A Detection Line Counting Method Based on Multi-Target Detection and Tracking for Precision Rearing and High-Quality Breeding of Young Silkworm (Bombyx mori)
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.1.1. Image Acquisition
2.1.2. Dataset of Young Silkworm Bodies
2.1.3. Challenges
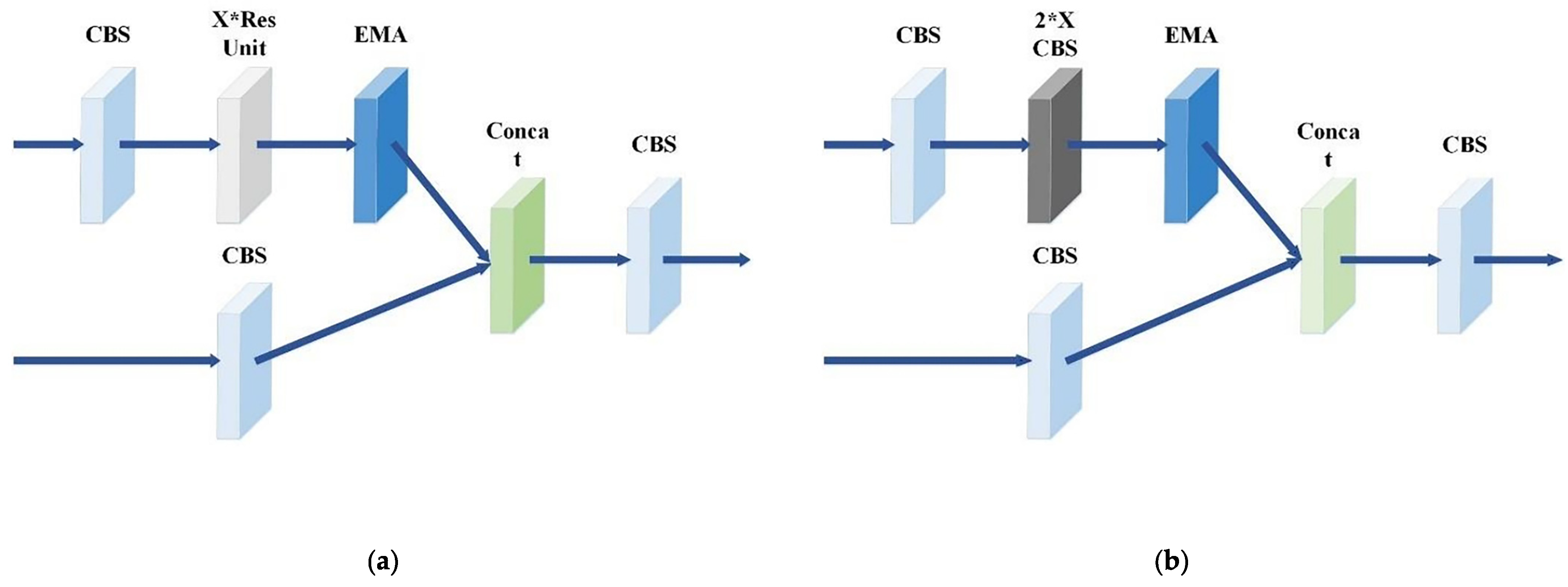
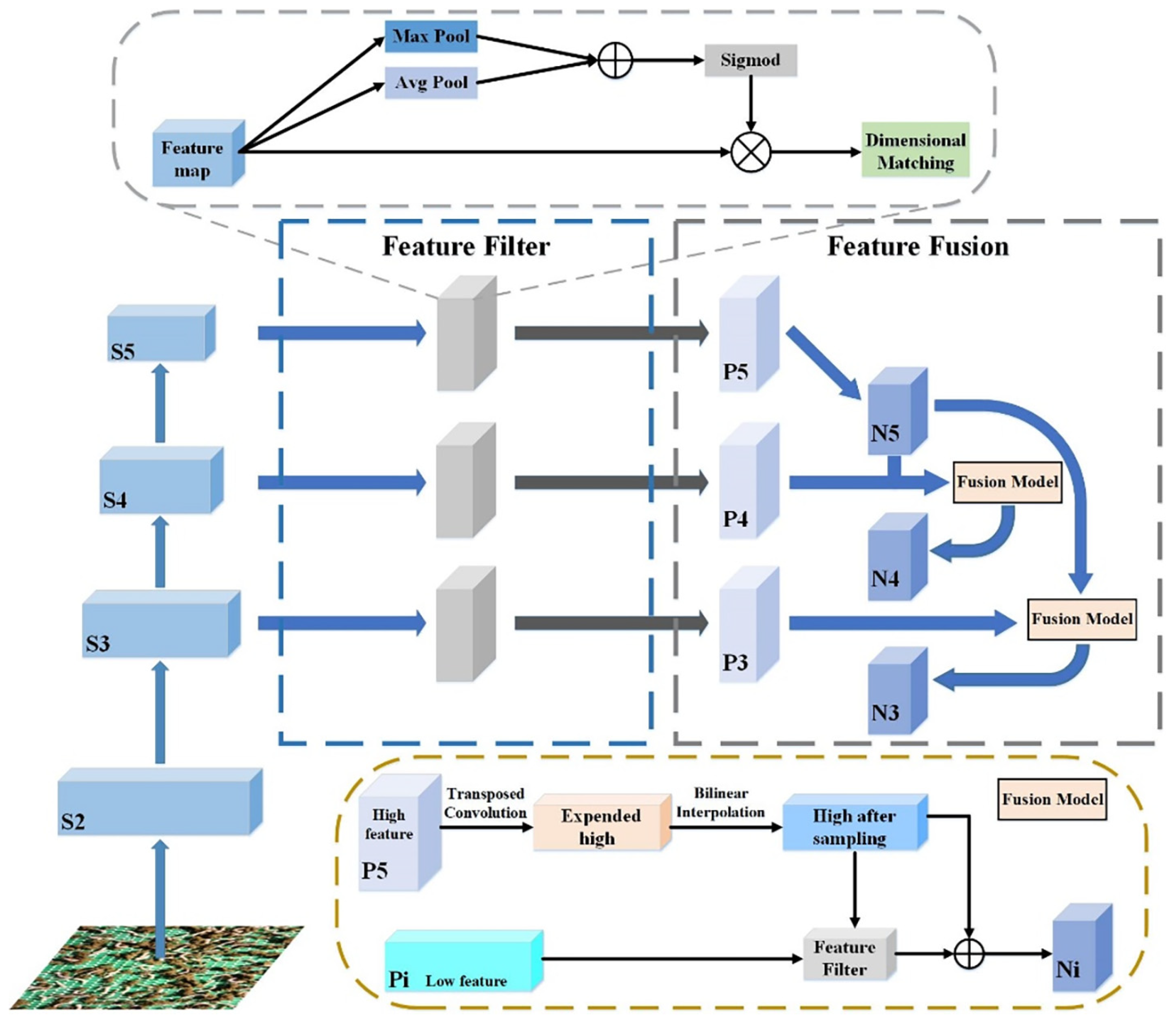
- Morphological Variability. As illustrated in Figure 2, the morphology of silkworms in the dataset varies due to multiple factors, primarily stemming from mutual occlusion among individual silkworms and occlusion by feed. These morphological differences contribute to the diversification of silkworm features. To address this issue, the PAFPN structure [25] of the original detector is proposed to be substituted with the multi-scale feature fusion (MSFF) structure, and the EMA mechanism [26] is introduced within the CSP module, aiming to enhance the detection accuracy in this study.
- ID Switching. The interleaved arrangement of silkworm individuals and occlusion by feed can result in a decrease in the score of tracking detection boxes, making it difficult to match them with tracking trajectories. This, in turn, triggers ID switching issues, compromising the accuracy of subsequent counting. To tackle this problem, an improved version of the ByteTrack [27] method is adopted in this study to reduce the probability of mismatching between trajectories and detection boxes. Additionally, by adjusting the matching strategy, the frequency of ID switching is further minimized.
- Duplicate Counting. Factors such as edge distortion and occlusion can lead to duplicate counting issues in the process of silkworm target tracking. To resolve this issue, a novel statistical approach specifically tailored for silkworm targets is introduced in this study. This method leverages constructed “detection lines” and target ID values within the statistical area to effectively mitigate the problem of duplicate counting.
2.2. Method for Detecting Young Silkworms
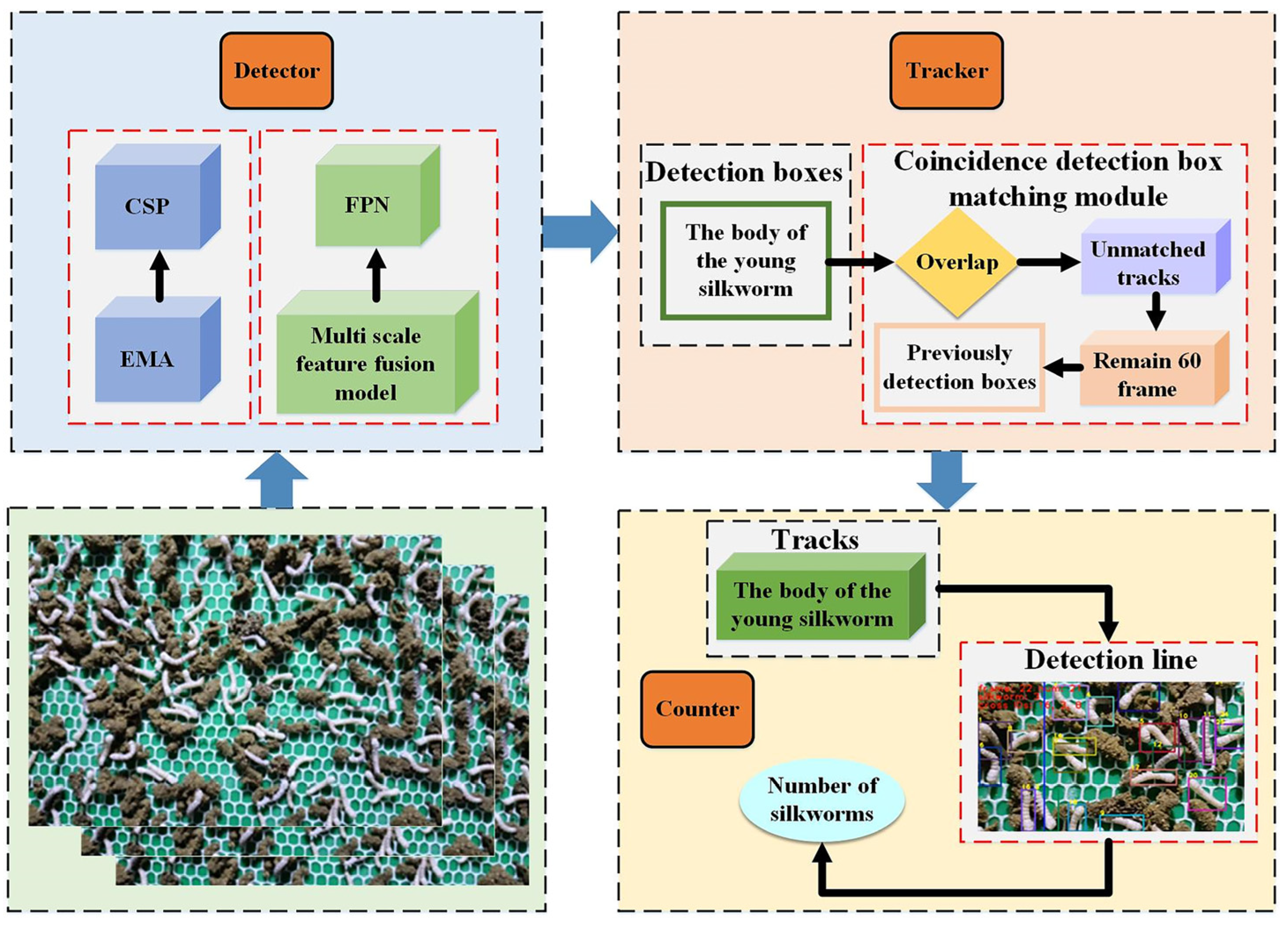
2.2.1. Overall Overview
2.2.2. Detector
- (1)
- EMA-CSP Detection Module
- (2)
- Multi-scale feature fusion method
2.2.3. Tracker
- (1)
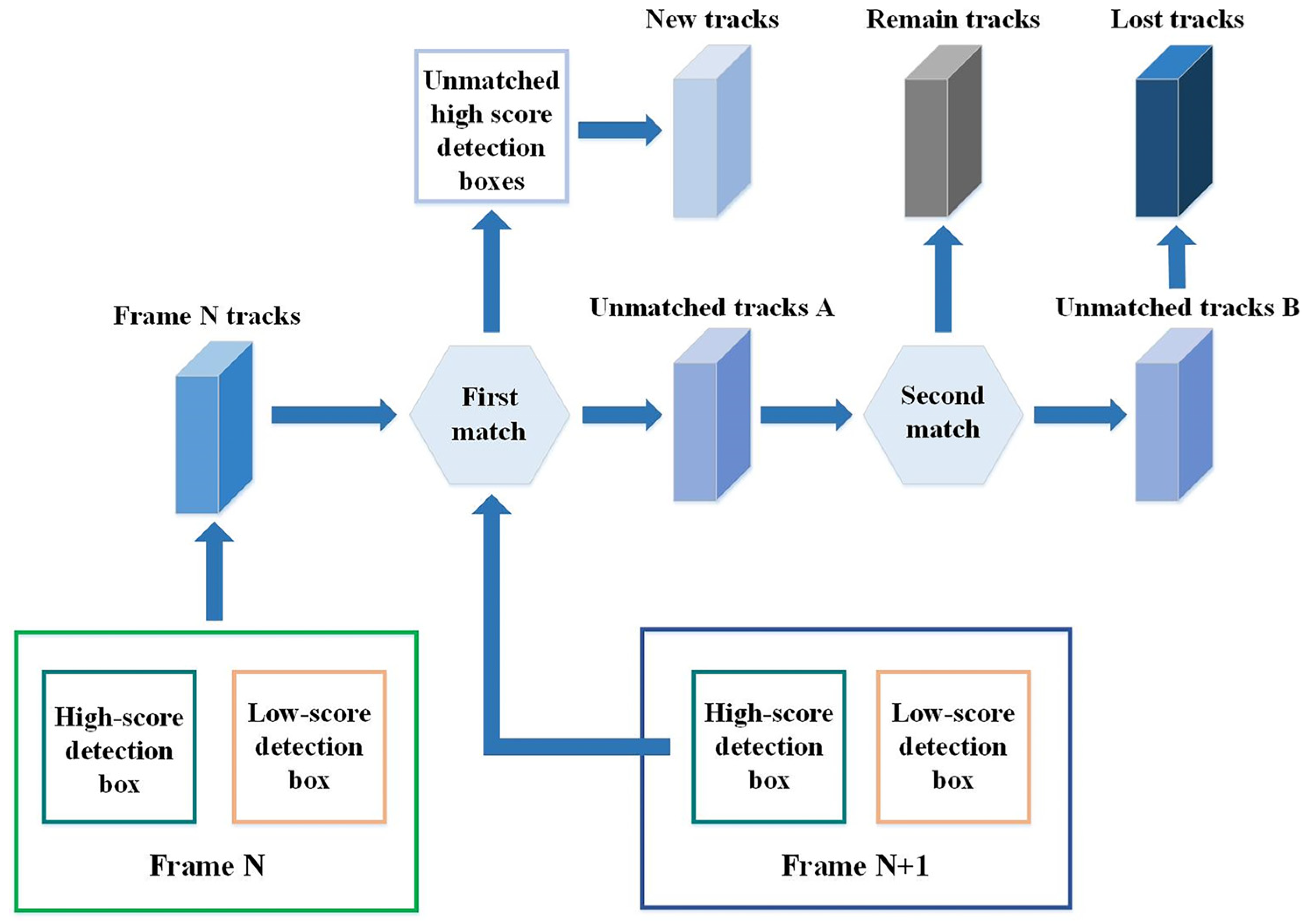
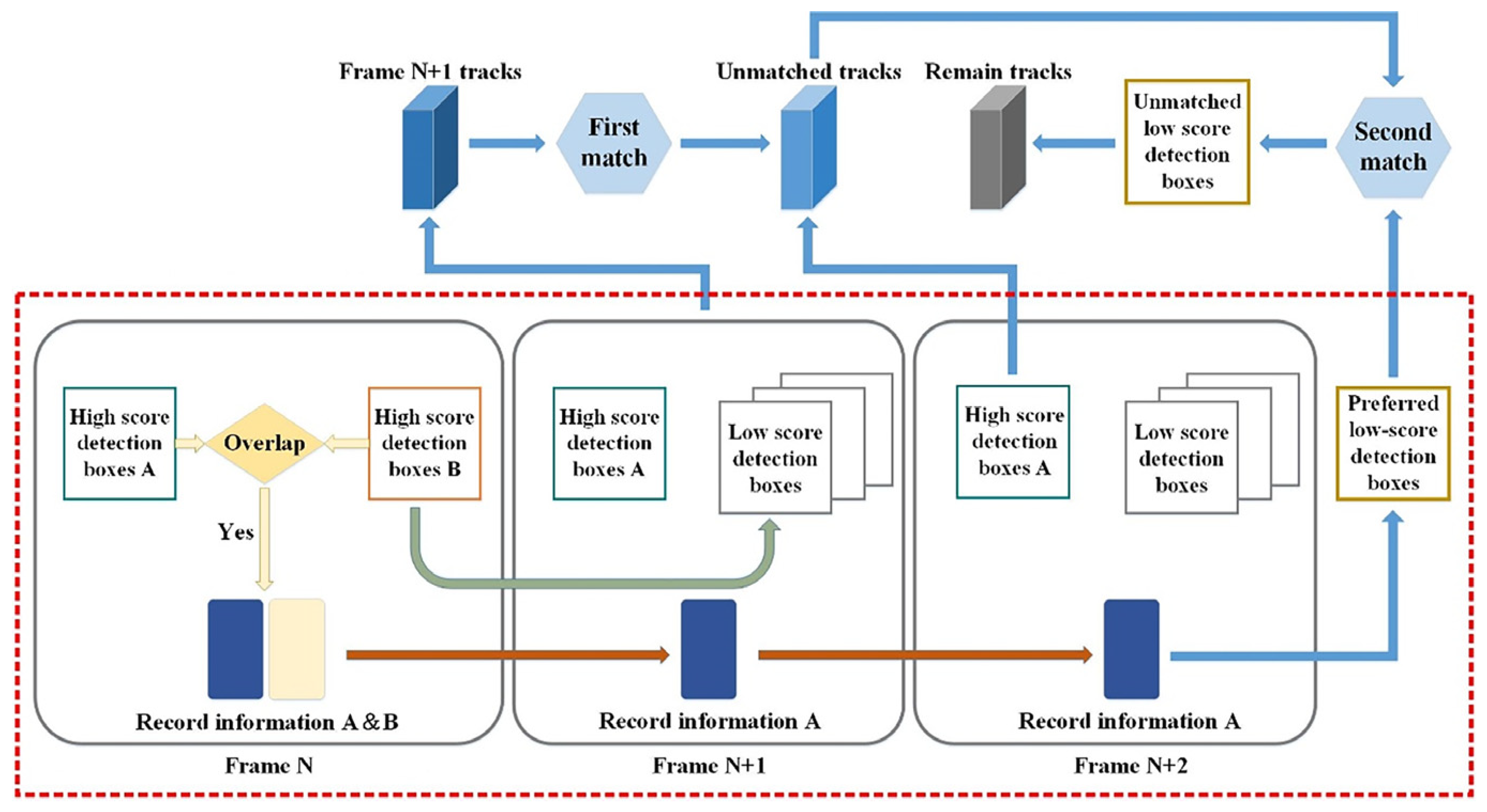
- To address redundant computations, the original ByteTrack strategy is modified to process only the low-scoring boxes within the minimum bounding rectangle of overlapping high-confidence boxes. This optimized strategy reduces the number of detection boxes to be processed, thereby shortening the processing time.
- (2)
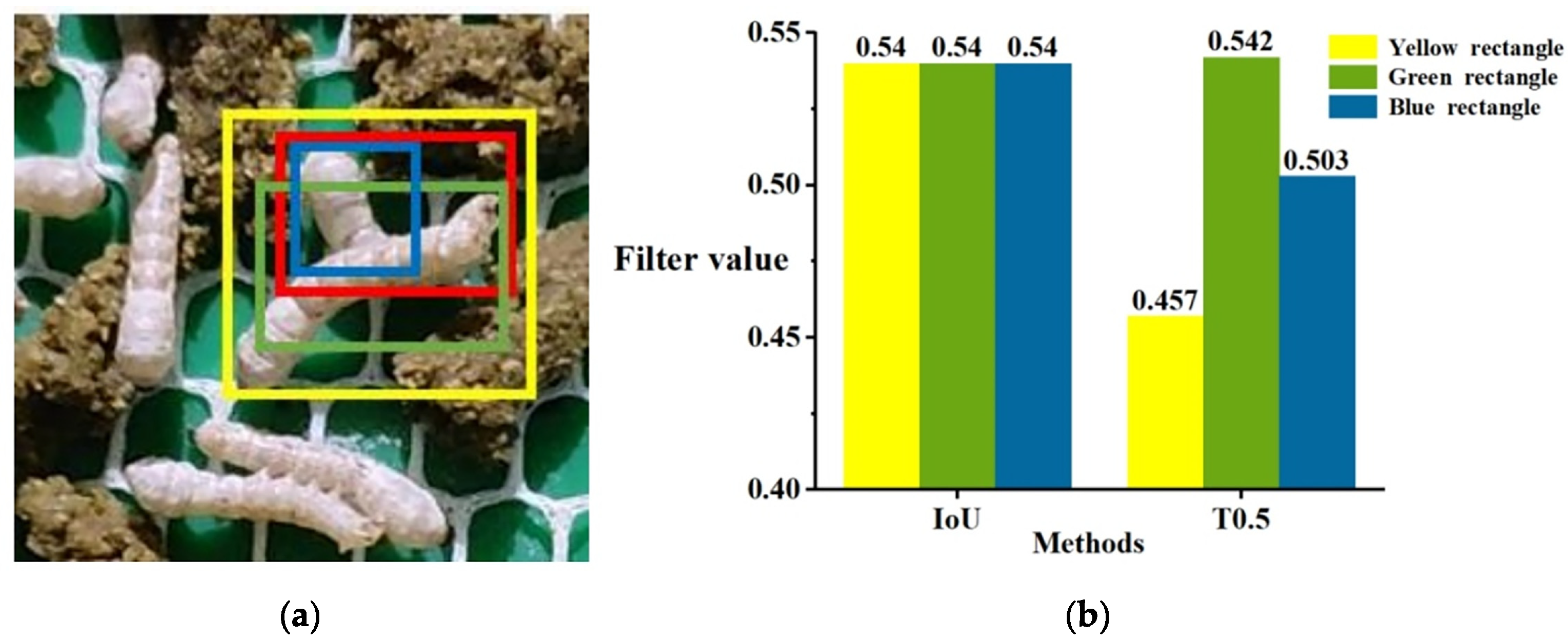
- To tackle the problem of mismatching, a pre-screening step is incorporated into the optimized method before trajectory matching. This step aims to select more suitable low-scoring detection boxes, which are then matched with the tracking trajectories that failed to match with high-scoring boxes in the first attempt. The schematic diagram of the screening process is illustrated in Figure 7. The threshold (T0.5) for low-scoring box selection is calculated using the formula provided, and a box is considered superior if T0.5 > 0.5, and it meets the specified condition.where IoU stands for intersection over union; α is a hyperparameter representing the influence of the distance between the center points, set to 0.4; β is a hyperparameter representing the influence of the difference in width and height, set to 0.45; ρ(cl, ch) is the Euclidean distance between the center points of low-scoring and high-scoring boxes; at and bt represent the width and height of the smallest detection rectangle encompassing the high-scoring and low-scoring detection boxes; ah and al represent the width of the high-scoring and low-scoring detection boxes; bh and bl represent the height of the high-scoring and low-scoring detection boxes.
- (3)
- To address the issue of trajectory loss due to prolonged unmatched periods, the retention time for trajectories that fail to match detection boxes was extended from 30 frames to 60 frames. Additionally, the criterion for creating a new trajectory was revised to consider a match between a superior low-scoring box and a trajectory with a score below 80%, and to discard the original trajectory and create a new one if no match is established within 60 frames.
2.2.4. Counter
2.3. Experimental Platform
2.4. Evaluation Metrics
3. Results
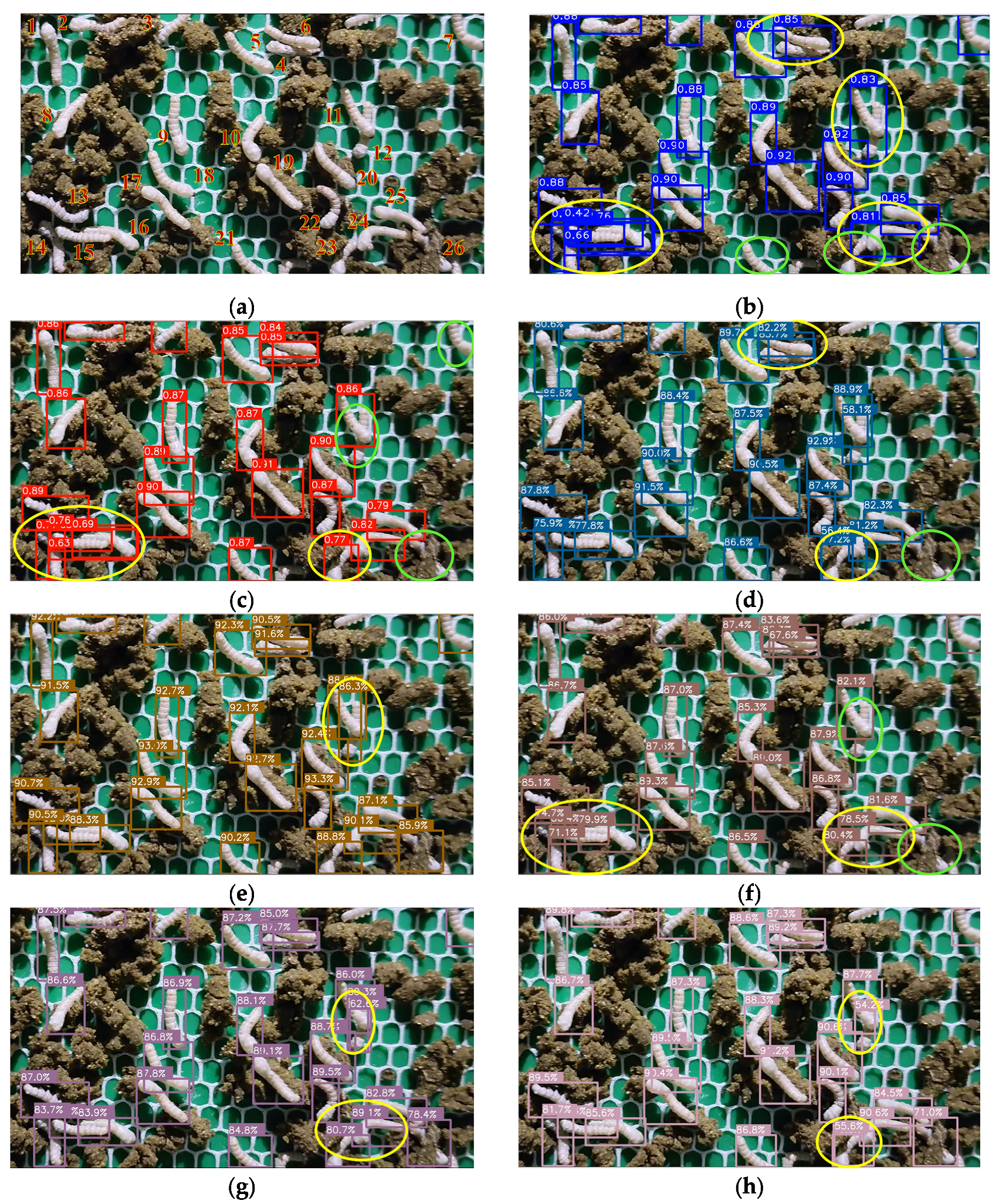
3.1. Detection Results
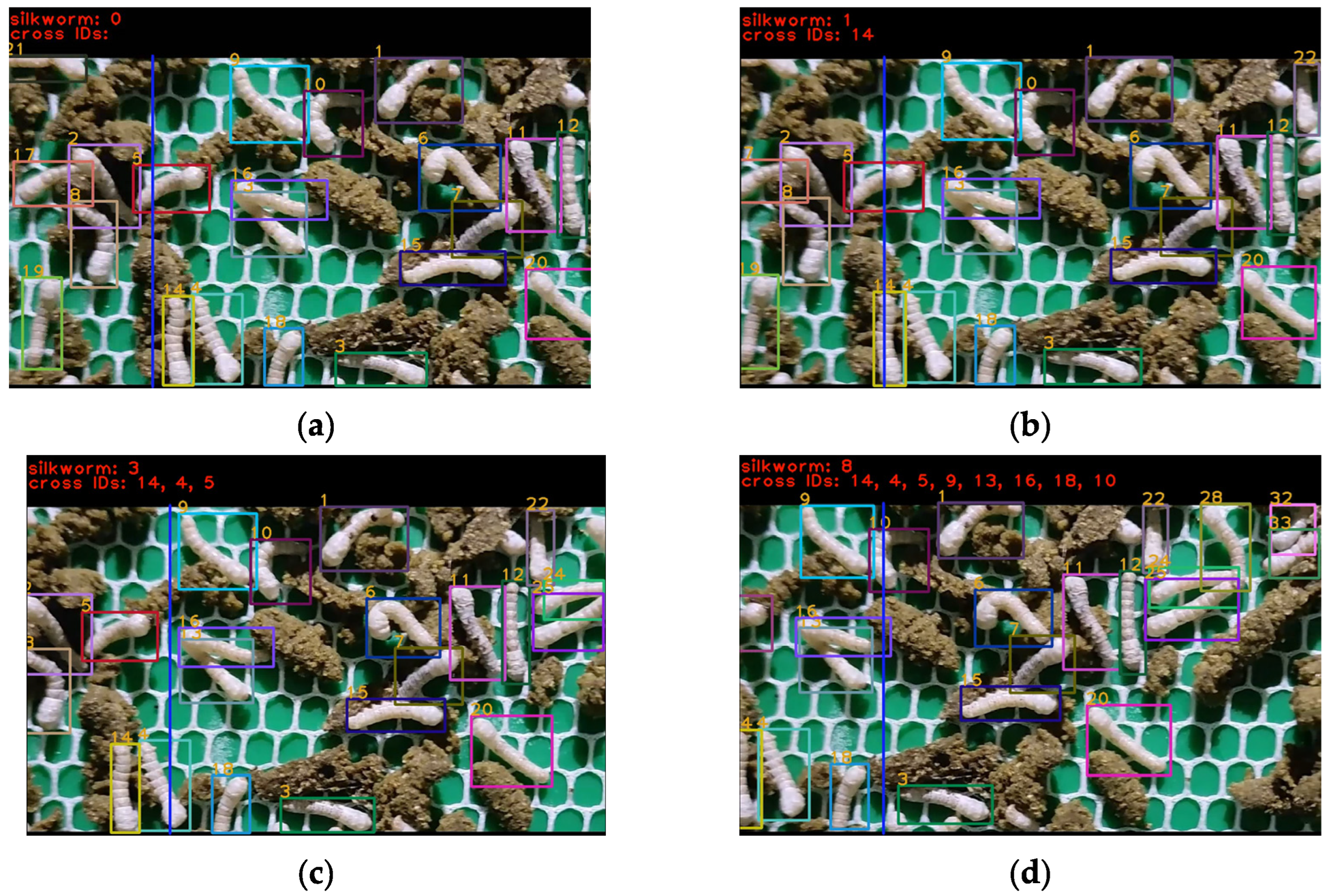
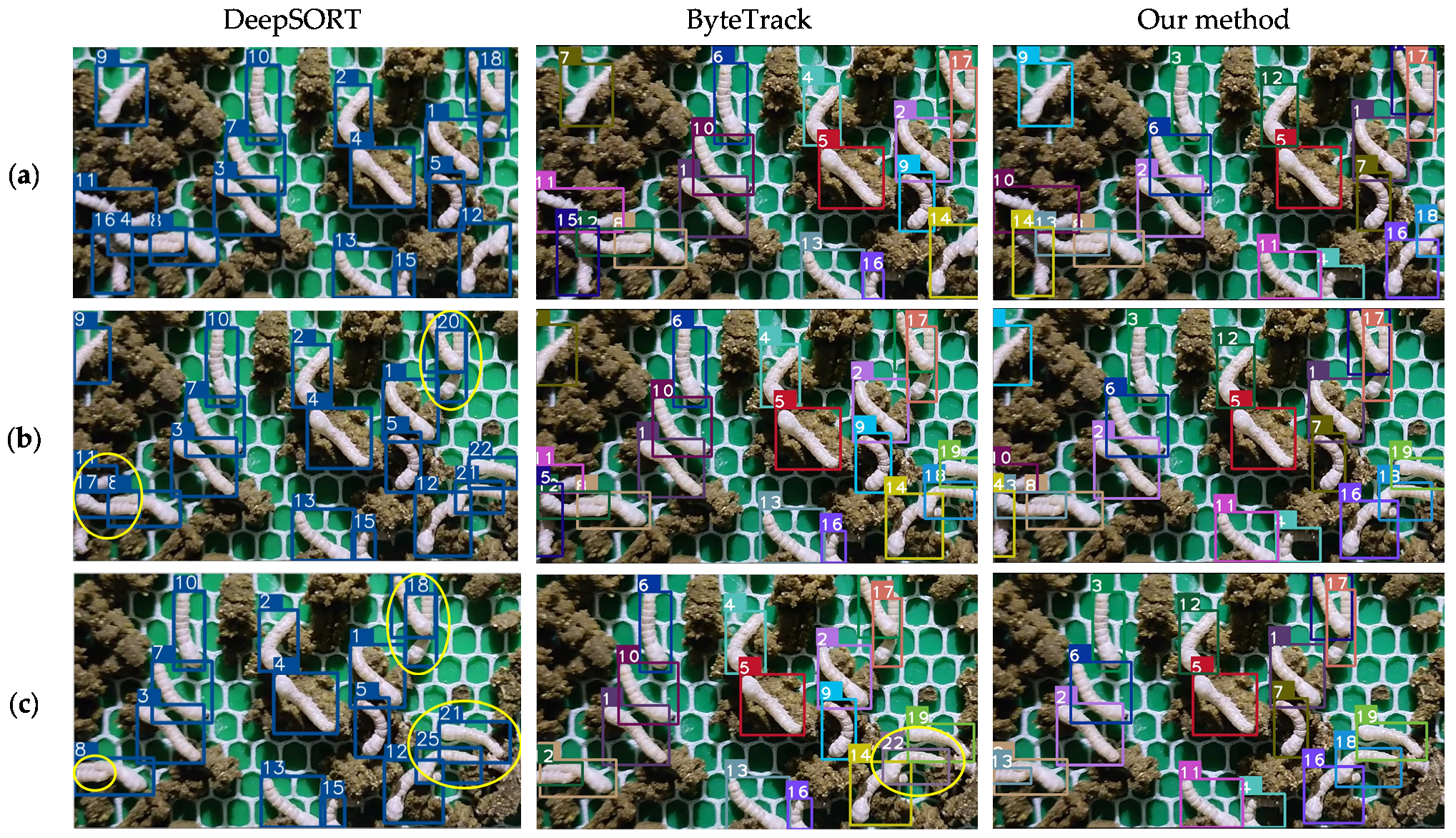
3.2. Tracking Results
3.3. Ablation Tests
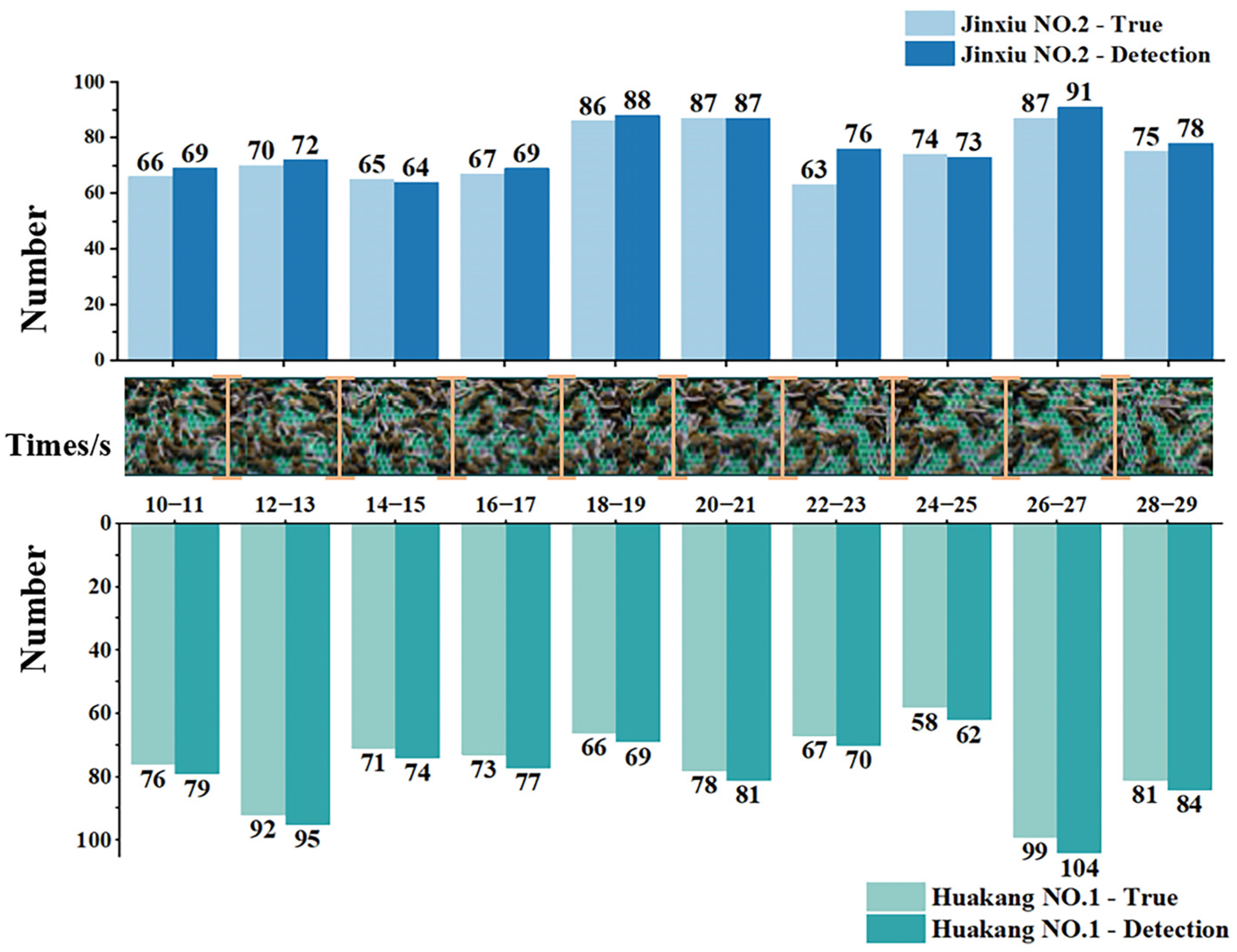
3.4. Counting Effectiveness for Young Silkworms
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Nomenclature
| A(c) | The association accuracy |
| Ac | The counting accuracy |
| AF-RCNN | Anchor-Free Region-based Convolutional Neural Network |
| ah | The width of the high-score detection boxes |
| al | The width of the low-score detection boxes |
| AP50 | The AP value when the IoU threshold is 50 |
| AP50:95 | The average AP value under different IoU thresholds ranging from 50 to 95 at intervals of 5 |
| AssA | Evaluate association accuracy |
| at | The width of the smallest detection rectangle encompassing the high and low score detection boxes |
| bh | The height of the high-score detection boxes |
| bl | The height of the low-score detection boxes |
| bt | The height of the smallest detection rectangle encompassing the high and low score detection boxes |
| C | A point that belongs to the set of True Positives (TP) |
| CAN | Context-Aware Network |
| CS-YOLOX-Byte | Counting of Silkworm-YOLOX Fusion ByteTrack |
| CSP | Cross Stage Partial |
| DeepSORT | Simple online and realtime tracking with a deep association metric |
| DetA | Assess detection accuracy |
| EMA | Efficient Multi-scale Attention |
| F1 | Harmonic mean of precision and recall |
| FN | False Negative |
| FNA(c) | The association prediction accuracy for unpredicted trajectories |
| FNt | The number of missed detections in the t-th frame |
| FP | False Positive |
| FPA(c) | The false association precision of trajectory predictions |
| FPS | Frames Per Second |
| FPt | The number of false detections in the t-th frame |
| frame | The total count of frames |
| GTt | The total number of tracked objects in the t-th frame |
| HOTA | High Order Tracking Accuracy |
| IDF1 | Identification Average Rate |
| IDFN | False negative IDs |
| IDFP | False positive IDs |
| IDs | Identity switches |
| IDSWt | The number of ID switches for tracked objects within the t-th frame |
| IDTP | True positive IDs |
| IoU | Intersection over Union |
| IPFC | In-field Pest in Food Crop |
| mAP | mean Average Precision |
| MOTA | Multiple Object Tracking Accuracy |
| MSFF | Multi-Scale Feature Fusion |
| Numc | The detected number of young silkworms |
| Numt | The true number of young silkworms |
| P | Precision |
| PAFPN | Path Aggregation Feature Pyramid Network |
| R | Recall |
| R-CNN | Regions with CNN features |
| RCS-OSA | Reparametrized Convolution based on channel Shuffle-One-Shot Aggregation |
| SORT | Simple Online and Realtime Tracking |
| timeelapsed | The duration of continuous operation |
| TN | True Negative |
| TP | True Positive |
| TPA(c) | The accuracy of correctly associated detections |
| α | A hyperparameter representing the influence of the distance between the centre points |
| β | A hyperparameter representing the influence of the difference in width and height |
| ρ(cl, ch) | The Euclidean distance between the centre points of low and high score box |
References
- Li, J.; Liang, Q.; Gu, G. The Impact of the New Crown Pneumonia Epidemic on China’s Silk Industry and Development Countermeasures. Chin. J. Anim. Sci. 2023, 59, 336–341. [Google Scholar] [CrossRef]
- Feng, H.; Li, J. The development history and characteristics of sericulture industry in 60 years of new China. China Seric. 2014, 35, 1–10. [Google Scholar] [CrossRef]
- Gu, G.; Li, J. Development of the Silk Industry and Industrialized Silkworm Rearing Using Artificial Diet for All Life Stages. Newsl. Sericult. Sci. 2019, 39, 46–48. [Google Scholar]
- Li, J.; Gu, G. Development Trends and Policy Recommendations of China’s Sericulture Industry in 2022. Chin. J. Anim. Sci. 2022, 58, 270–274. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, H.; Chen, Y.; Chen, S.; Jiang, Y.; Cui, C.; Liu, M.; Tang, H.; Fan, Y.; Yang, Q.; et al. Reflections on the co-rearing of commercialized young silkworms on artificial feed in Yunnan province. China Seric. 2023, 44, 38–43. [Google Scholar] [CrossRef]
- Cheng, Y.; Yuan, G.; Zhu, J.; Xiong, H.; Cao, N.; He, Y.; Xiao, H.; Yang, Z. Status and Prospects of Artificial Feed Rearing Technology for Silkworms in Sichuan Province. Sichuan Canye 2023, 49, 49–52. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, G. The large-scale artificial feed rearing of young silkworms in Guangxi has been successfully trialed. Guangxi Seric. 2023, 56, 41. [Google Scholar]
- Li, J.; Gu, G.; Cui, W. Progress and Prospects of Silkworm Rearing Using Artificial Feed. China Seric. 2021, 42, 46–52. [Google Scholar] [CrossRef]
- Jin, Q.; Feng, J.; Wu, H.; Zhan, P.; Feng, H. Artificial Feed Rearing for Young Silkworms + Leaf Strip Rearing for Larger Silkworms. China Seric. 2019, 40, 65–69. [Google Scholar] [CrossRef]
- Wang, L.; Hu, S. Promotion of the Babei Model for Factory-based Silkworm Rearing Using Artificial Feed for All Life Stages. Bull. Seric. 2020, 51, 37–38+54. [Google Scholar]
- Cheng, Y.; Cao, N.; Xiao, H.; He, Y. Suggestions on Prevention and Emergency Response Measures for Mold Formation in Artificial Feed for Bombyx mori. Sichuan Canye 2024, 52, 37–38. [Google Scholar] [CrossRef]
- Yan, Z.; Guan, S. Key technical measures for domestic silkworm rearing. Jiangsu Seric. 2017, 2017, 16–17. [Google Scholar]
- Lan, B.; Qin, M.; Qin, X. Technical measures to improve the quality of co-rearing of young silkworms in seed cocoon production. Guangdong Canye 2014, 48, 3–5+8. [Google Scholar] [CrossRef]
- Suo, X.; Liu, Z.; Lei, S.; Wang, J.; Zhao, Y. Aphid identification and counting based on smartphone and machine vision. J. Sens. 2017, 2017, 3964376. [Google Scholar] [CrossRef]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2015, 39, 1137–1149. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, D.; Li, M.; Gao, Y.; Wu, J.; Yang, X. Field detection of tiny pests from sticky trap images using deep learning in agricultural greenhouse. Comput. Electron. Agric. 2021, 183, 106048. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Wang, F.; Wang, R.; Xie, C.; Yang, P.; Liu, L. Fusing multi-scale context-aware information representation for automatic in-field pest detection and recognition. Comput. Electron. Agric. 2020, 169, 105222. [Google Scholar] [CrossRef]
- Jiao, L.; Dong, S.; Zhang, S.; Xie, C.; Wang, H. AF-RCNN: An anchor-free convolutional neural network for multi-categories agricultural pest detection. Comput. Electron. Agric. 2020, 174, 105522. [Google Scholar] [CrossRef]
- Ma, X.; Wang, M.; Kuang, H.; Tang, L.; Liu, X. Detecting and counting silkworms using improved YOLOv8n. Trans. Chin. Soc. Agric. Eng. 2024, 40, 143–151. [Google Scholar] [CrossRef]
- Zhang, R.; Lin, Y.; Wu, Y.; Deng, L.; Zhang, H.; Liao, M.; Peng, Y. MvMRL: A multi-view molecular representation learning method for molecular property prediction. Brief. Bioinform. 2024, 25, bbae298. [Google Scholar] [CrossRef]
- Wei, X.; Wu, H.; Wang, Z.; Zhu, J.; Wang, W.; Wang, J.; Wang, Y.; Wang, C. Rumen-protected lysine supplementation improved amino acid balance, nitrogen utilization and altered hindgut microbiota of dairy cows. Anim. Nutr. 2023, 15, 320–331. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Li, Q.; Chen, J. Highlight Removal from A Single Grayscale Image Using Attentive GAN. Appl. Artif. Intell. 2022, 36, 1988441. [Google Scholar] [CrossRef]
- Wang, B.; Yang, M.; Cao, P.; Liu, Y. A novel embedded cross framework for high-resolution salient object detection. Appl. Intell. 2025, 55, 277. [Google Scholar] [CrossRef]
- Liu, S.; Qi, L.; Qin, H.; Shi, J.; Jia, J. Path Aggregation Network for Instance Segmentation. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 8759–8768. [Google Scholar] [CrossRef]
- Ouyang, D.; He, S.; Zhan, J.; Guo, H.; Huang, Z.; Luo, M.; Zhang, G. Efficient Multi-Scale Attention Module with Cross-Spatial Learning. In Proceedings of the ICASSP 2023–2023 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Rhodes Island, Greece, 4–10 June 2023. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, P.; Jiang, Y.; Yu, D.; Weng, F.; Yuan, Z.; Luo, P.; Liu, W.; Wang, X. ByteTrack: Multi-Object Tracking by Associating Every Detection Box. In Proceedings of the Computer Vision–ECCV 2022, 17th European Conference, Tel Aviv, Israel, 23–27 October 2022; Springer Nature: Cham, Switzerland, 2022; pp. 1–21. [Google Scholar] [CrossRef]
- Ge, Z.; Liu, S.; Wang, F.; Li, Z.; Sun, J. YOLOX: Exceeding YOLO Series in 2021. arXiv 2021, arXiv:2107.08430. [Google Scholar] [CrossRef]
- Bochkovskiy, A.; Wang, C.-Y.; Liao, H.-Y.M. YOLOv4: Optimal Speed and Accuracy of Object Detection. arXiv 2020, arXiv:2004.10934. [Google Scholar] [CrossRef]
- Jocher, G.; Chaurasia, A.; Stoken, A.; Borovec, J.; NanoCode012; Kwon, Y.; Xie, T.; Fang, J.; Imyhxy; Michael, K.; et al. ultralytics/yolov5: V6.1–TensorRT, TensorFlow Edge TPU and OpenVINO Export and Inference; Zenodo: Geneva, Switzerland, 2022. [Google Scholar] [CrossRef]
- Bewley, A.; Ge, Z.; Ott, L.; Ramos, F.; Upcroft, B. Simple online and realtime tracking. In Proceedings of the 2016 IEEE International Conference on Image Processing (ICIP), Phoenix, AZ, USA, 25–28 September 2016; pp. 3464–3468. [Google Scholar] [CrossRef]
- Wojke, N.; Bewley, A.; Paulus, D. Simple online and realtime tracking with a deep association metric. In Proceedings of the 2017 IEEE International Conference on Image Processing (ICIP), Beijing, China, 17–20 September 2017; pp. 3645–3649. [Google Scholar] [CrossRef]
- Bereciartua-Pérez, A.; Gómez, L.; Picón, A.; Navarra-Mestre, R.; Klukas, C.; Eggers, T. Insect counting through deep learning-based density maps estimation. Comput. Electron. Agric. 2022, 197, 106933. [Google Scholar] [CrossRef]
- Saradopoulos, L.; Potamitis, L.; Konstantaras, A.; Eliopoulos, P.; Ntalampiras, S.; Rigakis, I. Image-Based Insect Counting Embedded in E-Traps That Learn without Manual Image Annotation and Self-Dispose Captured Insects. Information 2023, 14, 267. [Google Scholar] [CrossRef]



| Dataset Classification | Variety of Young Silkworms | Number of Images | Number of Samples |
|---|---|---|---|
| Training set | Liangguang No. 2 | 720 | 20,048 |
| Jingsong × Haoyue | 720 | 18,795 | |
| Validation set | Liangguang No. 2 | 240 | 5243 |
| Jingsong × Haoyue | 240 | 5104 | |
| Test set | Liangguang No. 2 | 240 | 5163 |
| Jingsong × Haoyue | 240 | 6323 | |
| Total | 2400 | 100,201 |
| Configuration | Parameter |
|---|---|
| CPU | Intel Xeon Gold 5218R |
| Memory | 128 G |
| GPU | GeForce RTX 3090 |
| Accelerated environment | CUDA 11.3 cuDNN 8.0.5 |
| Operating system | Windows 10.0 |
| Development environment | Python 3.8 Pytorch 1.12.0 |
| Algorithms | F1/% (↑) | Recall/% (↑) | Precision/% (↑) | AP50/% (↑) | AP50:95/% (↑) |
|---|---|---|---|---|---|
| YOLOv4 | 74.2 | 71.8 | 76.7 | 72.5% | 55.3 |
| YOLOv5s | 85.2 | 84.9 | 85.5 | 85.1 | 66.5 |
| YOLOX-S | 87.2 | 86.4 | 88.1 | 87.4 | 69.6 |
| Our method | 89.6 | 87.9 | 91.3 | 89.1 | 72.7 |
| YOLOv8s | 85.8 | 85.4 | 86.2 | 85.7 | 68.1 |
| YOLOv10s | 87.9 | 86.5 | 89.4 | 88.2 | 70.2 |
| YOLOv11s | 88.1 | 87.1 | 89.2 | 89.0 | 71.5 |
| Algorithms | MOTA/% (↑) | IDF1/% (↑) | HOTA/% (↑) | FP (↓) | FN (↓) | IDS (↓) | FPS (↑) |
|---|---|---|---|---|---|---|---|
| SORT | 81.6 | 83.0 | 67.2 | 554 | 77 | 55 | 51 |
| DeepSORT | 83.7 | 85.5 | 69.9 | 301 | 59 | 43 | 33 |
| ByteTrack | 85.6 | 87.9 | 74.3 | 224 | 37 | 33 | 38 |
| Our method | 88.3 | 90.2 | 78.1 | 113 | 21 | 18 | 29 |
| ByteTrack | MSFF | EMA | Overlapping Detection Box Matching | MOTA/% (↑) | IDF1/% (↑) | HOTA/% (↑) | FPS (↑) |
|---|---|---|---|---|---|---|---|
| 1 | 84.9 | 87.3 | 72.8 | 38 | |||
| 2 | ✔ | 86.4 | 88.5 | 74.6 | 35 | ||
| 3 | ✔ | 85.1 | 84.8 | 74.1 | 37 | ||
| 4 | ✔ | 86.6 | 88.6 | 75.2 | 33 | ||
| 5 | ✔ | ✔ | 87.4 | 87.9 | 74.9 | 34 | |
| 6 | ✔ | ✔ | 87.3 | 88.8 | 75.9 | 32 | |
| 7 | ✔ | ✔ | 87.2 | 89.5 | 76.4 | 30 | |
| 8 | ✔ | ✔ | ✔ | 88.3 | 90.4 | 76.7 | 29 |
| Methods | Test Videos | Numt | Numc | Max ID | MOTA/% | Ac/% |
|---|---|---|---|---|---|---|
| I YOLOv4-Byte | A | 2974 | 4892 | 5483 | 0.63 | 35.5 |
| B | 3088 | 5138 | 5733 | 0.70 | 33.6 | |
| C | 2682 | 4312 | 4948 | 0.64 | 39.2 | |
| II YOLOv5-Byte | A | 2974 | 3997 | 5196 | 0.67 | 65.6 |
| B | 3088 | 4093 | 5337 | 0.72 | 67.5 | |
| C | 2682 | 3666 | 4933 | 0.69 | 63.3 | |
| III YOLOX-Byte | A | 2974 | 3682 | 4543 | 0.82 | 76.2 |
| B | 3088 | 3661 | 4495 | 0.75 | 81.4 | |
| C | 2682 | 3234 | 4075 | 0.77 | 79.4 | |
| IV YSC (our method) | A | 2974 | 3230 | 3468 | 0.85 | 91.4 |
| B | 3088 | 3364 | 3482 | 0.80 | 91.1 | |
| C | 2682 | 2936 | 2921 | 0.82 | 90.5 |
| Test Video | Numt | Numc | Max ID | MOTA/% | Ac/% |
|---|---|---|---|---|---|
| A | 3123 | 3358 | 3552 | 0.84 | 92.5 |
| B | 2866 | 3063 | 3244 | 0.86 | 93.1 |
| C | 3049 | 3273 | 3431 | 0.84 | 92.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Chang, H.; Shang, M.; Song, Z.; Tian, F.; Li, F.; Zhang, G.; Sun, T.; Yan, Y.; Liu, M. A Detection Line Counting Method Based on Multi-Target Detection and Tracking for Precision Rearing and High-Quality Breeding of Young Silkworm (Bombyx mori). Agriculture 2025, 15, 1524. https://doi.org/10.3390/agriculture15141524
Li Z, Chang H, Shang M, Song Z, Tian F, Li F, Zhang G, Sun T, Yan Y, Liu M. A Detection Line Counting Method Based on Multi-Target Detection and Tracking for Precision Rearing and High-Quality Breeding of Young Silkworm (Bombyx mori). Agriculture. 2025; 15(14):1524. https://doi.org/10.3390/agriculture15141524
Chicago/Turabian StyleLi, Zhenghao, Hao Chang, Mingrui Shang, Zhanhua Song, Fuyang Tian, Fade Li, Guizheng Zhang, Tingju Sun, Yinfa Yan, and Mochen Liu. 2025. "A Detection Line Counting Method Based on Multi-Target Detection and Tracking for Precision Rearing and High-Quality Breeding of Young Silkworm (Bombyx mori)" Agriculture 15, no. 14: 1524. https://doi.org/10.3390/agriculture15141524
APA StyleLi, Z., Chang, H., Shang, M., Song, Z., Tian, F., Li, F., Zhang, G., Sun, T., Yan, Y., & Liu, M. (2025). A Detection Line Counting Method Based on Multi-Target Detection and Tracking for Precision Rearing and High-Quality Breeding of Young Silkworm (Bombyx mori). Agriculture, 15(14), 1524. https://doi.org/10.3390/agriculture15141524





