Genome-Wide Identification and Evolutionary Analysis of the SnRK2 Gene Family in Nicotiana Species
Abstract
1. Introduction
2. Methods
2.1. Identification and Physicochemical Analysis of SnRK2 Gene Family Members In Silico
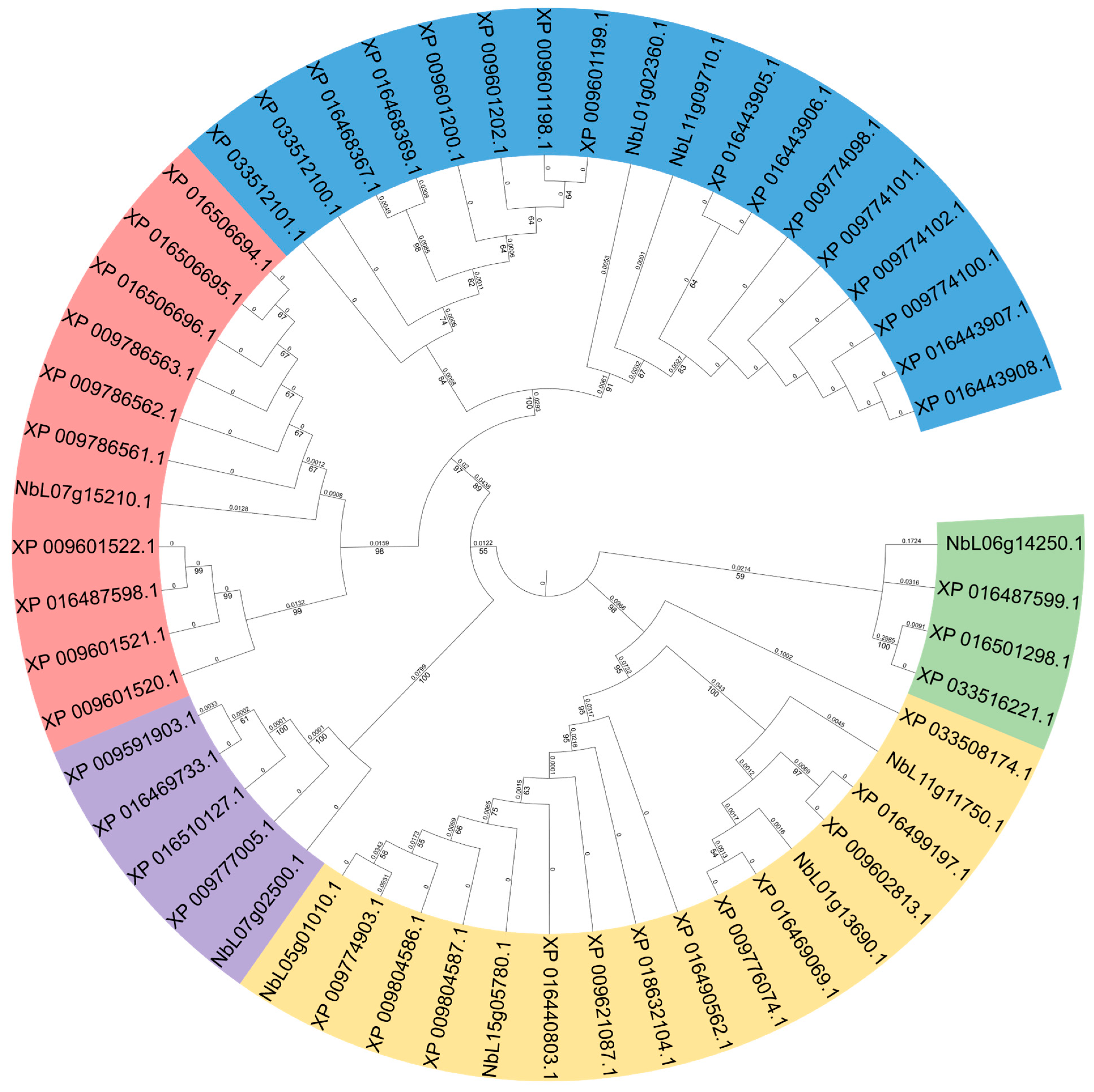
2.2. Phylogenetic Tree Construction of SnRK2 Proteins
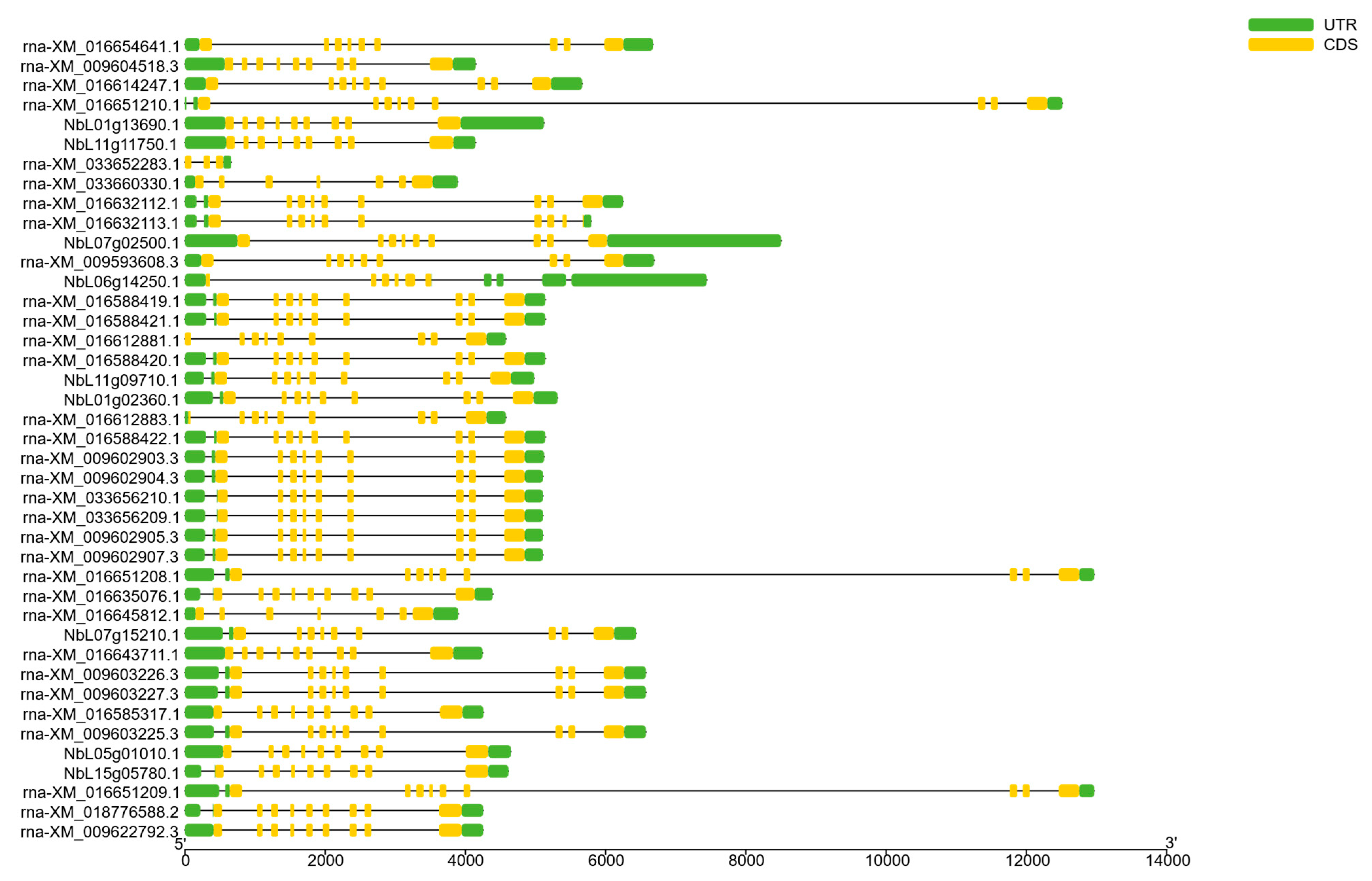
2.3. Conserved Motif and Gene Structure Analysis of SnRK2 Family
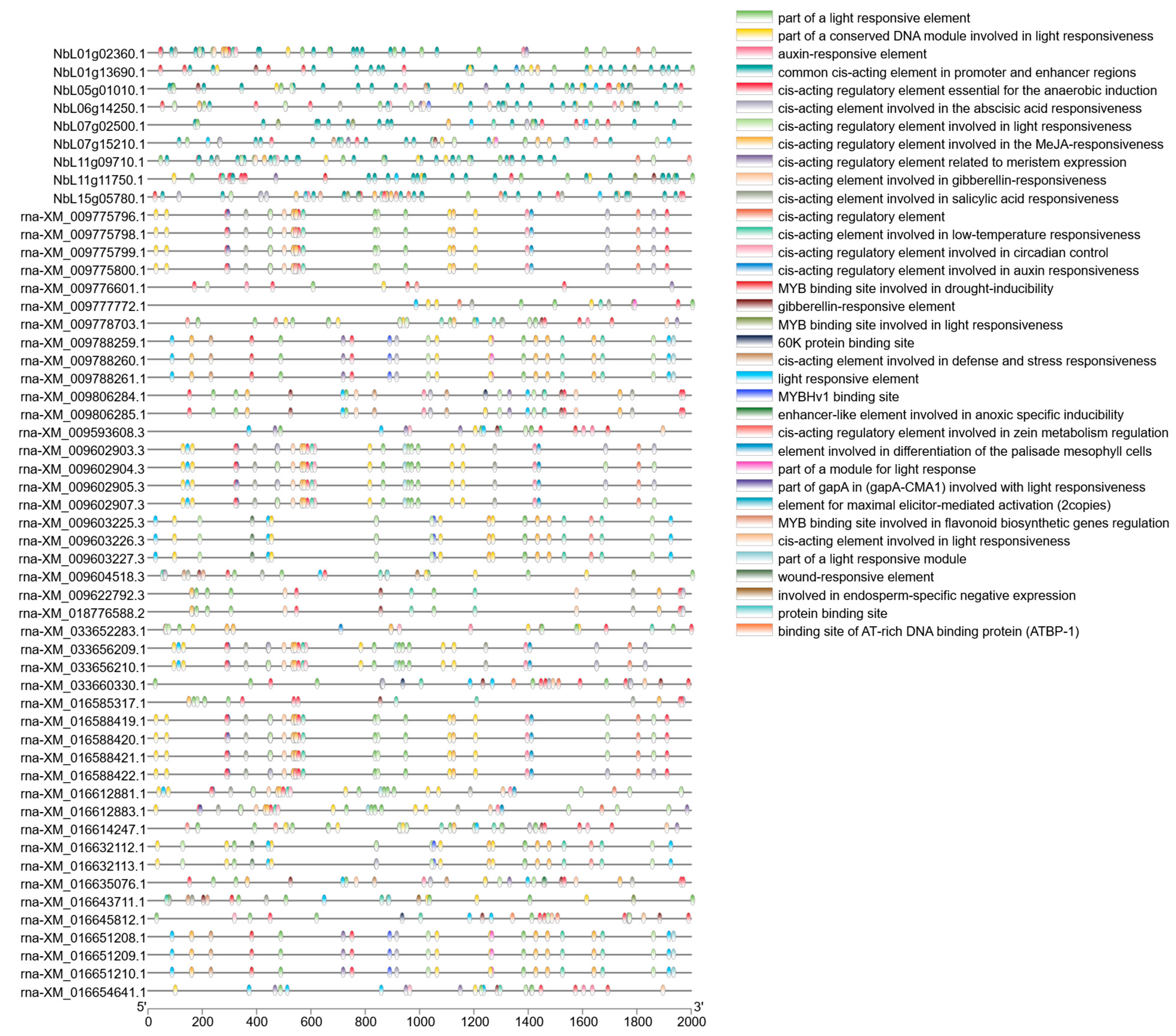
2.4. Cis-Acting Element Analysis of SnRK2 Promoters
3. Results
3.1. Identification and Physicochemical Properties Analysis of SnRK2 Gene Family Members
3.2. Phylogenetic Tree Analysis
3.3. The Conserved Motif and Gene Structure Analysis of the SnRK2 Family
3.4. Cis-Acting Element Analysis of the SnRK2 Family
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. Global Status of Salt-Affected Soils—Main Report; FAO: Rome, Italy, 2024. [Google Scholar] [CrossRef]
- Acosta-Motos, J.R.; Ortuño, M.F.; Bernal-Vicente, A.; Diaz-Vivancos, P.; Sanchez-Blanco, M.J.; Hernandez, J.A. Plant Responses to Salt Stress: Adaptive Mechanisms. Agronomy 2017, 7, 38. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Tunçturk, M.; Tunçturk, R.; Yasar, F. Changes in micronutrients, dry weight and plant growth of soybean (Glycine max L. Merrill) cultivars under salt stress. Afr. J. Biotechnol. 2008, 7, 1650–1654. [Google Scholar]
- Boopal, J.; Sathee, L.; Ramasamy, R.; Pandey, R.; Chinnusamy, V. Influence of Incremental Short Term Salt Stress at the Seedling Stage on Root Plasticity, Shoot Thermal Profile and Ion Homeostasis in Contrasting Wheat Genotypes. Agriculture 2023, 13, 20. [Google Scholar] [CrossRef]
- Duarte, B.; Sleimi, N.; Cacador, I. Biophysical and biochemical constraints imposed by salt stress: Learning from halophytes. Front. Plant Sci. 2014, 5, 746. [Google Scholar] [CrossRef]
- Zhao, C.Z.; Zhang, H.; Song, C.P.; Zhu, J.K.; Shabala, S. Mechanisms of Plant Responses and Adaptation to Soil Salinity. Innovation 2020, 1, 41. [Google Scholar] [CrossRef]
- Fujii, H.; Verslues, P.E.; Zhu, J.-K. Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proc. Natl. Acad. Sci. USA 2011, 108, 1717–1722. [Google Scholar] [CrossRef]
- Kulik, A.; Wawer, I.; Krzywinska, E.; Bucholc, M.; Dobrowolska, G. SnRK2 Protein Kinases-Key Regulators of Plant Response to Abiotic Stresses. Omics 2011, 15, 859–872. [Google Scholar] [CrossRef]
- Hrabak, E.M.; Chan, C.W.M.; Gribskov, M.; Harper, J.F.; Choi, J.H.; Halford, N.; Kudla, J.; Luan, S.; Nimmo, H.G.; Sussman, M.R.; et al. The Arabidopsis CDPK-SnRK Superfamily of Protein Kinases. Plant Physiol. 2003, 132, 666–680. [Google Scholar] [CrossRef]
- Anderberg, R.J.; Walker-Simmons, M.K. Isolation of a wheat cDNA clone for an abscisic acid-inducible transcript with homology to protein kinases. Proc. Natl. Acad. Sci. USA 1992, 89, 10183–10187. [Google Scholar] [CrossRef]
- Yoshida, T.; Mogami, J.; Yamaguchi-Shinozaki, K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 2014, 21, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Boudsocq, M.; Barbier-Brygoo, H.; Laurière, C. Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J. Biol. Chem. 2004, 279, 41758–41766. [Google Scholar] [CrossRef]
- Lim, C.W.; Baek, W.; Lee, S.C. Two pepper subclass II SnRK2 genes positively regulate drought stress response, with differential responsiveness to abscisic acid. Plant Physiol. Biochem. 2025, 220, 109477. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, X.; Zhu, X.; Zhang, D.; Wang, Y.; Wang, T.; Chen, L.; Wang, B.; Wei, X. Meta-analysis of SnRK2 gene overexpression in response to drought and salt stress. Physiol. Plant. 2024, 176, e14578. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.M.; Liu, X.-D.; Waseem, M.; Guang-Qian, Y.; Alabdallah, N.M.; Jahan, M.S.; Fang, X.-W. ABA activated SnRK2 kinases: An emerging role in plant growth and physiology. Plant Signal. Behav. 2022, 17, 2071024. [Google Scholar] [CrossRef]
- Long, T.; Xu, B.; Hu, Y.; Wang, Y.; Mao, C.; Wang, Y.; Zhang, J.; Liu, H.; Huang, H.; Liu, Y.; et al. Genome-wide identification of ZmSnRK2 genes and functional analysis of ZmSnRK2.10 in ABA signaling pathway in maize (Zea mays L). BMC Plant Biol. 2021, 21, 309. [Google Scholar] [CrossRef]
- Yin, S.; Ma, H.; Ye, Q.; Lu, H.; Wang, K.; Kong, S.; Hou, D.; Li, X.; Lin, X. Identification of SnRK2 family and functional study of PeSnRK2.2A and PeSnRK2.2B for drought resistance in Phyllostachys edulis. Ind. Crops Prod. 2024, 219, 119087. [Google Scholar] [CrossRef]
- Li, Q.; Hu, T.; Lu, T.; Yu, B.; Zhao, Y. Calcium-dependent protein kinases CPK3/4/6/11 and 27 respond to osmotic stress and activate SnRK2s in Arabidopsis. Dev. Cell 2025, 60, 1423–1438. [Google Scholar] [CrossRef]
- Sierro, N.; Battey, J.N.D.; Ouadi, S.; Bakaher, N.; Bovet, L.; Willig, A.; Goepfert, S.; Peitsch, M.C.; Ivanov, N.V. The tobacco genome sequence and its comparison with those of tomato and potato. Nat. Commun. 2014, 5, 3833. [Google Scholar] [CrossRef]
- Leitch, I.J.; Hanson, L.; Lim, K.Y.; Kovarik, A.; Chase, M.W.; Clarkson, J.J.; Leitch, A.R. The Ups and Downs of Genome Size Evolution in Polyploid Species of Nicotiana (Solanaceae). Ann. Bot. 2008, 101, 805–814. [Google Scholar] [CrossRef]
- Renny-Byfield, S.; Chester, M.; Kovařík, A.; Le Comber, S.C.; Grandbastien, M.-A.; Deloger, M.; Nichols, R.A.; Macas, J.; Novák, P.; Chase, M.W.; et al. Next Generation Sequencing Reveals Genome Downsizing in Allotetraploid Nicotiana tabacum, Predominantly through the Elimination of Paternally Derived Repetitive DNAs. Mol. Biol. Evol. 2011, 28, 2843–2854. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Pozo, N.; Menda, N.; Edwards, J.D.; Saha, S.; Tecle, I.Y.; Strickler, S.R.; Bombarely, A.; Fisher-York, T.; Pujar, A.; Foerster, H.; et al. The Sol Genomics Network (SGN)—From genotype to phenotype to breeding. Nucleic Acids Res. 2014, 43, D1036–D1041. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Ahmed, B.; Hasan, F.; Tabassum, A.; Ahmed, R.; Hassan, R.; Amin, M.R.; Alam, M. Genome-wide investigation of SnRK2 gene family in two jute species: Corchorus olitorius and Corchorus capsularis. J. Genet. Eng. Biotechnol. 2023, 21, 5. [Google Scholar] [CrossRef]
- Mathura, S.R.; Sutton, F.; Bowrin, V. Characterization and expression analysis of SnRK2, PYL, and ABF/ AREB/ ABI5 gene families in sweet potato. PLoS ONE 2023, 18, e0288481. [Google Scholar] [CrossRef]
- Zan, Y.; Chen, S.; Ren, M.; Liu, G.; Liu, Y.; Han, Y.; Dong, Y.; Zhang, Y.; Si, H.; Liu, Z.; et al. The genome and GeneBank genomics of allotetraploid Nicotiana tabacum provide insights into genome evolution and complex trait regulation. Nat. Genet. 2025, 57, 986–996. [Google Scholar] [CrossRef]
- Li, N.; Xu, C.; Zhang, A.; Lv, R.; Meng, X.; Lin, X.; Gong, L.; Wendel, J.F.; Liu, B. DNA methylation repatterning accompanying hybridization, whole genome doubling and homoeolog exchange in nascent segmental rice allotetraploids. New Phytol. 2019, 223, 979–992. [Google Scholar] [CrossRef]
- Edger, P.P.; Smith, R.; McKain, M.R.; Cooley, A.M.; Vallejo-Marin, M.; Yuan, Y.; Bewick, A.J.; Ji, L.; Platts, A.E.; Bowman, M.J.; et al. Subgenome Dominance in an Interspecific Hybrid, Synthetic Allopolyploid, and a 140-Year-Old Naturally Established Neo-Allopolyploid Monkeyflower. Plant Cell 2017, 29, 2150–2167. [Google Scholar] [CrossRef]
- Bird, K.A.; Niederhuth, C.E.; Ou, S.; Gehan, M.; Pires, J.C.; Xiong, Z.; VanBuren, R.; Edger, P.P. Replaying the evolutionary tape to investigate subgenome dominance in allopolyploid Brassica napus. New Phytol. 2021, 230, 354–371. [Google Scholar] [CrossRef] [PubMed]
- Burns, R.; Mandáková, T.; Gunis, J.; Soto-Jiménez, L.M.; Liu, C.; Lysak, M.A.; Novikova, P.Y.; Nordborg, M. Gradual evolution of allopolyploidy in Arabidopsis suecica. Nat. Ecol. Evol. 2021, 5, 1367–1381. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Jin, J.; Nifong, J.M.; Shew, D.; Lewis, R.S. Homoeologous chromosome exchange explains the creation of a QTL affecting soil-borne pathogen resistance in tobacco. Plant Biotechnol. J. 2022, 20, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Yuan, G.; Sun, K.; Yu, W.; Jiang, Z.; Jiang, C.; Liu, D.; Wen, L.; Si, H.; Wu, F.; Meng, H.; et al. Development of a MAGIC population and high-resolution quantitative trait mapping for nicotine content in tobacco. Front. Plant Sci. 2022, 13, 1086950. [Google Scholar] [CrossRef]
- Schulthess, A.W.; Kale, S.M.; Liu, F.; Zhao, Y.; Philipp, N.; Rembe, M.; Jiang, Y.; Beukert, U.; Serfling, A.; Himmelbach, A.; et al. Genomics-informed prebreeding unlocks the diversity in genebanks for wheat improvement. Nat. Genet. 2022, 54, 1544–1552. [Google Scholar] [CrossRef]
- Milner, S.G.; Jost, M.; Taketa, S.; Mazón, E.R.; Himmelbach, A.; Oppermann, M.; Weise, S.; Knüpffer, H.; Basterrechea, M.; König, P.; et al. Genebank genomics highlights the diversity of a global barley collection. Nat. Genet. 2019, 51, 319–326. [Google Scholar] [CrossRef]
- Du, L.; Ma, Z.; Mao, H. Duplicate Genes Contribute to Variability in Abiotic Stress Resistance in Allopolyploid Wheat. Plants 2023, 12, 2465. [Google Scholar] [CrossRef]
- Lin, Z.; Li, Y.; Wang, Y.; Liu, X.; Ma, L.; Zhang, Z.; Mu, C.; Zhang, Y.; Peng, L.; Xie, S.; et al. Initiation and amplification of SnRK2 activation in abscisic acid signaling. Nat. Commun. 2021, 12, 2456. [Google Scholar] [CrossRef]
- Suryawanshi, V.; Talke, I.N.; Weber, M.; Eils, R.; Brors, B.; Clemens, S.; Krämer, U. Between-species differences in gene copy number are enriched among functions critical for adaptive evolution in Arabidopsis halleri. BMC Genom. 2016, 17, 1034. [Google Scholar] [CrossRef]
- Liu, Z.; Ge, X.; Yang, Z.; Zhang, C.; Zhao, G.; Chen, E.; Liu, J.; Zhang, X.; Li, F. Genome-wide identification and characterization of SnRK2 gene family in cotton (Gossypium hirsutum L.). BMC Genet. 2017, 18, 54. [Google Scholar] [CrossRef]
- Chen, G.; Wang, J.; Qiao, X.; Jin, C.; Duan, W.; Sun, X.; Wu, J. Genome-wide survey of sucrose non-fermenting 1-related protein kinase 2 in Rosaceae and expression analysis of PbrSnRK2 in response to ABA stress. BMC Genom. 2020, 21, 781. [Google Scholar] [CrossRef] [PubMed]
- Wan, Z.; Luo, S.; Zhang, Z.; Liu, Z.; Qiao, Y.; Gao, X.; Yu, J.; Zhang, G. Identification and expression profile analysis of the SnRK2 gene family in cucumber. PeerJ 2022, 10, e13994. [Google Scholar] [CrossRef] [PubMed]


| Sequence ID | Tobacco Species | Number of Amino Acids | Molecular Weight | Theoretical pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|
| NbL01g02360.1 | N. benthamiana | 363 | 41,310.02 | 4.88 | 38.26 | 89.42 | −0.3 | cyto |
| NbL01g13690.1 | N. benthamiana | 355 | 40,881.36 | 5.79 | 53.28 | 79.38 | −0.578 | cysk |
| NbL05g01010.1 | N. benthamiana | 353 | 40,709.33 | 5.78 | 48.39 | 82.04 | −0.488 | cysk |
| NbL06g14250.1 | N. benthamiana | 170 | 19,773.52 | 9.08 | 43.05 | 81.94 | −0.46 | cyto |
| NbL07g02500.1 | N. benthamiana | 354 | 40,337.96 | 4.72 | 42.46 | 88.64 | −0.331 | cysk |
| NbL07g15210.1 | N. benthamiana | 362 | 41,001.65 | 4.94 | 34.78 | 87.54 | −0.284 | cyto |
| NbL11g09710.1 | N. benthamiana | 363 | 41,333 | 4.93 | 41.17 | 88.35 | −0.333 | cyto |
| NbL11g11750.1 | N. benthamiana | 359 | 41,475.91 | 5.45 | 58.24 | 78.77 | −0.607 | cysk |
| NbL15g05780.1 | N. benthamiana | 354 | 40,856.51 | 5.71 | 52.13 | 82.06 | −0.511 | cysk |
| XP_009591903.1 | N. tomentosiformis | 354 | 40,346.97 | 4.76 | 41.91 | 88.64 | −0.33 | cysk |
| XP_009601198.1 | N. tomentosiformis | 363 | 41,303.03 | 4.92 | 40.67 | 88.62 | −0.305 | cyto |
| XP_009601199.1 | N. tomentosiformis | 363 | 41,303.03 | 4.92 | 40.67 | 88.62 | −0.305 | cyto |
| XP_009601200.1 | N. tomentosiformis | 363 | 41,303.03 | 4.92 | 40.67 | 88.62 | −0.305 | cyto |
| XP_009601202.1 | N. tomentosiformis | 363 | 41,303.03 | 4.92 | 40.67 | 88.62 | −0.305 | cyto |
| XP_009601520.1 | N. tomentosiformis | 362 | 41,060.55 | 4.86 | 35.75 | 89.14 | −0.318 | cyto |
| XP_009601521.1 | N. tomentosiformis | 362 | 41,060.55 | 4.86 | 35.75 | 89.14 | −0.318 | cyto |
| XP_009601522.1 | N. tomentosiformis | 362 | 41,060.55 | 4.86 | 35.75 | 89.14 | −0.318 | cyto |
| XP_009602813.1 | N. tomentosiformis | 355 | 40,839.36 | 5.79 | 52.39 | 79.38 | −0.575 | cysk |
| XP_009621087.1 | N. tomentosiformis | 353 | 40,775.39 | 5.78 | 51.34 | 81.47 | −0.508 | cysk |
| XP_018632104.1 | N. tomentosiformis | 353 | 40,775.39 | 5.78 | 51.34 | 81.47 | −0.508 | cysk |
| XP_033508174.1 | N. tomentosiformis | 96 | 10,805.51 | 8.51 | 49.14 | 93.44 | 0.033 | nucl |
| XP_033512100.1 | N. tomentosiformis | 352 | 40,243.77 | 4.92 | 41.97 | 88.89 | −0.339 | cysk |
| XP_033512101.1 | N. tomentosiformis | 352 | 40,243.77 | 4.92 | 41.97 | 88.89 | −0.339 | cysk |
| XP_033516221.1 | N. tomentosiformis | 282 | 32,332.88 | 5.15 | 36.11 | 89.54 | −0.307 | cyto |
| XP_009774098.1 | N. sylvestris | 363 | 41,302.91 | 4.93 | 41.17 | 88.35 | −0.34 | cyto |
| XP_009774100.1 | N. sylvestris | 363 | 41,302.91 | 4.93 | 41.17 | 88.35 | −0.34 | cyto |
| XP_009774101.1 | N. sylvestris | 363 | 41,302.91 | 4.93 | 41.17 | 88.35 | −0.34 | cyto |
| XP_009774102.1 | N. sylvestris | 363 | 41,302.91 | 4.93 | 41.17 | 88.35 | −0.34 | cyto |
| XP_009774903.1 | N. sylvestris | 213 | 24,636.79 | 5.04 | 59.18 | 78.69 | −0.57 | cyto |
| XP_009776074.1 | N. sylvestris | 356 | 41,082.54 | 5.64 | 56.12 | 79.16 | −0.595 | cysk |
| XP_009777005.1 | N. sylvestris | 354 | 40,337.96 | 4.72 | 42.46 | 88.64 | −0.331 | cysk |
| XP_009786561.1 | N. sylvestris | 362 | 40,964.52 | 4.86 | 35.03 | 88.34 | −0.295 | cyto |
| XP_009786562.1 | N. sylvestris | 362 | 40,964.52 | 4.86 | 35.03 | 88.34 | −0.295 | cyto |
| XP_009786563.1 | N. sylvestris | 362 | 40,964.52 | 4.86 | 35.03 | 88.34 | −0.295 | cyto |
| XP_009804586.1 | N. sylvestris | 354 | 40,890.52 | 5.7 | 51.45 | 81.24 | −0.515 | cysk |
| XP_009804587.1 | N. sylvestris | 354 | 40,890.52 | 5.7 | 51.45 | 81.24 | −0.515 | cysk |
| XP_016440803.1 | N. tabacum | 353 | 40,789.42 | 5.78 | 51.89 | 81.47 | −0.508 | cysk |
| XP_016443905.1 | N. tabacum | 363 | 41,302.91 | 4.93 | 41.17 | 88.35 | −0.34 | cyto |
| XP_016443906.1 | N. tabacum | 363 | 41,302.91 | 4.93 | 41.17 | 88.35 | −0.34 | cyto |
| XP_016443907.1 | N. tabacum | 363 | 41,302.91 | 4.93 | 41.17 | 88.35 | −0.34 | cyto |
| XP_016443908.1 | N. tabacum | 363 | 41,302.91 | 4.93 | 41.17 | 88.35 | −0.34 | cyto |
| XP_016468367.1 | N. tabacum | 344 | 39,333.79 | 4.92 | 41.12 | 90.12 | −0.315 | cysk |
| XP_016468369.1 | N. tabacum | 315 | 35,951.04 | 4.85 | 46.89 | 94.1 | −0.225 | cyto |
| XP_016469069.1 | N. tabacum | 356 | 41,082.54 | 5.64 | 56.12 | 79.16 | −0.595 | cysk |
| XP_016469733.1 | N. tabacum | 354 | 40,337.96 | 4.72 | 42.46 | 88.64 | −0.331 | cysk |
| XP_016487598.1 | N. tabacum | 362 | 41,060.55 | 4.86 | 35.75 | 89.14 | −0.318 | cyto |
| XP_016487599.1 | N. tabacum | 291 | 32,941.69 | 5.87 | 36.42 | 89.42 | −0.234 | cyto |
| XP_016490562.1 | N. tabacum | 336 | 38,722.13 | 5.75 | 52.67 | 81.85 | −0.49 | cysk |
| XP_016499197.1 | N. tabacum | 355 | 40,839.36 | 5.79 | 52.39 | 79.38 | −0.575 | cysk |
| XP_016501298.1 | N. tabacum | 282 | 32,366.89 | 5.15 | 35.25 | 88.51 | −0.312 | cyto |
| XP_016506694.1 | N. tabacum | 362 | 40,964.52 | 4.86 | 35.03 | 88.34 | −0.295 | cyto |
| XP_016506695.1 | N. tabacum | 362 | 40,964.52 | 4.86 | 35.03 | 88.34 | −0.295 | cyto |
| XP_016506696.1 | N. tabacum | 362 | 40,964.52 | 4.86 | 35.03 | 88.34 | −0.295 | cyto |
| XP_016510127.1 | N. tabacum | 354 | 40,337.96 | 4.72 | 42.46 | 88.64 | −0.331 | cysk |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, Y.; Zhang, Y.; Hu, Z.; Yan, X.; Hu, R.; Fan, J. Genome-Wide Identification and Evolutionary Analysis of the SnRK2 Gene Family in Nicotiana Species. Agriculture 2025, 15, 1396. https://doi.org/10.3390/agriculture15131396
Tang Y, Zhang Y, Hu Z, Yan X, Hu R, Fan J. Genome-Wide Identification and Evolutionary Analysis of the SnRK2 Gene Family in Nicotiana Species. Agriculture. 2025; 15(13):1396. https://doi.org/10.3390/agriculture15131396
Chicago/Turabian StyleTang, Yu, Yangxin Zhang, Zhengrong Hu, Xuebing Yan, Risheng Hu, and Jibiao Fan. 2025. "Genome-Wide Identification and Evolutionary Analysis of the SnRK2 Gene Family in Nicotiana Species" Agriculture 15, no. 13: 1396. https://doi.org/10.3390/agriculture15131396
APA StyleTang, Y., Zhang, Y., Hu, Z., Yan, X., Hu, R., & Fan, J. (2025). Genome-Wide Identification and Evolutionary Analysis of the SnRK2 Gene Family in Nicotiana Species. Agriculture, 15(13), 1396. https://doi.org/10.3390/agriculture15131396






