Introducing Legumes into Wheat–Maize Rotation Complicates Soil Microbial Co-Occurrence Network and Reduces Soil Allelochemicals in Succeeding Wheat Season
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Site and Design
2.2. Soil Sampling
2.3. Soil Physical–Chemical Analysis
2.4. Soil DNA Extraction and Quantitative PCR Analysis
2.5. Amplicon High-Throughput Sequencing
2.6. Soil Metabolite Extraction and Untargeted Analysis
2.7. Statistical Analysis
3. Results
3.1. Soil Physical–Chemical Properties
3.2. Abundance and Alpha Diversity of Soil Microbial Community
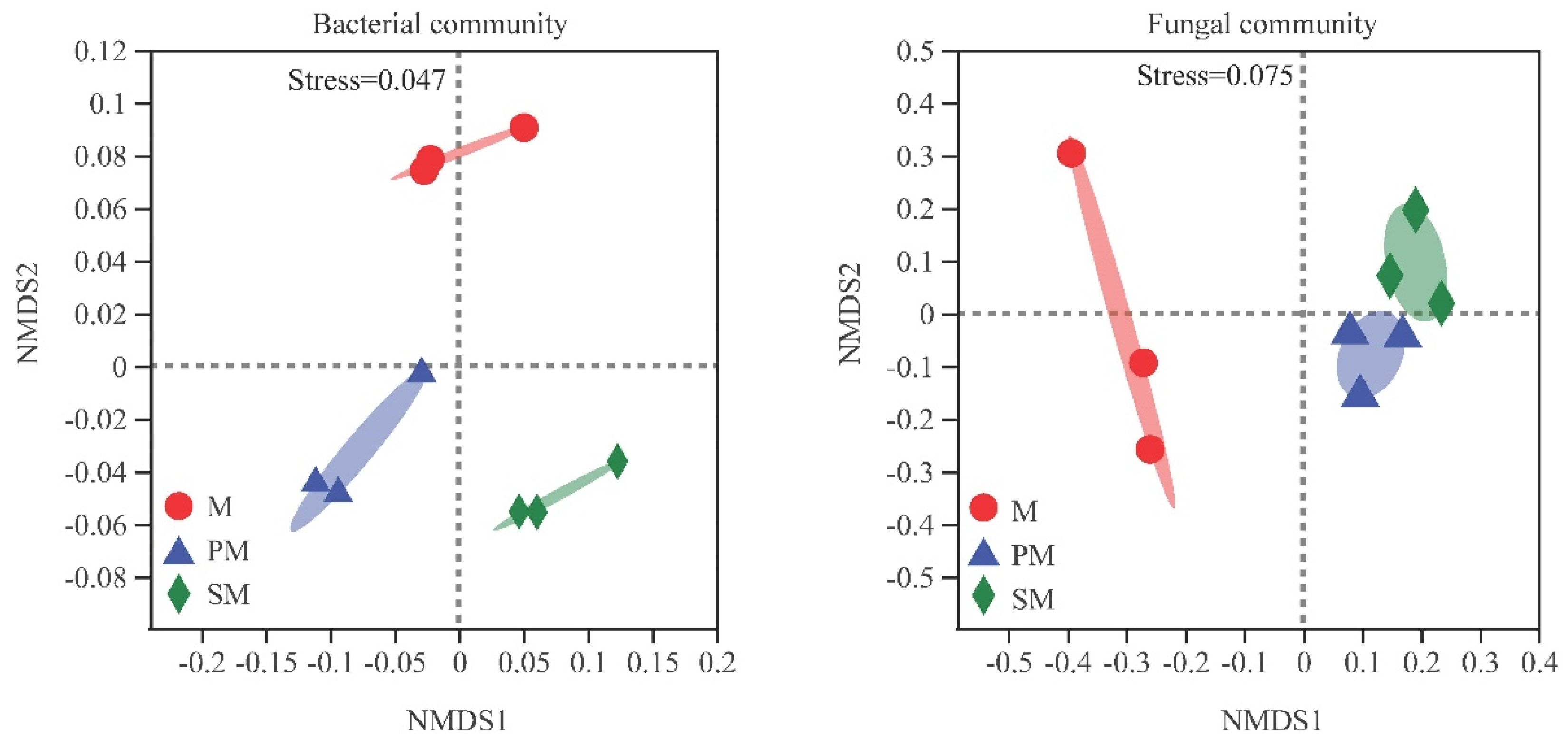
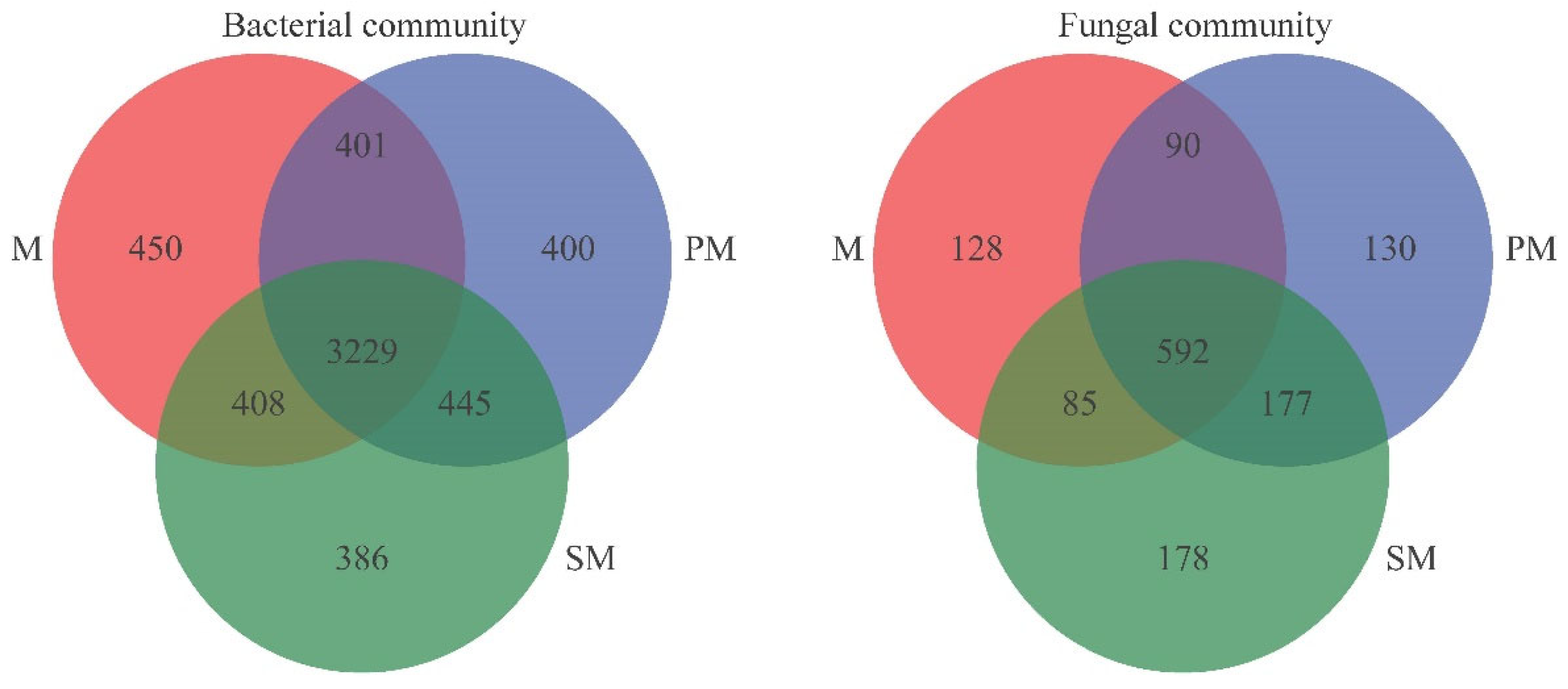
3.3. Beta Diversity and Composition of Soil Microbial Community
3.4. Co-Occurrence Network of Soil Microbial Community
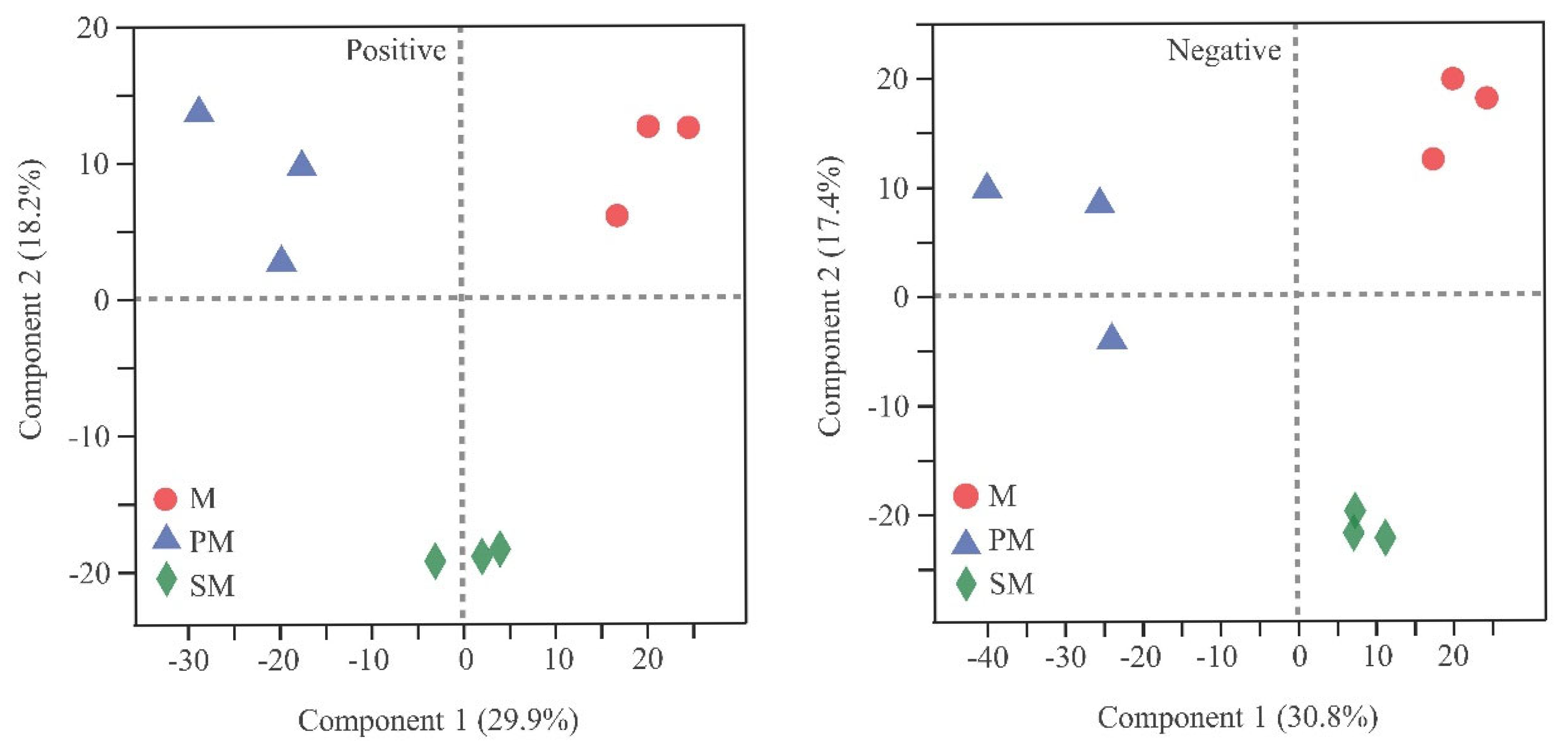
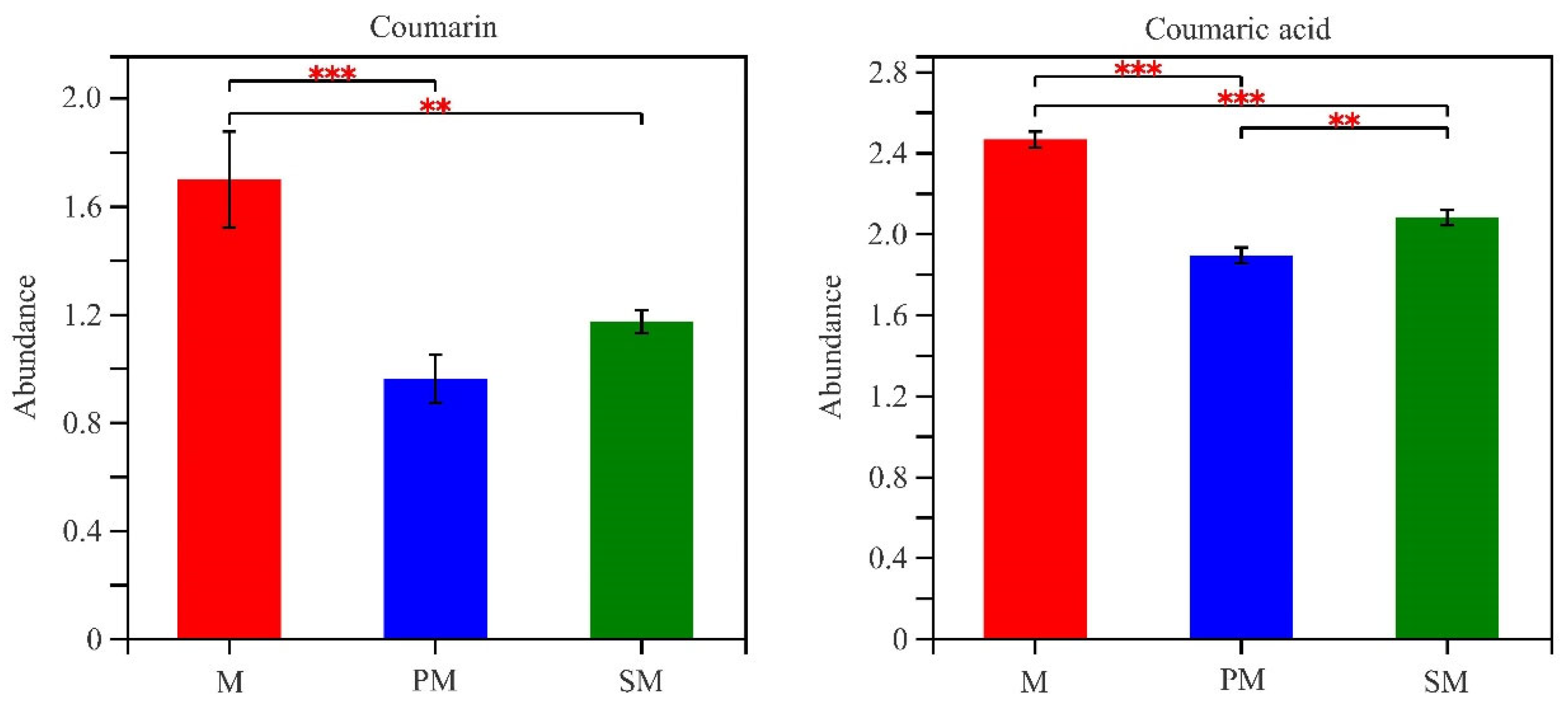
3.5. Soil Metabolites
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Cook, R.J. Toward cropping systems that enhance productivity and sustainability. Proc. Natl. Acad. Sci. USA 2006, 103, 18389–18394. [Google Scholar] [CrossRef] [PubMed]
- Gaudin, A.C.M.; Janovicek, K.; Deen, B.; Hooker, D.C. Wheat improves nitrogen use efficiency of maize and soybean-based cropping systems. Agric. Ecosyst. Environ. 2015, 210, 1–10. [Google Scholar] [CrossRef]
- McDaniel, M.D.; Grandy, A.S.; Tiemann, L.K.; Weintraub, M.N. Crop rotation complexity regulates the decomposition of high and low quality residues. Soil Biol. Biochem. 2014, 78, 243–254. [Google Scholar] [CrossRef]
- Li, X.G.; Ding, C.F.; Zhang, T.L.; Wang, X.X. Fungal pathogen accumulation at the expense of plant-beneficial fungi as a consequence of consecutive peanut monoculturing. Soil Biol. Biochem. 2014, 72, 11–18. [Google Scholar] [CrossRef]
- Drury, C.F.; Yang, X.M.; Reynolds, W.D.; McLaughlin, N.B. Nitrous oxide and carbon dioxide emissions from monoculture and rotational cropping of corn, soybean and winter wheat. Can. J. Soil. Sci. 2008, 88, 163–174. [Google Scholar] [CrossRef]
- Xiao, H.; van Es, H.M.; Amsili, J.P.; Shi, Q.Q.; Sun, J.B.; Chen, Y.Q.; Sui, P. Lowering soil greenhouse gas emissions without sacrificing yields by increasing crop rotation diversity in the North China Plain. Field Crops Res. 2022, 276, 108366. [Google Scholar] [CrossRef]
- Grover, K.K.; Karsten, H.D.; Roth, G.W. Corn grain yields and yield stability in four long-term cropping systems. Agron. J. 2009, 101, 940–946. [Google Scholar] [CrossRef]
- Mao, L.; Liu, Y.J.; Zhang, J.J.; Okerblad, J.; Chen, S.Y.; Johnson, N.C. Soil biota suppress maize growth and influence root traits under continuous monoculture. Plant Soil 2021, 461, 441–455. [Google Scholar] [CrossRef]
- Tiemann, L.K.; Grandy, A.S.; Atkinson, E.E.; Marin-Spiotta, E.; McDaniel, M.D. Crop rotational diversity enhances belowground communities and functions in an agroecosystem. Ecol. Lett. 2015, 18, 761–771. [Google Scholar] [CrossRef]
- Yang, T.; Lupwayi, N.; Marc, S.; Siddique, K.H.M.; Bainard, L.D. Anthropogenic drivers of soil microbial communities and impacts on soil biological functions in agroecosystems. Glob. Ecol. Conserv. 2021, 27, e01521. [Google Scholar] [CrossRef]
- Dias, T.; Dukes, A.; Antunes, P.M. Accounting for soil biotic effects on soil health and crop productivity in the design of crop rotations. J. Sci. Food Agric. 2015, 95, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.L.; Steenhuis, T.S.; Davis, K.F.; Werf, W.; Ritsema, C.J.; Pacenka, S.; Zhang, F.S.; Siddique, K.H.M.; Du, T.S. Diversified crop rotations enhance groundwater and economic sustainability of food production. Food Energy Secur. 2021, 10, e311. [Google Scholar] [CrossRef]
- Marini, L.; St-Martin, A.; Vico, G.; Baldoni, G.; Berti, A.; Blecharczyk, A.; Malecka-Jankowiak, I.; Morari, F.; Sawinska, Z.; Bommarco, R. Crop rotations sustain cereal yields under a changing climate. Environ. Res. Lett. 2020, 15, 124011. [Google Scholar] [CrossRef]
- Renard, D.; Tilman, D. National food production stabilized by crop diversity. Nature 2019, 571, 257–260. [Google Scholar] [CrossRef]
- Andert, S.; Burger, J.; Stein, S.; Gerowitt, B. The influence of crop sequence on fungicide and herbicide use intensities in North German arable farming. Eur. J. Agron. 2016, 77, 81–89. [Google Scholar] [CrossRef]
- Gossen, B.D.; McDonald, M.R. New technologies could enhance natural biological control and disease management and reduce reliance on synthetic pesticides. Can. J. Plant Pathol. 2020, 42, 30–40. [Google Scholar] [CrossRef]
- Snapp, S.S.; Blackie, M.J.; Gilbert, R.A.; Bezner-Kerr, R.; Kanyama-Phiri, G.Y. Biodiversity can support a greener revolution in Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 20840–20845. [Google Scholar] [CrossRef]
- Yu, Y.L.; Xue, L.H.; Yang, L.Z. Winter legumes in rice crop rotations reduces nitrogen loss, and improves rice yield and soil nitrogen supply. Agron. Sustain. Dev. 2014, 34, 633–640. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, S.Q.; Xing, G.X. Maintaining rice yield and reducing N pollution by substituting winter legume for wheat in a heavily-fertilized rice-based cropping system of southeast China. Agric. Ecosyst. Environ. 2015, 202, 79–89. [Google Scholar] [CrossRef]
- Reckling, M.; Bergkvist, G.; Watson, C.A.; Stoddard, F.L.; Zander, P.M.; Walker, R.L.; Pristeri, A.; Toncea, I.; Bachinger, J. Trade-offs between economic and environmental impacts of introducing legumes into cropping systems. Front. Plant Sci. 2016, 7, 669. [Google Scholar] [CrossRef]
- Sokol, N.W.; Slessarev, E.; Marschmann, G.L.; Nicolas, A.; Blazewicz, S.J.; Brodie, E.L.; Firestone, M.K.; Foley, M.M.; Hestrin, R.; Hungate, B.A.; et al. Life and death in the soil microbiome: How ecological processes influence biogeochemistry. Nat. Rev. Microbiol. 2022, 20, 415–430. [Google Scholar] [CrossRef] [PubMed]
- Compant, S.; Samad, A.; Faist, H.; Sessitsch, A. A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Adv. Res. 2019, 19, 29–37. [Google Scholar] [CrossRef] [PubMed]
- van der Heijden, M.G.A.; Bardgett, R.D.; van Straalen, N.M. The unseen majority: Soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 2008, 11, 296–310. [Google Scholar] [CrossRef] [PubMed]
- Espana, M.; Rasche, F.; Kandeler, E.; Brune, T.; Rodriguez, B.; Bending, G.D.; Cadisch, G. Assessing the effect of organic residue quality on active decomposing fungi in a tropical Vertisol using 15N-DNA stable isotope probing. Fungal Ecol. 2011, 4, 115–119. [Google Scholar] [CrossRef]
- Eilers, K.G.; Lauber, C.L.; Knight, R.; Fierer, N. Shifts in bacterial community structure associated with inputs of low molecular weight carbon compounds to soil. Soil Biol. Biochem. 2010, 42, 896–903. [Google Scholar] [CrossRef]
- Jiang, Y.J.; Liang, Y.T.; Li, C.M.; Wang, F.; Sui, Y.Y.; Suvannang, N.; Zhou, J.Z.; Sun, B. Crop rotations alter bacterial and fungal diversity in paddy soils across East Asia. Soil Biol. Biochem. 2016, 95, 250–261. [Google Scholar] [CrossRef]
- Wang, G.Z.; Bei, S.K.; Li, J.P.; Bao, X.G.; Zhang, J.D.; Schultz, P.A.; Li, H.G.; Li, L.; Zhang, F.S.; Bever, J.D.; et al. Soil microbial legacy drives crop diversity advantage: Linking ecological plant-soil feedback with agricultural intercropping. J. Appl. Ecol. 2021, 58, 496–506. [Google Scholar] [CrossRef]
- De Corato, U.; Patruno, L.; Avella, N.; Salimbeni, R.; Lacolla, G.; Cucci, G.; Crecchio, C. Soil management under tomato -wheat rotation increases the suppressive response against Fusarium wilt and tomato shoot growth by changing the microbial composition and chemical parameters. Appl. Soil Ecol. 2020, 154, 103601. [Google Scholar] [CrossRef]
- Nayyar, A.; Hamel, C.; Lafond, G.; Gossen, B.D.; Hanson, K.; Germida, J. Soil microbial quality associated with yield reduction in continuous-pea. Appl. Soil Ecol. 2009, 43, 115–121. [Google Scholar] [CrossRef]
- Fu, X.; Wang, J.; Sainju, U.M.; Liu, W.Z. Soil carbon fractions in response to long-term crop rotations in the Loess Plateau of China. Soil Sci. Soc. Am. J. 2017, 81, 503–513. [Google Scholar] [CrossRef]
- D’Acunto, L.; Andrade, J.F.; Poggio, S.L.; Semmartin, M. Diversifying crop rotation increased metabolic soil diversity and activity of the microbial community. Agric. Ecosyst. Environ. 2018, 257, 159–164. [Google Scholar] [CrossRef]
- Venter, Z.S.; Jacobs, K.; Hawkins, H.J. The impact of crop rotation on soil microbial diversity: A meta-analysis. Pedobiologia 2016, 59, 215–223. [Google Scholar] [CrossRef]
- Liu, H.; Pan, F.J.; Han, X.Z.; Song, F.B.; Zhang, Z.M.; Yan, J.; Xu, Y.L. A comprehensive analysis of the response of the fungal community structure to long-term continuous cropping in three typical upland crops. J. Integr. Agric. 2020, 19, 866–880. [Google Scholar] [CrossRef]
- Zhou, X.G.; Liu, J.; Wu, F.Z. Soil microbial communities in cucumber monoculture and rotation systems and their feedback effects on cucumber seedling growth. Plant Soil 2017, 415, 507–520. [Google Scholar] [CrossRef]
- Lu, S.; He, Y.H.; Chen, Y.Q.; Chen, L.J.; Wang, Z.Y.; Yuan, J.; Wu, L.C. Co-analysis of rhizosphere metabolomics and bacterial community structures to unfold soil ecosystem health in Camellia oleifera land under long-term cultivation. Appl. Soil Ecol. 2022, 171, 104336. [Google Scholar] [CrossRef]
- Withers, E.; Hill, P.W.; Chadwick, D.R.; Jones, D.L. Use of untargeted metabolomics for assessing soil quality and microbial function. Soil Biol. Biochem. 2020, 143, 107758. [Google Scholar] [CrossRef]
- Li, X.N.; Song, Y.; Bian, Y.R.; Gu, C.G.; Yang, X.L.; Wang, F.; Jiang, X. Insights into the mechanisms underlying efficient Rhizodegradation of PAHs in biochar-amended soil: From microbial communities to soil metabolomics. Environ. Int. 2020, 144, 105995. [Google Scholar] [CrossRef]
- Cheng, H.Y.; Yuan, M.S.; Tang, L.; Shen, Y.F.; Yu, Q.; Li, S.Q. Integrated microbiology and metabolomics analysis reveal responses of soil microorganisms and metabolic functions to phosphorus fertilizer on semiarid farm. Sci. Total Environ. 2022, 817, 152878. [Google Scholar] [CrossRef]
- Fan, D.M.; Zhao, Z.M.; Wang, Y.; Ma, J.H.; Wang, X.C. Crop-type-driven changes in polyphenols regulate soil nutrient availability and soil microbiota. Front. Microbiol. 2022, 13, 964039. [Google Scholar] [CrossRef]
- Zhang, J.X.; Zhou, D.P.; Yuan, X.Q.; Xu, Y.H.; Chen, C.B.; Zhao, L. Soil microbiome and metabolome analysis reveals beneficial effects of ginseng-celandine rotation on the rhizosphere soil of ginseng-used fields. Rhizosphere 2022, 23, 100559. [Google Scholar] [CrossRef]
- Soil Survey Staff. Keys to Soil Taxonomy, 12th ed.; USDA—Natural Resources Conservation Service: Washington, DC, USA, 2014.
- Lu, R.K. Method of Agricultural Chemistry and Soil; China Agricultural Science and Technology Press: Beijing, China, 2000. [Google Scholar]
- Sun, Y.; Shi, Y.; Tang, Y.; Tian, J.; Wu, X. Correlation between Plant Diversity and the Physicochemical Properties of Soil Microbes. Appl. Ecol. Env. Res. 2019, 17, 10371–10388. [Google Scholar] [CrossRef]
- Song, H.J.; Peng, L.; Li, Z.Y.; Deng, X.Z.; Shao, J.H.; Gu, J.D. Metal distribution and biological diversity of crusts in paddy fields polluted with different levels of cadmium. Ecotoxicol. Environ. Saf. 2019, 184, 109620. [Google Scholar] [CrossRef] [PubMed]
- Gade, L.; Scheel, C.M.; Pham, C.D.; Lindsley, M.D.; Iqbal, N.; Cleveland, A.A.; Whitney, A.M.; Lockhart, S.R.; Brandt, M.E.; Litvintseva, A.P. Detection of fungal DNA in human body fluids and tissues during a multistate outbreak of fungal meningitis and other infections. Eukaryot. Cell 2013, 12, 677–683. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Ren, Y.; Yu, G.; Shi, C.; Liu, L.; Guo, Q.; Han, C.; Zhang, D.; Zhang, L.; Liu, B.; Gao, H. Majorbio Cloud: A one-stop, comprehensive bioinformatic platform for multiomics analyses. iMeta 2022, 1, e12. [Google Scholar] [CrossRef]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.R.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glöckner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, R.H.; Larsson, K.-H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019, 47, 259–264. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Ma, F.; Wang, Y.Z.; Quan, H.Y.; Wang, Z.P.; Zhao, C.Y.; Li, X.; Liang, B.R.; Zhang, H.; Hao, L.Y.; Zhu, T. Exploring the humification process of municipal sludge in hyperthermophilic composting through metagenomic and untargeted metabolomic. Bioresour. Technol. 2023, 387, 129575. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D. Reintroducing mothur: 10 years later. Appl. Environ. Microbiol. 2020, 86, e02343-19. [Google Scholar] [CrossRef]
- Good, I.J. The Population Frequencies of Species and the Estimation of Population Parameters. Biometrika 1953, 40, 237–264. [Google Scholar] [CrossRef]
- Faust, K.; Raes, J. CoNet app: Inference of biological association networks using Cytoscape. F1000Res 2016, 5, 1519. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Assenov, Y.; Ramírez, F.; Schelhorn, S.-E.; Lengauer, T.; Albrecht, M. Computing topological parameters of biological networks. Bioinformatics 2008, 24, 282–284. [Google Scholar] [CrossRef]
- Butterly, C.R.; Baldock, J.A.; Tang, C. The contribution of crop residues to changes in soil pH under field conditions. Plant Soil 2013, 366, 185–198. [Google Scholar] [CrossRef]
- Smagacz, J.; Koziel, M.; Martyniuk, S. Soil properties and yields of winter wheat after long-term growing of this crop in two contrasting rotations. Plant Soil Environ. 2016, 62, 566–570. [Google Scholar] [CrossRef]
- Geisseler, D.; Scow, K.M. Long-term effects of mineral fertilizers on soil microorganisms—A review. Soil Biol. Biochem. 2014, 75, 54–63. [Google Scholar] [CrossRef]
- Neugschwandtner, R.W.; Száková, J.; Pachtrog, V.; Tlustos, P.; Cerny, J.; Kulhánek, M.; Kaul, H.P.; Euteneuer, P.; Moitzi, G.; Wagentristl, H. Basic soil chemical properties after 15 years in a long-term tillage and crop rotation experiment. Int. Agrophys 2020, 34, 133–140. [Google Scholar] [CrossRef]
- Nyambo, P.; Thengeni, B.; Cornelius, C.; Araya, T. Tillage, crop rotation, residue management and biochar influence on soil chemical and biological properties. South Afr. J. Plant Soil 2021, 38, 390–397. [Google Scholar] [CrossRef]
- Liu, R.; Liu, Y.; Gao, Y.; Zhao, F.Z.; Wang, J. The Nitrogen Cycling Key Functional Genes and Related Microbial Bacterial Community α-Diversity Is Determined by Crop Rotation Plans in the Loess Plateau. Agronomy 2023, 13, 1769. [Google Scholar] [CrossRef]
- Wang, H.; Chen, J.P.; Ruan, Y.H.; Sun, W.; Wang, S.L.; Wang, H.T.; Zhang, Y.L.; Guo, J.M.; Wang, Y.C.; Guo, H.Y.; et al. Metagenomes reveal the effect of crop rotation systems on phosphorus cycling functional genes and soil phosphorus avail-ability. Agr. Ecosyst. Environ. 2024, 364, 108886. [Google Scholar] [CrossRef]
- Xia, M.M.; Ma, X.L.; Liu, J.; Wu, M.; Li, Z.P.; Liu, M. Potential effect of key soil bacterial taxa on the increase of rice yield under milk vetch rotation. Front. Microbiol. 2023, 14, 1150505. [Google Scholar] [CrossRef]
- Hassan, H.M.; Marschner, P.; McNeill, A.; Tang, C.X. Grain legume pre-crops and their residues affect the growth, P uptake and size of P pools in the rhizosphere of the following wheat. Biol. Fert. Soils 2012, 48, 775–785. [Google Scholar] [CrossRef]
- Rose, T.J.; Wood, R.H.; Gleeson, D.B.; Rose, M.T.; Van Zwieten, L. Removal of phosphorus in residues of legume or cereal plants determines growth of subsequently planted wheat in a high phosphorus fixing soil. Biol. Fert. Soils 2016, 52, 1085–1092. [Google Scholar] [CrossRef]
- Batterman, S.A.; Wurzburger, N.; Hedin, L.O. Nitrogen and phosphorus interact to control tropical symbiotic N fixation: A test in. J. Ecol. 2013, 101, 1400–1408. [Google Scholar] [CrossRef]
- Zheng, M.H.; Chen, H.; Li, D.J.; Zhu, X.M.; Zhang, W.; Fu, S.L.; Mo, J.M. Biological nitrogen fixation and its response to nitrogen input in two mature tropical plantations with and without legume trees. Biol. Fert. Soils 2016, 52, 665–674. [Google Scholar] [CrossRef]
- Samaddar, S.; Schmidt, R.; Tautges, N.E.; Scow, K. Adding alfalfa to an annual crop rotation shifts the composition and functional responses of tomato rhizosphere microbial communities. Appl. Soil Ecol. 2021, 167, 104102. [Google Scholar] [CrossRef]
- Ma, L.Y.; Zhang, J.J.; Li, H.; Xu, M.W.; Zhao, Y.G.; Shi, X.Y.; Shi, Y.; Wan, S.Q. Key microbes in wheat maize rotation present better promoting wheat yield effect in a variety of crop rotation systems. Agric. Ecosyst. Environ. 2025, 379, 109370. [Google Scholar] [CrossRef]
- Wang, Q.; Liang, A.Z.; Chen, X.W.; Zhang, S.X.; Zhang, Y.; McLaughlin, N.B.; Gao, Y.; Jia, S.X. The impact of cropping system, tillage and season on shaping soil fungal community in a long-term field trial. Eur. J. Soil Biol. 2021, 102, 103253. [Google Scholar] [CrossRef]
- Wallander, H.; Nilsson, L.O.; Hagerberg, D.; Rosengren, U. Direct estimates of C:N ratios of ectomycorrhizal mycelia collected from Norway spruce forest soils. Soil Biol. Biochem. 2003, 35, 997–999. [Google Scholar] [CrossRef]
- de Boer, W.; Folman, L.B.; Summerbell, R.C.; Boddy, L. Living in a fungal world: Impact of fungi on soil bacterial niche development. FEMS Microbiol. Rev. 2005, 29, 795–811. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, X.; Zhang, L.; Zeng, L.; Liu, Y.; Wang, X.; He, P.; Li, S.; Liang, G.; Zhou, W.; et al. The stronger impact of inorganic nitrogen fertilization on soil bacterial community than organic fertilization in short-term condition. Geoderma 2021, 382, 114752. [Google Scholar] [CrossRef]
- Hu, X.J.; Liu, J.J.; Yu, Z.H.; Yao, Q.; Zhang, W.; Mi, G.; Liang, A.Z.; Li, L.J.; Chen, X.L.; Jin, J.; et al. Continuous cropping of soybean induced a more fluctuating fungal network and intensive pathogenic fungal interactions in a Mollisol of Northeast China. Soil Sci. Soc. Am. J. 2020, 84, 775–783. [Google Scholar] [CrossRef]
- Hu, X.J.; Liu, J.J.; Wang, X.Z.; Yu, Z.H.; Yao, Q.; Jin, J.; Liu, X.B.; Wang, G.H. Dramatic changes in bacterial co-occurrence patterns and keystone taxa responses to cropping systems in Mollisols of Northeast China. Arch. Agron. Soil Sci. 2021, 67, 426–434. [Google Scholar] [CrossRef]
- Yang, T.; Evans, B.; Bainard, L.D. Pulse frequency in crop rotations alters soil microbial community networks and the relative abundance of fungal plant pathogens. Front. Microbiol. 2021, 12, 667394. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Li, W.X.; Yang, Y.H.; Ye, F.; Lu, H.Y.; Chen, X.Y.; Chen, F.; Wen, X.Y. The impact of different rotation regime on the soil bacterial and fungal communities in an intensively managed agricultural region. Arch. Microbiol. 2022, 204, 142. [Google Scholar] [CrossRef] [PubMed]
- Ye, G.P.; Banerjee, S.; He, J.Z.; Fan, J.B.; Wang, Z.H.; Wei, X.Y.; Hu, H.W.; Zheng, Y.; Duan, C.J.; Wan, S.; et al. Manure application increases microbiome complexity in soil aggregate fractions: Results of an 18-year field experiment. Agric. Ecosyst. Environ. 2021, 307, 107249. [Google Scholar] [CrossRef]
- White, R.A.; Rivas-Ubach, A.; Borkum, M.I.; Koberl, M.; Bilbao, A.; Colby, S.M.; Hoyt, D.W.; Bingol, K.; Kim, Y.M.; Wendler, J.P.; et al. The state of rhizospheric science in the era of multi-omics: A practical guide to omics technologies. Rhizosphere 2017, 3, 212–221. [Google Scholar] [CrossRef]
- Song, Y.; Li, X.N.; Yao, S.; Yang, X.L.; Jiang, X. Correlations between soil metabolomics and bacterial community structures in the pepper rhizosphere under plastic greenhouse cultivation. Sci. Total Environ. 2020, 728, 138439. [Google Scholar] [CrossRef]
- Mamude, C.; Asfaw, Z. Allelopathic effects of Oldeania alpina (K. Schum.) Stapleton leaf aqueous extract on seed germination and initial seedling growth of two selected crops. Adv. Bamboo Sci. 2023, 4, 100034. [Google Scholar] [CrossRef]
- Niro, E.; Marzaioli, R.; De Crescenzo, S.; D’Abrosca, B.; Castaldi, S.; Esposito, A.; Fiorentino, A.; Rutigliano, F.A. Effects of the allelochemical coumarin on plants and soil microbial community. Soil Biol. Biochem. 2016, 95, 30–39. [Google Scholar] [CrossRef]
- Gnonlonfin, G.J.B.; Sanni, A.; Brimer, L. Review scopoletin -a coumarin phytoalexin with medicinal properties. Crit. Rev. Plant Sci. 2012, 31, 47–56. [Google Scholar] [CrossRef]
- Voges, M.J.E.E.E.; Bai, Y.; Schulze-Lefert, P.; Sattely, E.S. Plant-derived coumarins shape the composition of an Arabidopsis synthetic root microbiome. Proc. Natl. Acad. Sci. USA 2019, 116, 12558–12565. [Google Scholar] [CrossRef]
- Harbort, C.J.; Hashimoto, M.; Inoue, H.; Niu, Y.L.; Guan, R.; Rombola, A.D.; Kopriva, S.; Voges, M.J.E.E.E.; Sattely, E.S.; Garrido-Oter, R.; et al. Root-secreted coumarins and the microbiota interact to improve iron nutrition in Arabidopsis. Cell Host Microbe 2020, 28, 825–837. [Google Scholar] [CrossRef]
- Yang, Y.H.; Xu, J.; Li, Y.; He, Y.C.; Yang, Y.Q.; Liu, D.L.; Wu, C.X. Effects of Coumarin on Rhizosphere Microbiome and Metabolome of. Plants 2023, 12, 1096. [Google Scholar] [CrossRef] [PubMed]
- Stringlis, I.A.; Yu, K.; Feussner, K.; de Jonge, R.; van Bentum, S.; Van Verk, M.C.; Berendsen, R.L.; Bakker, P.A.H.M.; Feussner, I.; Pieterse, C.M.J. MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc. Natl. Acad. Sci. USA 2018, 115, E5213–E5222. [Google Scholar] [CrossRef]
- Vismans, G.; van Bentum, S.; Spooren, J.; Song, Y.; Goossens, P.; Valls, J.; Snoek, B.L.; Thiombiano, B.; Schilder, M.; Dong, L.M.; et al. Coumarin biosynthesis genes are required after foliar pathogen infection for the creation of a microbial soil-borne legacy that primes plants for SA-dependent defenses. Sci. Rep. 2022, 12, 22473. [Google Scholar] [CrossRef]
- Kumar, G.A.; Kumar, S.; Bhardwaj, R.; Swapnil, P.; Meena, M.; Seth, C.S.; Yadav, A. Recent advancements in multifaceted roles of flavonoids in plant-rhizomicrobiome interactions. Front. Plant Sci. 2024, 14, 1297706. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, J.; Schmidt, S.; Chutia, R.; Muller, J.; Bottcher, C.; Strehmel, N.; Scheel, D.; Abel, S. Non-targeted profiling of semi-polar metabolites in Arabidopsis root exudates uncovers a role for coumarin secretion and lignification during the local response to phosphate limitation. J. Exp. Bot. 2016, 67, 1421–1432. [Google Scholar] [CrossRef]
- Rosenkranz, T.; Oburger, E.; Baune, M.; Weber, G.; Puschenreiter, M. Root exudation of coumarins from soil-grown Arabidopsis thaliana in response to iron deficiency. Rhizosphere 2021, 17, 100296. [Google Scholar] [CrossRef]
- Ran, L.Y.; Li, J.F.; Xing, Y.Y.; Zhang, J.Y.; Zhou, X.G. Effects of p-coumaric acid on the structure and abundance of soil Pseudomonas spp. community. Allelopath. J. 2021, 53, 211–218. [Google Scholar] [CrossRef]
- Jin, X.; Shi, Y.J.; Tan, S.C.; Ma, C.L.; Wu, F.Z.; Pan, K.; Zhou, X.G. Effects of cucumber root exudates components on soil bacterial community structure and abundance. Allelopath. J. 2019, 48, 167–174. [Google Scholar] [CrossRef]
- Zhou, X.G.; Wu, F.Z. p-Coumaric acid influenced cucumber rhizosphere soil microbial communities and the growth of Fusarium oxysporum f.sp. cucumerinum Owen. PLoS ONE 2012, 7, e48288. [Google Scholar] [CrossRef]
- Zhou, X.G.; Zhang, J.H.; Pan, D.D.; Ge, X.; Jin, X.; Chen, S.C.; Wu, F.Z. p-Coumaric acid can alter the composition of cucumber rhizosphere microbial communities and induce negative plant-microbial interactions. Biol. Fert. Soils 2018, 54, 363–372. [Google Scholar] [CrossRef]
- Wang, R.; Liu, J.; Jiang, W.; Ji, P.; Li, Y. Metabolomics and microbiomics reveal impacts of rhizosphere metabolites on alfalfa continuous cropping. Front. Microbiol. 2022, 13, 833968. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.Y.; Yue, J.Q.; Yan, Y.Q.; Zhang, D.Q.; Yang, C.; LI, X.D.; Shao, Y.H.; Fang, B.T. Soil bacterial communities of different crop rotations and yield of succeeding wheat. Chin. J. Appl. Ecol. 2022, 33, 2954–2962. [Google Scholar]

| 2015–2016 | 2016–2017 | 2017–2018 | 2018–2019 | 2019–2020 | 2020–2021 | |
|---|---|---|---|---|---|---|
| M | Maize–Wheat | Maize–Wheat | Maize–Wheat | Maize–Wheat | Maize–Wheat | Maize–Wheat |
| PM | Peanut–Wheat | Maize–Wheat | Peanut–Wheat | Maize–Wheat | Peanut–Wheat | Peanut–Wheat |
| SM | Soybean–Wheat | Maize–Wheat | Soybean–Wheat | Maize–Wheat | Soybean–Wheat | Soybean–Wheat |
| pH | SOC (g kg−1) | TN (g kg−1) | TP (g kg−1) | TK (g kg−1) | Nmin (mg kg−1) | AP (mg kg−1) | AK (mg kg−1) | |
|---|---|---|---|---|---|---|---|---|
| M | 8.54 ± 0.02 a | 5.77 ± 0.25 a | 0.375 ± 0.005 a | 0.780 ± 0.017 b | 18.69 ± 0.10 a | 69.72 ± 2.50 ab | 8.91 ± 1.06 b | 165.67 ± 7.55 a |
| PM | 8.51 ± 0.01 a | 4.82 ± 0.28 b | 0.342 ± 0.016 b | 0.818 ± 0.013 a | 18.70 ± 0.29 a | 67.66 ± 3.17 b | 11.36 ± 0.77 a | 116.67 ± 10.35 b |
| SM | 8.51 ± 0.02 a | 5.32 ± 0.12 a | 0.357 ± 0.007 ab | 0.839 ± 0.012 a | 18.82 ± 0.16 a | 73.02 ± 2.15 a | 10.68 ± 0.81 ab | 119.35 ± 2.95 b |
| Bacterial Community | Fungal Community | |||||
|---|---|---|---|---|---|---|
| 16S rRNA Gene Copy Number (109 Copies g−1 Dry Soil) | Chao 1 Index | Shannon Index | ITS Sequence Copy Number (107 Copies g−1 Dry Soil) | Chao 1 Index | Shannon Index | |
| M | 7.58 ± 0.97 a | 4386 ± 108 a | 6.79 ± 0.04 a | 7.06 ± 1.22 a | 655 ± 74 a | 3.55 ± 0.24 b |
| PM | 4.71 ± 0.91 b | 4345 ± 140 a | 6.75 ± 0.05 a | 5.66 ± 0.36 a | 700 ± 64 a | 4.28 ± 0.24 a |
| SM | 4.40 ± 1.27 b | 4309 ± 61 a | 6.76 ± 0.03 a | 5.24 ± 1.18 a | 721 ± 19 a | 4.38 ± 0.12 a |
| Topological Parameter | M | PM | SM |
|---|---|---|---|
| Number of nodes | 254 | 256 | 264 |
| Number of edges | 1679 | 1873 | 1834 |
| Number of copresence edges | 850 | 925 | 917 |
| Number of mutual exclusion edges | 829 | 948 | 917 |
| Average degree | 13.220 | 14.136 | 13.894 |
| Average number of neighbors | 15.527 | 16.200 | 15.694 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, Y.; Jin, H.; Zheng, F.; Yang, X.; Song, H.; Wang, J.; Fang, B.; Cheng, H.; Li, X.; He, D. Introducing Legumes into Wheat–Maize Rotation Complicates Soil Microbial Co-Occurrence Network and Reduces Soil Allelochemicals in Succeeding Wheat Season. Agriculture 2025, 15, 1307. https://doi.org/10.3390/agriculture15121307
Yan Y, Jin H, Zheng F, Yang X, Song H, Wang J, Fang B, Cheng H, Li X, He D. Introducing Legumes into Wheat–Maize Rotation Complicates Soil Microbial Co-Occurrence Network and Reduces Soil Allelochemicals in Succeeding Wheat Season. Agriculture. 2025; 15(12):1307. https://doi.org/10.3390/agriculture15121307
Chicago/Turabian StyleYan, Yaqian, Haiyang Jin, Fei Zheng, Xiwen Yang, Hang Song, Jiarui Wang, Baoting Fang, Hongjian Cheng, Xiangdong Li, and Dexian He. 2025. "Introducing Legumes into Wheat–Maize Rotation Complicates Soil Microbial Co-Occurrence Network and Reduces Soil Allelochemicals in Succeeding Wheat Season" Agriculture 15, no. 12: 1307. https://doi.org/10.3390/agriculture15121307
APA StyleYan, Y., Jin, H., Zheng, F., Yang, X., Song, H., Wang, J., Fang, B., Cheng, H., Li, X., & He, D. (2025). Introducing Legumes into Wheat–Maize Rotation Complicates Soil Microbial Co-Occurrence Network and Reduces Soil Allelochemicals in Succeeding Wheat Season. Agriculture, 15(12), 1307. https://doi.org/10.3390/agriculture15121307





