol. 2025, 25, 422. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Hong, H.; Zha, B.; Lamlom, S.F.; Qiu, H.; Cao, Y.; Sun, R.; Wang, H.; Ma, J.; Zhang, H. Soybean productivity can be enhanced by understanding rhizosphere microbiota: Evidence from metagenomics analysis from diverse agroecosystems. Microbiome 2025, 13, 105. [Google Scholar] [CrossRef]
- McDonald, B.A.; Linde, C. Pathogen population genetics, evolutionary potential, and durable resistance. Annu. Rev. Phytopathol. 2002, 40, 349–379. [Google Scholar] [CrossRef]
- Lurá, M.C.; Latorre Rapela, M.G.; Vaccari, M.C.; Maumary, R.; Soldano, A.; Mattio, M.; González, A.M. Genetic diversity of Cercospora kikuchii isolates from soybean cultured in Argentina as revealed by molecular markers and cercosporin production. Mycopathologia 2011, 171, 361–371. [Google Scholar] [CrossRef]
- Brown, J.K. Durable resistance of crops to disease: A Darwinian perspective. Annu. Rev. Phytopathol. 2015, 53, 513–539. [Google Scholar] [CrossRef]
- Patil, G.; Mian, R.; Vuong, T.; Pantalone, V.; Song, Q.; Chen, P.; Shannon, G.J.; Carter, T.C.; Nguyen, H.T. Molecular mapping and genomics of soybean seed protein: A review and perspective for the future. Theor. Appl. Genet. 2017, 130, 1975–1991. [Google Scholar] [CrossRef]
- Singer, W.M.; Lee, Y.C.; Shea, Z.; Vieira, C.C.; Lee, D.; Li, X.; Cunicelli, M.; Kadam, S.S.; Khan, M.A.W.; Shannon, G. Soybean genetics, genomics, and breeding for improving nutritional value and reducing antinutritional traits in food and feed. Plant Genome 2023, 16, e20415. [Google Scholar] [CrossRef] [PubMed]
- Usigbe, M.J.; Uyeh, D.D.; Park, T.; Ha, Y.; Mallipeddi, R. Many objective optimization and decision support for dairy cattle feed formulation. Sci. Rep. 2025, 15, 13451. [Google Scholar] [CrossRef] [PubMed]
- Wachong Kum, S.; Voccia, D.; Grimm, M.; Froldi, F.; Suciu, N.A.; Lamastra, L. Reducing the Environmental Impacts of Pig Production Through Feed Reformulation: A Multi-Objective Life Cycle Assessment Optimisation Approach. Sustainability 2025, 17, 8509. [Google Scholar] [CrossRef]
| Trait | Source | df | Sum Sq | Mean Sq | F Value | Pr (>F) | Sig | CV (%) | Mean |
|---|---|---|---|---|---|---|---|---|---|
| Plant Height (cm) | Environment | 6 | 1.45 × 103 | 241.61 | 3.0 | 0.020 | * | 10.55 | 88.7 |
| Rep (Env) | 14 | 980.8 | 17.51 | 1.22 | 0.184 | NS | |||
| Season | 3 | 954 | 318 | 3.6 | 0.018 | * | |||
| Season × Env | 18 | 7.23 × 103 | 401.42 | 4.6 | <0.001 | *** | |||
| Bottom Pod Height (cm) | Environment | 6 | 332.57 | 55.43 | 20.0 | <0.001 | *** | 9.43 | 17.8 |
| Rep (Env) | 14 | 31.6 | 0.56 | 1.22 | 0.184 | NS | |||
| Season | 3 | 75.75 | 25.25 | 8.9 | <0.001 | *** | |||
| Season × Env | 18 | 588 | 32.67 | 11.6 | <0.001 | *** | |||
| Main Stem Nodes | Environment | 6 | 83.57 | 13.93 | 5.0 | <0.001 | *** | 10.75 | 15.3 |
| Rep (Env) | 14 | 30.4 | 0.54 | 1.22 | 0.184 | NS | |||
| Season | 3 | 8.89 | 2.96 | 1.1 | 0.360 | NS | |||
| Season × Env | 18 | 69.86 | 3.88 | 1.4 | 0.154 | NS | |||
| Lodging Rate (%) | Environment | 6 | 2.86 × 104 | 4.77 × 103 | 188.0 | <0.001 | *** | 19.08 | 26.4 |
| Rep (Env) | 14 | 284.8 | 5.09 | 1.22 | 0.184 | NS | |||
| Season | 3 | 3.69 × 104 | 1.23 × 104 | 483.7 | <0.001 | *** | |||
| Season × Env | 18 | 5.04 × 104 | 2.80 × 103 | 110.1 | <0.001 | *** | |||
| Pods per Plant | Environment | 6 | 250.07 | 41.68 | 6.0 | <0.001 | *** | 10.44 | 25.9 |
| Rep (Env) | 14 | 82.0 | 1.46 | 1.22 | 0.184 | NS | |||
| Season | 3 | 539.57 | 179.86 | 24.6 | <0.001 | *** | |||
| Season × Env | 18 | 1.50 × 103 | 83.11 | 11.4 | <0.001 | *** | |||
| Seeds per Plant | Environment | 6 | 3.08 × 103 | 513.36 | 14.0 | <0.001 | *** | 10.08 | 59.2 |
| Rep (Env) | 14 | 399.2 | 7.13 | 1.22 | 0.184 | NS | |||
| Season | 3 | 2.85 × 103 | 950.43 | 26.7 | <0.001 | *** | |||
| Season × Env | 18 | 1.11 × 104 | 618.6 | 17.4 | <0.001 | *** | |||
| Hundred-seed Weight (g) | Environment | 6 | 24.5 | 4.08 | 1.0 | 0.642 | NS | 10.57 | 22.7 |
| Rep (Env) | 14 | 64.27 | 1.15 | 1.22 | 0.184 | NS | |||
| Season | 3 | 44.57 | 14.86 | 2.6 | 0.062 | NS | |||
| Season × Env | 18 | 144.26 | 8.01 | 1.4 | 0.170 | NS | |||
| Plot Yield (kg) | Environment | 6 | 572.79 | 95.46 | 4.0 | <0.001 | *** | 13.47 | 34.3 |
| Rep (Env) | 14 | 239.07 | 4.27 | 1.22 | 0.184 | NS | |||
| Season | 3 | 6.99 × 104 | 2.33 × 104 | 1091.9 | <0.001 | *** | |||
| Season × Env | 18 | 966.36 | 53.69 | 2.5 | 0.004 | ** | |||
| Yield (kg ha−1) | Environment | 6 | 5.55 × 106 | 9.25 × 105 | 11.0 | <0.001 | *** | 9.82 | 2888.6 |
| Rep (Env) | 14 | 9.02 × 105 | 1.61 × 104 | 1.22 | 0.184 | NS | |||
| Season | 3 | 5.81 × 106 | 1.94 × 106 | 24.0 | <0.001 | *** | |||
| Season × Env | 18 | 5.78 × 106 | 3.21 × 105 | 4.0 | <0.001 | *** | |||
| Protein Content (%) | Environment | 6 | 2.95 | 0.49 | 1.0 | 0.661 | NS | 2.01 | 42.0 |
| Rep (Env) | 14 | 8.03 | 0.14 | 1.22 | 0.184 | NS | |||
| Season | 3 | 0.05 | 0.02 | 0.0 | 0.995 | NS | |||
| Season × Env | 18 | 8.38 | 0.47 | 0.6 | 0.844 | NS | |||
| Oil Content (%) | Environment | 6 | 1.27 | 0.21 | 0.0 | 0.900 | NS | 3.84 | 19.9 |
| Rep (Env) | 14 | 6.53 | 0.12 | 1.22 | 0.184 | NS | |||
| Season | 3 | 0.22 | 0.07 | 0.1 | 0.943 | NS | |||
| Season × Env | 18 | 5.00 | 0.28 | 0.5 | 0.958 | NS |
| Location | Sowing Date | Growth Days | Plant Height (cm) | Bottom Pod Height (cm) | Main Stem Nodes | Effective Branches |
|---|---|---|---|---|---|---|
| Beian Branch Research Institute | 16 May | 108 | 90.2 b | 13 d | 15 ab | 1 |
| Beian Dalong Seed Industry | 25 May | 104 | 83 e | 18 b | 15 ab | 0 |
| Heihe Seed Division | 14 May | 131 | 88.7 c | 20 a | 16 a | 0 |
| Heshan Farm | 11 May | 123 | 88.7 c | 17 bc | 16 a | 0 |
| Nenjiang County Far East Seed | 25 May | 106 | 87.7 cd | 16 c | 14 b | 0 |
| Nenjiang Farm | 16 May | 116 | 97.2 a | 14 d | 16 a | 0 |
| Wudalianchi Seed Station | 16 May | 116 | 85.2 d | 15 c | 14 b | 0 |
| Mean | - | 114 | 88.7 | 17.6 | 15.1 | 0.1 |
| CV (%) | - | 8.7 | 16.4 | 26.1 | 5.3 | 316.2 |
| Location | Flower Color | Pod Color | Seed Shape | Seed Color | Hilum Color | Lodging Resistance (Grade) | Lodging Rate (%) |
|---|---|---|---|---|---|---|---|
| Beian Branch Research Institute | Purple | Yellow brown | Round | Yellow | Yellow | 3 | 30 |
| Beian Dalong Seed Industry | Purple | Brown | Round | Yellow | Yellow | 0 | 0 |
| Heihe Seed Division | Purple | Brown | Oval | Yellow | Yellow | 1 | 70 |
| Heshan Farm | Purple | Brown | Round | Yellow | Yellow | 0 | 0 |
| Nenjiang County Far East Seed | Purple | Brown | Round | Yellow | Yellow | 0 | 0 |
| Nenjiang Farm | Purple | Brown | Round | Yellow | Yellow | 0 | 0 |
| Wudalianchi Seed Station | Purple | Brown | Round | Yellow | Yellow | 0 | 0 |
| Uniformity (%) | 100 | 85.7 | 85.7 | 100 | 100 | Variable | Variable |
| Locations | Yield kg/ha | HSW | Seeds no./Plant | Plot Yield/kg | Compared to Control Percent % |
|---|---|---|---|---|---|
| Beian Branch Research Institute | 2696.5 e | 22.9 ab | 62 b | 34.2 bc | 12 |
| Beian Dalong Seed Industry | 2723 e | 22.2 c | 65.25 a | 30 c | 8.9 |
| Heihe Seed Division | 2449 f | 22.6 bc | 57.25 | 30.8 c | 13.4 |
| Heshan Farm | 3104 b | 22.9 ab | 65.25 a | 36.8 a | 11.9 |
| Nenjiang County Far East Seed | 3042 c | 22.2 c | 47 d | 35.2 b | 13.5 |
| Nenjiang Farm | 2953 d | 23.7 a | 62.25 b | 35.3 b | 11 |
| Wudalianchi Seed Station | 3250 a | 21.9 d | 55.5 c | 36.9 a | 10.5 |
| Level | Protein_Content | Oil_Content |
|---|---|---|
| Beian Branch Research Institute | 41.95 a | 19.92 a |
| Beian Dalong Seed Industry | 41.69 a | 20.13 a |
| Heihe Seed Division | 42.19 a | 19.74 a |
| Heshan Farm | 42.2 a | 19.74 a |
| Nenjiang County Far East Seed | 41.95 a | 19.86 a |
| Nenjiang Farm | 42.25 a | 19.82 a |
| Wudalianchi Seed Station | 41.95 a | 19.89 a |
| Location | Gray Spot Disease (Grade) | SMV Virus Disease (Grade) | Cyst Nematode (Grade) | Diseased Seeds (%) | Insect Damage (%) | Perfect Seeds (%) |
|---|---|---|---|---|---|---|
| Beian Branch Research Institute | 1 | 0 | 0 | 2 | 3 | 97 |
| Beian Dalong Seed Industry | 0 | 0 | 0 | 0 | 1 | 99 |
| Heihe Seed Division | 1 | 0 | 0 | 0 | 0 | 100 |
| Heshan Farm | 0 | 0 | 0 | 0 | 0 | 100 |
| Nenjiang County Far East Seed | 1 | 0 | 0 | 1 | 1 | 98 |
| Nenjiang Farm | 0 | 0 | 0 | 0 | 0 | 100 |
| Wudalianchi Seed Station | 0 | 0 | 0 | 0 | 0 | 98 |
| Mean | 0.4 | 0 | 0 | 0.4 | 0.7 | 99.1 |
| Range | 0–1 | 0 | 0 | 0–2 | 0–3 | 97–100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, D.; Yan, X.; Li, W.; Jia, H.; Ren, H.; Lu, W. Multi-Environment Evaluation of Soybean Variety Heike 88: Transgressive Segregation and Regional Adaptation in Northern China. Agriculture 2025, 15, 2106. https://doi.org/10.3390/agriculture15202106
Han D, Yan X, Li W, Jia H, Ren H, Lu W. Multi-Environment Evaluation of Soybean Variety Heike 88: Transgressive Segregation and Regional Adaptation in Northern China. Agriculture. 2025; 15(20):2106. https://doi.org/10.3390/agriculture15202106
Chicago/Turabian StyleHan, Dezhi, Xiaofei Yan, Wei Li, Hongchang Jia, Honglei Ren, and Wencheng Lu. 2025. "Multi-Environment Evaluation of Soybean Variety Heike 88: Transgressive Segregation and Regional Adaptation in Northern China" Agriculture 15, no. 20: 2106. https://doi.org/10.3390/agriculture15202106
APA StyleHan, D., Yan, X., Li, W., Jia, H., Ren, H., & Lu, W. (2025). Multi-Environment Evaluation of Soybean Variety Heike 88: Transgressive Segregation and Regional Adaptation in Northern China. Agriculture, 15(20), 2106. https://doi.org/10.3390/agriculture15202106






