Abstract
Beekeeping is facing a serious crisis due to climate change and diseases such as bee mites (Varroa destructor), which have led to declining populations, collapsing colonies, and reduced beekeeping productivity. Bee mites are small, reddish-brown in color, and difficult to distinguish from bees. Rapid bee mite detection techniques are essential for overcoming this crisis. This study developed a technology for recognizing bee mites and beekeeping objects in beecombs using the You Only Look Once (YOLO) object detection algorithm. The dataset was constructed by acquiring RGB images of beecombs containing mites. Regions of interest with a size of 640 × 640 pixels centered on the bee mites were extracted and labeled as seven classes: bee mites, bees, mite-infected bees, larvae, abnormal larvae, and cells. Image processing, data augmentation, and stratified data distribution methods were applied to enhance the object recognition performance. Four datasets were constructed using different augmentation and distribution strategies, including random and stratified sampling. The datasets were partitioned into training, testing, and validation sets in a 7:2:1 ratio, respectively. A YOLO-based model for the detection of bee mites and seven beekeeping-related objects was developed for each dataset. The F1 scores for the detection of bee mites and seven beekeeping-related objectives using the YOLO model based on original datasets were 94.1% and 91.9%, respectively. The model applied data augmentation, and stratified sampling achieved the highest performance, with F1 scores of 97.4% and 96.4% for the detection of bee mites and seven beekeeping-related objects, respectively. The results underscore the efficacy of using the YOLO architecture on RGB images of beecombs for simultaneously detecting bee mites and various beekeeping-related objects. This advanced mite detection method is expected to contribute significantly to the early identification of pests and disease outbreaks, offering a valuable tool for enhancing beekeeping practices.
1. Introduction
Bees contribute to ecosystem maintenance and provide important products such as honey, beeswax, and royal jelly. Bee mites parasitize larvae and honeybees; the infected bee larvae do not grow into normal adults and present symptoms such as wing deformities, abdominal shrinkage, and death, which result in weight loss, deformities, and pupal mortality [1,2,3]. Bee mite populations are estimated to increase by 35% considering future climate change scenarios [4]. Thus, bee mites pose a major risk to the global beekeeping industry, causing significant economic damage to farmers because of the need for more miticides and the expenses to control them being higher than for any other bee disease [5,6,7]
Several methods have been used to identify the presence of apiary pests such as bee mites, for example, visual inspection of the beecomb, the application of powdered sugar to dislodge mites from adult bees, opening the cells to examine larvae, and inspection of the bottom of the hive. The visual inspection method that involves manually inspecting every bee in the hive is time consuming and can lead to inconsistencies between workers [8,9]. Therefore, objective and rapid detection technology for bee mites was deemed essential for quantification, occurrence frequency monitoring, and comprehensive damage assessment [10,11].
Object detection algorithms would be used to identify multiple objects within an image simultaneously. These algorithms include You Only Look Once (YOLO) architectures, region-based convolutional network methods (RCNNs), and fast RCNNs [12,13,14].
In accordance with these emerging trends, investigations are being conducted that integrate imaging technologies and object detection algorithms for the purpose of pest surveillance and monitoring [15,16]. Previous research has focused on using deep learning methods such as object detection algorithms to detect beekeeping-related objects. Visually distinguishable objects, such as bees and mites, exist in beekeeping images. Artificial intelligence (AI) models trained on beekeeping images for automatically detecting bee mites were developed. Thakker et al. developed models for distinguishing bees from the background in bee images using Mask-RCNNs, faster R-CNNs, and domain-adaptive faster R-CNNs. Faster R-CNNs achieved an average precision (AP) of 0.8773, suggesting the potential of object recognition algorithms to be applied to beekeeping data [17]. However, these image segmentation-based models generally require significant computational resources and extended inference times, which potentially limit their practical deployment in field environments. This computational burden limits real-time applications in beekeeping environments.
Other studies have also attempted to detect bees and bee-attracting objects using YOLO and single-shot multi-box detectors [18]. A beecomb measurement system was constructed and implemented to identify bee mites utilizing a convolutional neural network architecture [19]. Previous studies used image data obtained under limited conditions. Therefore, conducting an image-based study that measures beecomb in the same manner as in an actual beekeeping environment and simultaneously identifies the beekeeping objects that exist in the beecomb is required to ensure that it is applicable to a beekeeping environment.
Research on multi-object detection has been conducted in the agricultural field, as various objects can coexist [20]. In the apiculture field, pest objects such as bee mites, deformed wing bees, and abnormal larvae are less abundant than bees and cells. An imbalance of objects and lack of data on objects can indicate biased deep learning training results. Various data sampling and augmentation methods are available to overcome the imbalance of objects and lack of data [21,22]. The data sampling methods included random, stratified, and reservoir sampling. Random and reservoir sampling randomly select a portion of the total data, whereas stratified sampling maintains a specified proportion of the target class [23]. Stratified sampling ensures the uniformity of a dataset in a statistically precise manner [24]. In this study, random and stratified sampling were applied to compare the performance of bee mite recognition based on different sampling methods.
Detecting small pests in agricultural fields has been one of the most significant challenges [25,26]. In particular, as small objects, bee mite detection challenges stem from limited features, requiring enhanced architecture and data. Modified architectures include optimization function refinement and attention mechanism integration into existing YOLO models to better address the challenges inherent in small objects [27,28]. Data augmentation can enhance the robustness of AI models [29,30]. Synthesis and generative datasets were demonstrated to be efficacious for capabilities in small objects [31,32,33]. However, indiscriminate data augmentation and data augmentation using generative data can lead to general performance degradation and overfitting [34]. In contrast, data augmentation can be achieved through image processing techniques [35], and in the case of bee mites, feature enhancement was made possible using histogram equalization-based image processing [36]. Accordingly, this study considers data augmentation using histogram equalization techniques as a necessary approach to enhance the performance of beekeeping object detection.
This study aimed to develop a YOLO model for simultaneously detecting bee mites and seven other beekeeping objects within the beecomb. Four kinds of datasets were constructed by applying image processing-based data augmentation and two dataset splitting methods (random and stratified sampling). These datasets were used to train models for detecting bee mites and other beekeeping objects, and their performance was comparatively analyzed. Specifically, the effectiveness of the image-based data augmentation technique was verified, and the impact of stratified and random sampling on mitigating dataset imbalances was validated. Through this process, the optimal method for bee mite detection was identified.
2. Materials and Methods
2.1. Building Datasets for Bee Mite and Beekeeping Objects
2.1.1. Measurement of Beecomb Images with Beekeeping Objects
Images of the beekeeping objects used in this study were acquired by measuring the RGB images of the beecombs. The beecomb images were acquired from bee farms in Chuncheon-si, Gangwon-do, and Wanju-Gun, Jeollabuk-do, Republic of Korea, and the National Institute of Agricultural Sciences in September 2021, as well as in June and August 2022. A total of 26 bee hives (Apis mellifera) were used for image acquisition. The images were taken during the spring to fall seasons, from morning to evening, reflecting the seasonal characteristics of the Republic of Korea.
A schematic representation of the imaging apparatus implemented for beecomb image measurement is presented in Figure 1. The beecomb was held by a support frame. The angles of the camera and beecomb were such that they prevented saturation. A RGB camera (BlackflySGifE, FLIR, Wilsonville, OR, USA) with a resolution of 2048 × 1536 pixels was used to acquire the images. Previous research has found that bee mite identification is effective at this resolution when the camera is positioned 300 mm from the beecomb [36]. Therefore, the measurement distance was set to 300 mm and an additional correction model was applied to mitigate the distortion caused by the lens. Images have been taken of both the front-facing and rear-facing surfaces of the beecomb, replicating the visual inspection protocols employed by apiculturists during standard hive management procedures. A total of 40,222 images were measured, and 4613 images were selected for use after excluding the potential duplicates and blurred images.
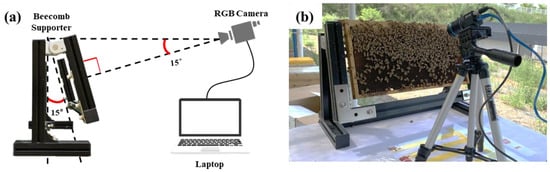
Figure 1.
Beecomb image measurement system. (a) Schematic of overall configuration, (b) image data acquisition in the field.
Seven beekeeping objects that could be present in the beecombs were selected: bees, bee mites, mite-infected bees, deformed wing bees, abnormal larvae, normal larvae, and cells. A bee with a deformed wing refers to a bee exhibiting symptoms of deformed wing virus. Abnormal larvae showed signs of infection with diseases, such as stonebrood, chalkbrood, or foulbrood, and they had a watery appearance, sagging in the direction of gravity. An image of the actual shape of the measured objects is shown in Figure 2 and Figure 3.
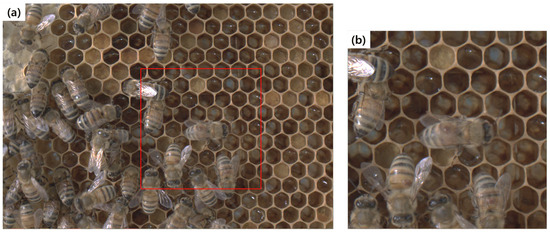
Figure 2.
Region of interest (ROI) cropping: (a) Original image, (b) extracted ROI image. Selecting a point on the original image with a mouse click creates a red box of the specified size.
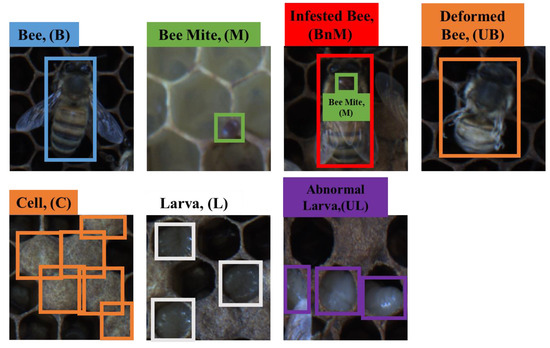
Figure 3.
Seven beekeeping objects and box annotation data. Annotation boxes were created to contain the features of an object.
2.1.2. Extracting Region of Interest Images for Building Datasets
The disproportionate ratio of bees to parasitic bee mites in beecomb imagery poses a significant class imbalance challenge for parasite detection systems. This distributional asymmetry compromises model training efficacy and predictive accuracy for the minority class of bee mites. The quantitative overrepresentation of host organisms relative to mites as a main target becomes particularly pronounced when employing high-resolution (2048 × 1536 pixel) full-frame imagery for annotation purposes, potentially undermining classification performance metrics. Therefore, a Region of Interest (ROI) approach was necessary to control object counts and data for the target objects.
Furthermore, utilizing large-size images with high resolution can significantly increase computational resource demands. To improve the efficiency of model development, regions of interest (ROI) were extracted by limiting the image size for training. From the cropped segments of the whole image, those containing honeybee mites and beekeeping objects were specifically selected for model training. Therefore, to effectively verify the whole image using an object detection model, the complete image must be cropped to sizes appropriate for the model and divided into multiple images. In this study, the whole image was verified using this approach. Among these segments, ROI images without objects of interest were excluded from the verification process.
The process of extracting ROI from the full-resolution images is shown in Figure 2. The ROI had a size of 640 × 640 pixels and contained at least one of the following: bee mites, deformed wing bees, or abnormal larvae. Various images were extracted as target objects if multiple target objects existed in a single image. The extraction of ROI centered on them was not considered because bees, normal larvae, and cells were present together in the ROI. In addition, the images were sampled to extract discontinuous data. For the ROI data, 1463 images were extracted from 4613 sampled beecomb images.
2.1.3. Bee Mite and Beekeeping Object Annotation Rules and Distribution Methods
Object detection algorithms learn the features of objects through supervised learning, which requires annotated data as reference values [37]. Annotation data were created for bee mites and beekeeping objects in the extracted ROI. Object labeling was applied to the data under the same conditions for each object in the images.
The annotation guidelines for each object are presented in Figure 3. For the label classes, the class names were set as B, UB, BnM, M, UL, L, and C representing normal bees, deformed wing bees, mite-infected bees, bee mites, abnormal larvae, normal larvae, and cells, respectively. For a normal bee, the entire bee body is specified, and for discriminating a bee with deformed wings, the wing parts are included. The annotation data for mite-infested bees are generated in the same manner as for normal bees, but are only applied to bees parasitized by bee mites. The bee mites are labeled regardless of the object to which they are attached (bee, larvae, or cell). Each object is annotated separately if a bee mite is attached to a bee with a deformed wing. The label data of the normal and abnormal larvae include the entire body, and in the case of cells, the beecomb structure is considered, even when it is densely packed.
Based on these aforementioned rules, 13,090 normal bees, 1651 mite-infested bees, 1784 bee mites, 179 deformed wing bees, 311 normal larvae, 388 abnormal larvae, and 5762 cells were labeled and used for training. The annotation software Labelme (version 4.6.0, https://github.com/wkentaro/labelme, accessed on 6 June 2022) was used to label and create the bounding boxes.
During the process of splitting a dataset (for training, testing, and validation), some classes could be biased towards particular datasets, leading to overfitting or underfitting. Therefore, in this study, by considering the effect of dataset distribution, a dataset was constructed by applying random and stratified sampling to the main identification target, i.e., bee mites. Object detection models were developed based on this dataset, and the results were compared and analyzed.
A random sampling method was used to divide the data into specified ratios without considering the distribution of the target (bee mite). The data were split in a ratio of 7:2:1 and used as the training, testing, and validation datasets, respectively. The training and testing datasets were utilized for model learning and development, with the validation dataset being employed to verify the developed model performance using arbitrary data. Stratified sampling divides a target population into strata and randomly selects samples from each stratum. This method offers the advantage of distributing selected classes to each dataset at a chosen ratio [38]. A stratified sampling method was used to create the training, test, and validation datasets to ensure the consistent representation of bee mites. The data were distributed in a 7:2:1 ratio and aligned with the ratio used for random sampling.
Data augmentation was used to increase the amount of training data [30]. The robustness of the object detection model was enhanced by training on images of varying quality and achieved by applying different image processing techniques [18,35]. In this study, additional training data were generated by augmenting the data using image processing techniques such as histogram normalization and equalization, which can effectively enhance the distinction between bee mites and bees [36]. Image normalization was applied using a linear transformation based on the maximum and minimum pixel intensity values. For contrast enhancement, the Contrast Limited Adaptive Histogram Equalization (CLAHE) technique was implemented. These image processing methodologies can enhance object boundary definition and improve algorithm robustness to variations in imaging conditions.
2.2. Development of Bee Mite and Beekeeping Object Detection Models
2.2.1. YOLO Architecture and Hyperparameters for Bee Mite Beekeeping Object Detection
A multi-object YOLO detection model was developed to simultaneously identify bee mites and other beekeeping objects from RGB images of a beecomb to enable the early diagnosis of pests and diseases in beekeeping. A dataset of seven beekeeping-related objects was constructed and two data distribution methods were applied to the datasets processed using image techniques optimized for bee mite detection and augmented datasets.
All models were initially developed using original and preprocessed datasets, and the model performance was evaluated systematically. Furthermore, YOLO models were trained on augmented datasets, comparing the detection performance between datasets with randomly distributed training, test, and validation splits, and those with splits were determined by stratified sampling. Object detection accuracy and robustness were subsequently assessed through a comprehensive analysis based on the applied data distribution methodologies.
Building on the concepts of the “Bag of Freebies” and “Bag of Specials”, numerous object detection algorithms employ advanced image processing techniques and deeper neural network architectures to enhance both accuracy and computational efficiency [39]. YOLOv7 incorporates a trainable “Bag of Freebies” optimization module, which increases computational demands during the training phase while preventing an escalation of the inference computational costs. Furthermore, this module includes several integrated augmentation methods such as mosaic augmentation, random affine transformations, MixUp, and CutOut techniques that were applied [14]. The YOLOv7 structure was used to learn how to recognize bee mites and beekeeping objects. The computational framework of the backbone and head in YOLOv7 is illustrated in Figure 4.
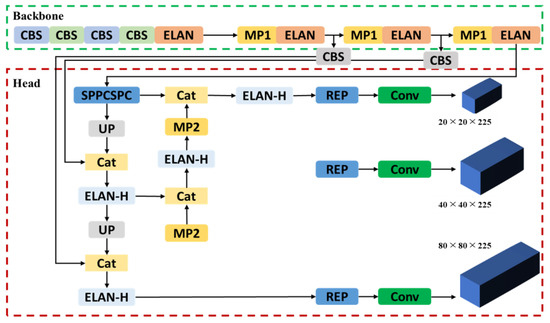
Figure 4.
YOLOv7 architecture [14].
All methodologies, including the models and image processing methods, were developed using Python 3.10.9 (Python Software Foundation, Wilmington, DE, USA). Deep learning operations were executed using the PyTorch CUDA 11.4 framework, while dataset partitioning was performed utilizing scikit-learn’s data splitting functionality. Image processing and various utility tasks were developed in Python using OpenCV. The detailed parameters and environments are listed in Table 1. The image size was set to 640 × 640 pixels, which matches the size of the training data. The batch size, which refers to the number of samples used for each weight update, was set to 34, and 1200 training iterations were performed. A cache function that stores image files in the RAM was applied to improve the computation speed. For computations using two GPUs, the number of devices was set to zero and one, and the neural network architecture was modified to accommodate seven input classes for training.

Table 1.
Hyperparameters used for developing bee mite detection model.
2.2.2. Performance Evaluation Methods for Bee Mite and Beekeeping Object Detection Models
The performances of the bee mite and beekeeping object detection models were assessed using accuracy, precision, recall, F1 score, and mean average precision (mAP). Accuracy was shown as the proportion of correct predictions, where True was predicted as true and False was predicted as false by the model, as demonstrated in Equation (1), indicating the overall correctness of the model’s predictions. TruePositive represents instances where the model correctly identifies the presence of the target object, demonstrating accurate positive classification. FalsePositive denotes scenarios where the model erroneously indicates the presence of the target object when it is absent. FalseNegative describes cases where the model fails to detect the presence of the target object when it is actually present. TrueNegative encompasses situations where the model correctly identifies the absence of the target object. In this study, the accuracy metric was interpreted as a quantitative assessment of the model’s ability to correctly identify each class, representing a fundamental measure of the system’s overall classification performance. The accuracy metric was directly reflected by the model’s recognition capability to capture intrinsic patterns in the input data and assign the correct class labels.
Precision was measured as the accuracy of the detected objects corresponding to the actual objects, indicating the proportion of correctly identified positive instances out of all instances predicted as positive. In other words, it was used to assess how many of the predicted positives were truly positive, as demonstrated in Equation (2).
Recall was reflected as the proportion of true positives identified by the model relative to the total number of actual instances. This metric was particularly useful for evaluating the detection capabilities of the model, and it was based on Equation (3).
Precision and recall can exhibit imbalances, particularly in cases of overfitting, where high precision may be accompanied by low recall, resulting in a skewed evaluation of performance. This makes using precision and recall alone insufficient for assessing overall performance in object detection. The F1 score, by taking the harmonic means of precision and recall, provides a more balanced and comprehensive evaluation of the model’s performance, as demonstrated through Equation (4).
The mAP was defined as the mean of all average precision (AP) values, with the AP calculated by integrating the step function of the precision–recall (PR) curve, as demonstrated in Figure 5. The mAP [0.5] was considered to represent correct predictions when the intersection over union (IoU) was greater than or equal to 0.5, indicating that predicted and ground truth objects overlapped by at least half. Furthermore, mAP [0.5:0.95] was defined as the average of mAP values, calculated by increasing the IoU threshold from 0.5 to 0.95 in increments of 0.05. Throughout the 1200 training iterations, the model weights were saved each time an improved mAP score [0.5:0.95] was observed. The performance of the saved weights was evaluated subsequently using the mAP [0.5:0.95] metric on a separate validation dataset that was not used in the training process. Finally, the set of weights that achieved the highest mAP score [0.5:0.95] in the validation dataset was selected as the final model.
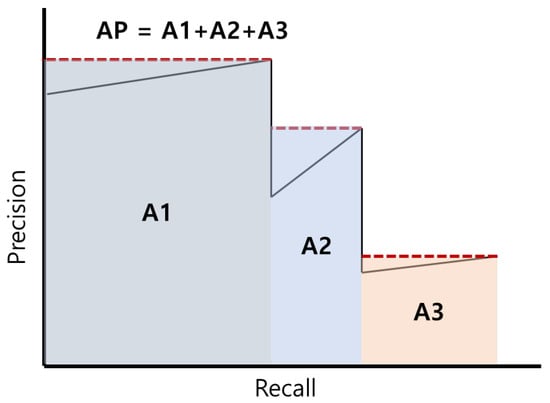
Figure 5.
Average precision (AP) calculation method.
3. Results and Discussion
3.1. Results of Building Dataset for Bee Mites and Beekeeping Object Detection
Table 2 presents the composition of the datasets. “Original data” refers to the raw RGB image data, while “Image-processed data” indicates images that underwent histogram normalization and smoothing techniques. Datasets A and B were randomly divided into training, testing, and validation sets irrespective of the class. Datasets C and D, which underwent data augmentation, were distributed using random sampling for Dataset C, whereas stratified sampling was applied to Dataset D to maintain a consistent proportion of bee mites.

Table 2.
Dataset construction by applying random and stratified data split methods to original and augmented images.
Figure 6 illustrates representative effects of the implemented image processing methodologies. Following normalization application, significant enhancement was observed in the chromatic information of apicultural specimens, including bees and bee mites. However, Figure 6(a3,b3) exhibited no discernible enhancement in chromatic properties, attributable to the original image’s pre-existing dynamic range spanning the complete 8-bit spectrum (minimum value 0, maximum value 255). Thus, the augmentation of intrinsic morphological characteristics necessitated the subsequent implementation of advanced histogram equalization techniques.
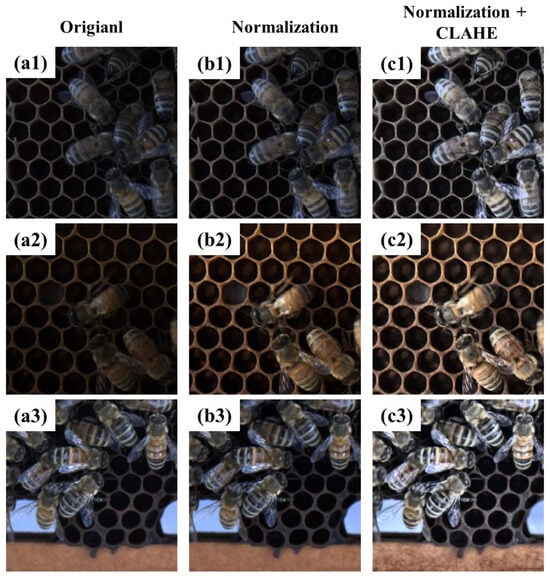
Figure 6.
Comparative analysis of image enhancement techniques for improved features of bees and mites: (a1–a3) Original image, (b1–b3) normalization image, and (c1–c3) normalization and CLAHE image.
Figure 7 shows the frequencies of the seven classes in Datasets A, B, C, and D built from the extracted images. The percentages of each class in each dataset are listed in Table 3. Datasets A and B, which were built with random sampling, had average ratios of 70.81:20.10:9.09 and 70.67:20.14:9.19 between training, testing, and validation for the seven classes.
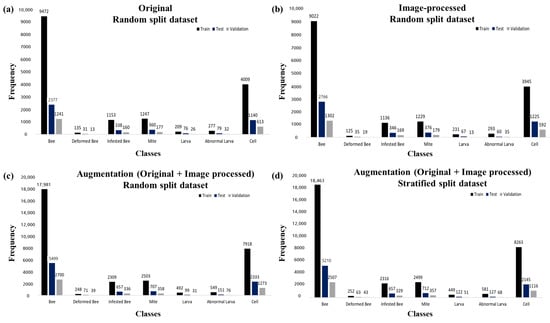
Figure 7.
The number of training, testing, and validation data of seven classes randomly split into Datasets A, B, C, and D. (a–d) represent information about datasets A, B, C, and D. The training, testing, and validation data were split into a ratio of 7:2:1. The number at the top of the bar chart was the frequency of that class in the dataset.

Table 3.
Proportions of training, testing, and validation data of seven classes randomly split in Datasets A, B, C, and D.
Dataset C, which was built with random sampling of the augmented data (Original + Image-processed data), had an average ratio of 70.94:19.45:9.60 between training, testing, and validation for the seven classes. Dataset D, which was built using stratified sampling of the augmented data, was divided 71.41:18.85:9.74 on average across the seven classes. When the dataset was constructed using stratified sampling based on the proportion of bee mites, the distribution ratios for the training, testing, and validation sets were 70.04, 19.96, and 10.01%, respectively, providing a more precise allocation compared to that with random sampling. For the remaining categories (deformed wing bees, abnormal larvae, and cells), Dataset C exhibited a more refined and balanced distribution.
3.2. Evaluation of Model Performance
3.2.1. Performance of Bee Mite and Beekeeping Object Detection Models
Models were developed based on the object detection algorithm YOLOv7 to recognize bee mites and six beekeeping objects. Datasets A (original and random sampling), B (image processing and random sampling), C (data augmentation (original and image processing) and random sampling), and D (data augmentation (original and image processing) and stratified sampling) were used. The YOLO models developed using each dataset were referred to as YOLO-DA, YOLO-DB, YOLO-DC, and YOLO-DD, respectively.
Figure 8 shows an example of the four YOLO-based bee mite and six beekeeping object-based detection models. All four models could recognize seven beekeeping objects, including bee mites, on the beecomb. The inference time required for the detection of the bee mites and six beekeeping objects for individual images was below 0.02 s on the GPU and 0.2 s on the CPU. The performance of the model was validated after the whole image had been cropped to the ROI size; therefore, it was anticipated that no significant difference in identification performance would be observed between the whole image and the ROI image, although a difference in the processing speed was expected.
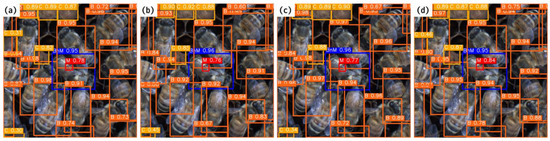
Figure 8.
Results of predicted images from (a) YOLO-DA, (b) YOLO-DB, (c) YOLO-DC, and (d) YOLO-DD.
Figure 9 shows the confusion matrix for the accuracy of the developed object detection models. The average accuracies of YOLO-DC and YOLO-DD with data augmentation were 0.97 and 0.98, respectively, showing a clear improvement compared to the 0.93 and 0.89 achieved by YOLO-DA and YOLO-DB, respectively without augmentation. The detection accuracy for bee mites increased from 92.3% with the original data to 98.0% after data augmentation, reflecting a 5.7% improvement. In addition, key performance metrics such as accuracy, the F1 score, and mAP improved across all beekeeping object classes, except for normal larvae (Table 4 and Table 5). These results highlight the effectiveness of data augmentation techniques for improving the detection and classification of bee mites and other beekeeping-related objects.
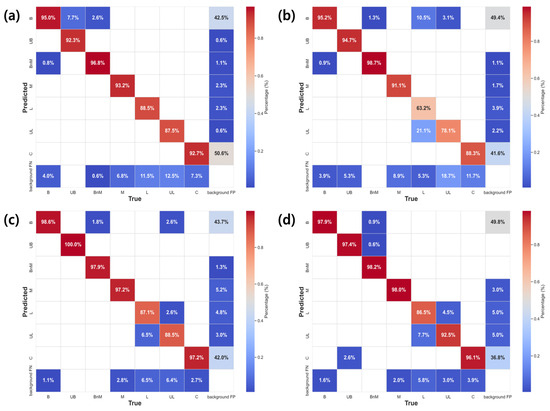
Figure 9.
Confusion matrix of YOLO models: (a) YOLO-DA, (b) DB, (c) DC, and (d) DD models. The numbers in the figure represent accuracy, expressed as percentages (%). B, UB, BnM, L, UL, and C refer to Bee, Deformed bee, Infested bee, Larvae, Abnormal larvae, and Cell, respectively.

Table 4.
Results of the object detection evaluations of the YOLO-DA and YOLO-DB models.

Table 5.
Results of multi-object detection evaluation of the YOLO-DC and YOLO-DD models.
Significant performance variation was observed across different dataset configurations, highlighting how class distribution impacted not only the detection of primary targets, but also of secondary objects within multi-object detection systems. When multi-object detection datasets were constructed with emphasis on one primary object (bee mites), the distribution ratio for the other objects was found to be potentially insufficient. This was clearly demonstrated in the larvae detection case, where lower representation percentages in validation sets, particularly in Dataset B at only 4.18%, directly corresponded with decreased detection accuracy 63.2% (Table 3, Figure 9b). Cross-class interference patterns were observed in the confusion matrices, revealing specific misclassification patterns (such as the 21.1% confusion between larvae and UL in Figure 9b). The analysis of these patterns revealed that certain classes exhibited visual characteristic overlaps that contributed to detection challenges. These findings indicated that even when a study had a designated primary target object, maintaining adequate representation across all classes remained crucial for model robustness.
The relationships between the F1 score and confidence thresholds were analyzed across four models (Figure 10). While the Bee, Deformed bee, Bee infected with mite, and Mite classes exhibited high F1 scores (>0.9), the Larvae and Abnormal larvae classes consistently underperformed. The F1 scores remained stable across low (0.1) to moderately high (0.8) confidence thresholds, but declined rapidly beyond 0.85. These findings suggested that thresholds in the range of 0.6–0.8 might offer an optimal trade-off between precision and recall for field applications. The YOLO-DC configuration achieved the highest overall average F1 score (0.96) at a confidence threshold of 0.720. However, YOLO-DD demonstrated a superior capability in detecting mites, which constituted the primary focus of this investigation. Although YOLO-DD achieved the highest accuracy for mite detection, YOLO-DC remained more suitable for applications requiring the simultaneous detection of all seven apicultural classes (Figure 9 and Figure 10). The suboptimal larvae detection was attributed to the limited validation dataset (Table 3). The Cell class consistently demonstrated high performance across all configurations. For practical deployment, employing class-specific confidence thresholds—rather than a single global threshold—may enhance the overall detection performance, particularly in classes with imbalanced representation.
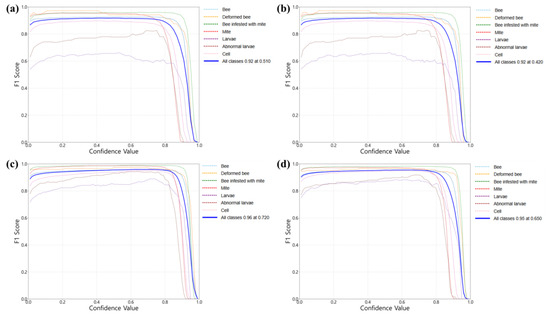
Figure 10.
F1 score–confidence threshold curves for different YOLO models: (a) YOLO-DA, (b) YOLO-DB, (c) YOLO-DC, and (d) YOLO-DD. B, UB, BnM, M, L, UL, and C refer to Bee, Deformed bee, Bee with mite, Bee mite, Larvae, Abnormal larvae, and Cell, respectively.
Figure 11 presents the loss curves observed during the training processes of the four YOLO models. All models exhibited typical convergence patterns, characterized by an initial rapid decline followed by the gradual stabilization of the training loss. The gap between the training and validation loss showed minimal divergence, indicating effective overfitting prevention through early stopping implementation. The validation loss initially decreased, reaching minimum values before slight increases and subsequent stabilization were noted, with particularly stable trends being demonstrated in the data-augmented YOLO-DC and YOLO-DD models. YOLO-DA and YOLO-DB converged efficiently at epochs 474 and 616, respectively, while the augmented models (YOLO-DC and YOLO-DD) required extended training (947 and 580 epochs, respectively), but achieved superior stability. The extended training requirements for augmented models could indicate a trade-off between enhanced robustness and training efficiency. The Best epochs for each model were determined based on mAP [0.5:0.95] metrics, with detailed performance scores being presented in Section 3.2.2 and Table 4 and Table 5.
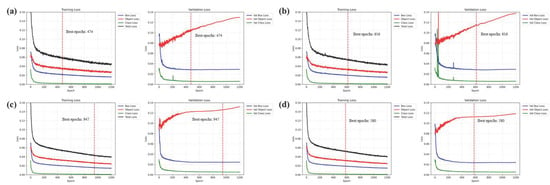
Figure 11.
Training and validation loss curves of the bee mite and beekeeping object detection models: (a) YOLO-DA, (b) YOLO-DB, (c) YOLO-DC, and (d) YOLO-DD. Red vertical lines indicate the best epoch for each model.
3.2.2. Performance Comparison of Bee Mite and Beekeeping Object Detection Models Based on Original and Image-Processed Data
Table 4 summarizes the comparative performances of the YOLO-DA and YOLO-DB models for the detection of bee mites and six beekeeping objects based on the image-processing techniques. The YOLO-DA model demonstrated superior performance in recognizing normal bees, bee mites, normal and abnormal larvae, and cells. However, the YOLO-DB model outperformed the other models in identifying bees infected with mites and those with deformed wings.
The YOLO-DA model based on the original dataset achieved the highest accuracy of 96.8% in recognizing infested bees. The accuracy of detecting the primary target (bee mites) was 93.2%. The average F1 score across all classes was 90.4%, with a notable F1 score of 94.0% for bee mites. The training iteration that recorded the highest mAP [0.5:0.95] occurred during 474 epochs.
The YOLO-DB model achieved an F1 score of 93.6% for identifying infected bees, which is higher than the 93.3% achieved with the original data-based model. The F1 score improved compared to the original, with precision increased and recall decreased. Consequently, accuracy was enhanced from 96.8% to 98.7%. Despite being an image-processed model, the accuracy for detecting bee mites was 91.1%, which is 2.1% lower than that of the original model. The highest F1 score in the performance evaluation of the YOLO-DB model was 96.7% for deformed wing bees, representing an improvement of 13.2% compared with that for YOLO-DA. The average mAP [0.5] and mAP [0.5:0.95] values across all classes were 89.6% and 63.5%, respectively. The highest mAP score [0.5:0.95] was recorded at the 616th training iteration.
When comparing YOLO-DA and YOLO-DB, notable differences in larvae detection performance were observed, which were attributed to dataset distribution variations (Table 3). The results suggested that features distinguishing normal bees from deformed wing bees could be enhanced through histogram normalization. However, the histogram normalization process was found to potentially reduce the accuracy of cell detection, indicating that future research should focus on identifying object-appropriate normalization methods, determining optimal preprocessing techniques that comprehensively enhance all objects, and developing differentiated image processing approaches based on image data characteristics.
3.2.3. Performance Comparison of Bee Mite and Beekeeping Object Detection Models Based on Random and Stratified Sampling Methods
Table 5 compares the object detection performances of the YOLO-DC and YOLO-DD models for each object category. For the YOLO-DC model, the mAP [0.5] exceeded 0.9 across all classes; however, 947 training iterations were required to achieve the highest mAP [0.5:0.95], which is greater than that for any other model. The average F1 score of 93.7% was higher than those of the YOLO-DA and YOLO-DB models. Furthermore, the average mAP [0.5:0.95] reached 72.0%, making it the most effective model among the four developed models. These results demonstrate that an increase in the training data through data augmentation enables the development of a more effective object detection model. Furthermore, F1 scores above 80% were achieved across all classes, with the primary target (bee mites) recording an F1 score of 96.8% and infected bees achieving 97.8%. The precision of the YOLO-DC model for detecting deformed wing bees was 100%, with a recall of 95.4% and an F1 score of 97.6%. The 100% precision indicates that all deformed wing bees were detected with 100% accuracy. The average mAP [0.5] and mAP [0.5:0.95] were calculated as 96.4% and 72.0%, respectively. This implies that during the inference process, when the confidence level was set at 0.5, a recognition performance of ~96.4% was achieved by the model.
The YOLO-DC and YOLO-DD model demonstrated the highest average accuracy of 95.2% among all developed models. An overall F1 score of 92.9% was obtained for the YOLO-DD model. The YOLO-DD model achieved an impressive 98.2% and 98.0% accuracy in detecting both infected bees and bee mites, respectively. The YOLO-DD model achieved the best performance for bee mites, with an F1 score of 97.4%, mAP [0.5] of 97.4%, and mAP [0.5:0.95] of 61.7%. These results underscore the exceptional accuracy of the model in detecting bee mites compared to that of the other developed models. These results surpassed the performance of the YOLO-DC model, which did not employ stratified sampling, with an F1 score of 96.8%, mAP [0.5] of 97.3%, and mAP [0.5:0.95] of 61.7%. Thus, in the case of stratified sampling, the even distribution of target objects across datasets can be considered an effective method for improving the model’s training performance.
The YOLO-DC model demonstrated exceptional detection capability with deformed bees, achieving 100% accuracy and precision, while it showed a comparatively lower performance with larvae detection, at 87.1% accuracy. This performance disparity suggested that certain morphological characteristics of deformed bees were inherently more detectable than others. For classes like abnormal larvae, notable differences between precision (94.3%) and recall (86.9%) with the YOLO-DC model indicated challenges in balancing false positives and false negatives. Additionally, mite detection required substantially more training epochs (947 for DC) compared to other categories, indicating a more complex learning challenge likely attributable to their small size, variable positioning on host bees, or subtle visual characteristics that differentiate them from background elements. Bee mite detection achieved strong mAP [0.5] performance (97.4%) with the YOLO-DC model, but exhibited a considerable decrease at the stricter mAP [0.5:0.95] threshold (61.7%), suggesting difficulties with precise localization across varying confidence thresholds.
3.2.4. Determining the Best Models for Bee Mite Detection
This study aimed to develop a bee mite detection model that can be implemented in real beekeeping environments. RGB image data were collected from beecombs in actual beekeeping fields, and key beekeeping objects, such as bees, deformed wing bees, infected bees, bee mites, larvae, abnormal larvae, and cells, were selected for model development. A YOLO model was developed for the detection of bee mites and other six beekeeping objects, attaining an F1 score of 97.4% and mAP [0.5] of 97.4%. Data augmentation techniques including histogram normalization and equalization were applied to improve the mAP [0.5] of bee mites and other beekeeping objects from 91.9% to 96.4%. Furthermore, applying stratified sampling for bee mite distribution further improved the detection performance, with mAP [0.5:0.95] increasing by 2.5% compared to random sampling.
For bee mite detection, the YOLO-DC and YOLO-DD models, utilizing augmented data, achieved accuracies of 97.2% and 98.0%, respectively. These results outperform the 93.2% and 91.1% accuracies recorded for the YOLO-DA and YOLO-DB models, as shown in Figure 9. Models with data augmentation outperformed the YOLO-DA model, which relied solely on the original data, and the YOLO-DB model, which used only the processed data. This demonstrates that combining image-processed data with the original data leads to the development of more robust and accurate object detection models than using the original data alone.
Figure 12 is a comparison of the F1 scores for each model. In the comparison between YOLO-DA and YOLO-DB, it can be noticed that the F1 scores of Larvae and Abnormal Larvae were decreased considerably when the preprocessing was applied. Furthermore, the accuracy for Larvae decreased from 88.5% to 63.2% with the processed data. However, applying optimal image processing to distinguish between normal bees and bee mites did not improve the detection of normal larvae, suggesting that different object classes may require tailored image-processing methods for optimal performance.
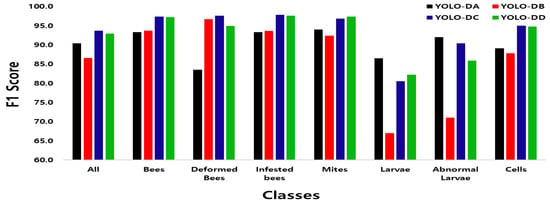
Figure 12.
Comparison of F1 scores for each object using developed model. Black: YOLO-DA, red: YOLO-DB, blue: YOLO-DC, and green: YOLO-DD.
Table 6 summarizes the best-performing models, underscoring the superior performance of the YOLO-DC and YOLO-DD models. The YOLO-DC model demonstrated the highest performance across most evaluation metrics, followed by the YOLO-DD model. The YOLO-DD model delivered the highest detection performance in terms of accuracy (98.0%), F1 score (97.4%), and mAP [0.5] (97.4%) for the primary target, the bee mite. YOLO-DC recorded an average F1 score of 93.7%, indicating its capability to classify key beekeeping objects, including normal bees, bees with deformed wings, infected bees, bee mites, abnormal larvae, normal larvae, and cells.

Table 6.
Performance comparison of four YOLO models for each object detection.
Overall, YOLO-DD exhibited higher detection accuracy, specifically for bee mites, compared to that for the YOLO-DC model. The performance improvement of the YOLO-DD model can be interpreted as a result of balancing rare classes (e.g., bee mites, infested bees) within the dataset through stratified sampling. Thus, a stratified sampling distribution enables a more precise object allocation, which, in turn, enhances the learning outcomes. Moreover, it was confirmed that when objects are randomly distributed, the proportion of class instances can vary, potentially affecting the accuracy of the model during the training, testing, and validation phases. This results in the potential influence of dataset distribution on learning performance and suggests that adjusting the amount of data or fine tuning the computational processes for classes with insufficient instances can significantly affect the learning outcomes. Therefore, additional data collection for underperforming objects is recommended to improve the recognition performance across classes.
For normal larvae detection, the YOLO-DA model trained on the original data exhibited the best performance. For YOLO-DA, the proportion of normal larvae in the test dataset was the highest, at 24%. This suggests that increasing the proportion of test data, such as adopting a 6.5:2.5:1 ratio instead of a 7:2:1 ratio when the data were limited, can enhance object recognition accuracy. Alternatively, fine tuning the object recognition algorithm to place greater weight on the loss extracted during the test phase may also be effective.
The YOLO-DB model, trained solely on the processed data, did not achieve the highest scores for any object category. When image processing is applied, shallow features in deep learning models may be reinforced, which can inadvertently enhance the background features. Consequently, this processing approach may have led to a decline in performance. Compared with the results based on the original dataset, although image processing improved the detection performance for infected bees, it decreased the detection accuracy for larvae. This suggests that different image processing methods may be more suitable for specific objects.
3.3. Comparative Analysis with Previous Research
Table 7 compares the performance of the developed bee mite detection models with that of previous research models. The comparison included two previous studies that utilized RGB images and object detection algorithms. The results of this study demonstrated superior bee mite identification performance, with an F1 score of 97.4% compared to 97.0% and 52.9% reported in prior research [18]. In addition, previous studies did not consider real beekeeping environments or use background-subtracted or individual object images, which may have limited their practical applicability.

Table 7.
Comparing bee mite detection performance with previous research models.
In contrast, this study utilized RGB images captured from actual beekeeping environments, suggesting greater field applicability. Furthermore, the capability of the model to recognize other beekeeping objects, including bees and bee mites, may reduce errors caused by other objects in real-world applications.
The developed multi-object beekeeping detection model for detecting seven distinct objects on beecomb holds promise for effectively identifying pests and diagnosing hive conditions. The model demonstrated high detection performance, particularly for bee mites. For successful deployment in beekeeping environments, it is crucial that the dataset accurately reflects the conditions encountered in real-world settings and that the limitations related to RGB camera measurement conditions are addressed. To overcome these challenges, data augmentation techniques such as image rotation and flipping can be utilized to enhance robustness across different angles. In addition, introducing noise and filters that mimic common environmental conditions can further adapt the model and ensure its effectiveness in diverse settings.
4. Conclusions
The beekeeping industry is currently facing significant challenges, including reduced productivity, population decline, and colony collapse, which is further exacerbated by climate change and pests. Among these, bee mites pose a major threat due to their small size and reddish-brown color, making them difficult to distinguish from bees. This study addresses the need for rapid and accurate detection technologies by developing a YOLO-based object detection method tailored to detect bee mites and other beekeeping-related objects.
Beekeeping image data captured at distances of less than 300 mm were used with data augmentation techniques, random sampling, and stratified sampling. Four YOLO models (YOLO-DA, YOLO-DB, YOLO-DC, and YOLO-DD), each employing different data augmentation or sampling strategies, were developed and systematically evaluated. The results indicated that models trained on augmented datasets (with the data increased from 1463 to 2926 images) exhibited superior performance compared to those trained on original or processed data alone. The YOLO-DC model achieved the highest average F1 score of 93.7% and demonstrated an effective classification of key beekeeping objects with 95.2% accuracy. YOLO-DD achieved the highest performance in detecting bee mites (F1 score: 97.4%, mAP [0.5]: 97.4%), outperforming the other models in this task. Stratified sampling effectively mitigated distribution bias, resulting in improved detection performance across multiple classes.
This research focused specifically on Western honeybees (Apis mellifera) and was confined to the detection of seven critical apicultural objects: bees, deformed bees, infested bees, mites, larvae, abnormal larvae, and cells. The model exhibits limitations in its capacity to detect beekeeping elements beyond these specified classes. For more comprehensive apicultural object detection systems, future research should incorporate additional hive components and environmental factors present in diverse beekeeping contexts.
Despite these limitations, this investigation makes a significant contribution to the pressing agricultural challenge of varroa mite diagnosis in commercial apiaries. The developed model demonstrates potential for practical implementation in bee health monitoring systems, addressing a critical need in contemporary apiculture.
This research establishes a foundation for future work, which will focus on model refinement and the development of field-deployable devices for apiary management. Subsequent investigations will aim to translate these computational approaches into practical technological solutions that can be readily integrated into existing beekeeping operations, thereby enhancing disease monitoring capabilities and supporting sustainable apicultural practices.
Author Contributions
Conceptualization, H.-G.L., S.-B.K. and C.M.; methodology, H.-G.L., J.-Y.S. and C.M.; software, H.-G.L. and H.L.; validation, H.-G.L. and S.-B.K.; formal analysis, H.-G.L.; investigation, H.-G.L., J.-Y.S. and M.-J.K.; resources, S.-B.K.; data curation, H.-G.L. and M.-J.K.; writing—original draft preparation, H.-G.L.; writing—review and editing, C.M.; visualization, H.-G.L. and J.-Y.S.; supervision, C.M. and M.S.K.; project administration, C.M.; funding acquisition, C.M. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Rural Development Administration as part of the Cooperative Research Program for Agriculture Science and Technology Development [Project No. RS-2023-00232224].
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The authors do not have permission to share data.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Abu, E.S. The use of smart apiculture management system. Asian J. Adv. Res. 2020, 5, 6–16. [Google Scholar]
- Sammataro, D.; Gerson, U.; Needham, G. Parasitic mites of honey bees: Life history, implications, and impact. Annu. Rev. Entomol. 2000, 45, 519–548. [Google Scholar] [CrossRef] [PubMed]
- Wilfert, L.; Long, G.; Leggett, H.; Schmid-Hempel, P.; Butlin, R.; Martin, S.; Boots, M. Deformed wing virus is a recent global epidemic in honeybees driven by Varroa mites. Science 2016, 351, 594–597. [Google Scholar] [CrossRef] [PubMed]
- Chuleui, J. Simulation Study of Varroa Population under the Future Climate Conditions. J. Apic. 2015, 30, 349–358. [Google Scholar] [CrossRef]
- Boecking, O.; Genersch, E. Varroosis—The ongoing crisis in bee keeping. J. Für Verbraucherschutz Lebensmittelsicherheit 2008, 3, 221–228. [Google Scholar] [CrossRef]
- Hristov, P.; Shumkova, R.; Palova, N.; Neov, B. Factors associated with honey bee colony losses: A mini-review. Vet. Sci. 2020, 7, 166. [Google Scholar] [CrossRef]
- Kane, T.R.; Faux, C.M. Honey Bee Medicine for the Veterinary Practitioner; John Wiley & Sons: Hoboken, NY, USA, 2021; pp. 229–234. [Google Scholar] [CrossRef]
- Braga, A.R.; Gomes, D.G.; Rogers, R.; Hassler, E.E.; Freitas, B.M.; Cazier, J.A. A method for mining combined data from in-hive sensors, weather and apiary inspections to forecast the health status of honey bee colonies. Comput. Electron. Agric. 2020, 169, 105161. [Google Scholar] [CrossRef]
- Gregorc, A.; Sampson, B. Diagnosis of Varroa Mite (Varroa destructor) and sustainable control in honey bee (Apis mellifera) colonies—A review. Diversity 2019, 11, 243. [Google Scholar] [CrossRef]
- Delaplane, K.S.; Berry, J.A.; Skinner, J.A.; Parkman, J.P.; Hood, W.M. Integrated pest management against Varroa destructor reduces colony mite levels and delays treatment threshold. J. Apic. Res. 2005, 44, 157–162. [Google Scholar] [CrossRef]
- Jack, C.J.; Ellis, J.D. Integrated pest management control of Varroa destructor (Acari: Varroidae), the most damaging pest of (Apis mellifera L. (Hymenoptera: Apidae)) colonies. J. Insect Sci. 2021, 21, 6. [Google Scholar] [CrossRef]
- Girshick, R. Fast r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 1440–1448. Available online: https://openaccess.thecvf.com/content_iccv_2015/html/Girshick_Fast_R-CNN_ICCV_2015_paper.html (accessed on 1 March 2023).
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You only look once: Unified, real-time object detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 779–788. Available online: https://www.cv-foundation.org/openaccess/content_cvpr_2016/html/Redmon_You_Only_Look_CVPR_2016_paper.html (accessed on 1 April 2023).
- Wang, C.-Y.; Bochkovskiy, A.; Liao, H.-Y.M. YOLOv7: Trainable bag-of-freebies sets new state-of-the-art for real-time object detectors. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 18–22 June 2023; pp. 7464–7475. [Google Scholar] [CrossRef]
- Badgujar, C.M.; Poulose, A.; Gan, H. Agricultural object detection with You Only Look Once (YOLO) Algorithm: A bibliometric and systematic literature review. Comput. Electron. Agric. 2024, 223, 109090. [Google Scholar] [CrossRef]
- Suto, J. Codling moth monitoring with camera-equipped automated traps: A review. Agriculture 2022, 12, 1721. [Google Scholar] [CrossRef]
- Thakker, M.; Anand, S.; Purandare, S. Assessment of Honey Bee Colony Health Using Computer Vision and Machine Learning; Indraprastha Institute of Information Technology New Delhi: New Delhi, India, 2019; p. 2016159. Available online: http://repository.iiitd.edu.in/xmlui/handle/123456789/901 (accessed on 20 October 2022).
- Bilik, S.; Kratochvila, L.; Ligocki, A.; Bostik, O.; Zemcik, T.; Hybl, M.; Horak, K.; Zalud, L. Visual diagnosis of the varroa destructor parasitic mite in honeybees using object detector techniques. Sensors 2021, 21, 2764. [Google Scholar] [CrossRef] [PubMed]
- Voudiotis, G.; Moraiti, A.; Kontogiannis, S. Deep Learning Beehive Monitoring System for Early Detection of the Varroa Mite. Signals 2022, 3, 506–523. [Google Scholar] [CrossRef]
- Jiao, L.; Xie, C.; Chen, P.; Du, J.; Li, R.; Zhang, J. Adaptive feature fusion pyramid network for multi-classes agricultural pest detection. Comput. Electron. Agric. 2022, 195, 106827. [Google Scholar] [CrossRef]
- Kaur, P.; Khehra, B.S.; Mavi, E.B.S. Data augmentation for object detection: A review. In Proceedings of the 2021 IEEE International Midwest Symposium on Circuits and Systems (MWSCAS), Lansing, MI, USA, 9–11 August 2021; pp. 537–543. [Google Scholar]
- Rendon, E.; Alejo, R.; Castorena, C.; Isidro-Ortega, F.J.; Granda-Gutierrez, E.E. Data sampling methods to deal with the big data multi-class imbalance problem. Appl. Sci. 2020, 10, 1276. [Google Scholar] [CrossRef]
- Mahmud, M.S.; Huang, J.Z.; Salloum, S.; Emara, T.Z.; Sadatdiynov, K. A survey of data partitioning and sampling methods to support big data analysis. Big Data Min. Anal. 2020, 3, 85–101. [Google Scholar] [CrossRef]
- Al-Kateb, M.; Lee, B.S. Stratified reservoir sampling over heterogeneous data streams. In International Conference on Scientific and Statistical Database Management; Springer: Berlin/Heidelberg, Germany, 2010; pp. 621–639. [Google Scholar]
- Dong, S.; Wang, R.; Liu, K.; Jiao, L.; Li, R.; Du, J.; Teng, Y.; Wang, F. CRA-Net: A channel recalibration feature pyramid network for detecting small pests. Comput. Electron. Agric. 2021, 191, 106518. [Google Scholar] [CrossRef]
- Wang, R.; Jiao, L.; Xie, C.; Chen, P.; Du, J.; Li, R. S-RPN: Sampling-balanced region proposal network for small crop pest detection. Comput. Electron. Agric. 2021, 187, 106290. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, S.; Chen, L.; Wu, W.; Wang, H.; Liu, X.; Fan, Z.; Wang, B. Microscopic Insect Pest Detection in Tea Plantations: Improved YOLOv8 Model Based on Deep Learning. Agriculture 2024, 14, 1739. [Google Scholar] [CrossRef]
- Ye, R.; Gao, Q.; Qian, Y.; Sun, J.; Li, T. Improved yolov8 and sahi model for the collaborative detection of small targets at the micro scale: A case study of pest detection in tea. Agronomy 2024, 14, 1034. [Google Scholar] [CrossRef]
- Kamilaris, A.; Prenafeta-Boldú, F.X. Deep learning in agriculture: A survey. Comput. Electron. Agric. 2018, 147, 70–90. [Google Scholar] [CrossRef]
- Rebuffi, S.-A.; Gowal, S.; Calian, D.A.; Stimberg, F.; Wiles, O.; Mann, T.A. Data augmentation can improve robustness. Adv. Neural Inf. Process. Syst. 2021, 34, 29935–29948. [Google Scholar]
- Bosquet, B.; Cores, D.; Seidenari, L.; Brea, V.M.; Mucientes, M.; Del Bimbo, A. A full data augmentation pipeline for small object detection based on generative adversarial networks. Pattern Recognit. 2023, 133, 108998. [Google Scholar] [CrossRef]
- Kisantal, M.; Wojna, Z.; Murawski, J.; Naruniec, J.; Cho, K. Augmentation for small object detection. arXiv 2019, arXiv:1902.07296. [Google Scholar] [CrossRef]
- Mahmoud, H.; Kurniawan, I.F.; Aneiba, A.; Asyhari, A.T. Enhancing detection of remotely-sensed floating objects via Data Augmentation for Maritime SAR. J. Indian Soc. Remote Sens. 2024, 52, 1285–1295. [Google Scholar] [CrossRef]
- Kumar, T.; Brennan, R.; Mileo, A.; Bendechache, M. Image data augmentation approaches: A comprehensive survey and future directions. IEEE Access 2024, 12, 187536–187571. [Google Scholar] [CrossRef]
- Shorten, C.; Khoshgoftaar, T.M. A survey on image data augmentation for deep learning. J. Big data 2019, 6, 60. [Google Scholar] [CrossRef]
- Lee, H.G.; Kim, M.-J.; Kim, S.-B.; Lee, S.; Lee, H.; Sin, J.Y.; Mo, C. Identifying an image-processing method for detection of bee mite in honey bee based on keypoint analysis. Agriculture 2023, 13, 1511. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Chaudhuri, S.; Das, G.; Narasayya, V. Optimized stratified sampling for approximate query processing. ACM Trans. Database Syst. (TODS) 2007, 32, 9. [Google Scholar] [CrossRef]
- Bochkovskiy, A.; Wang, C.-Y.; Liao, H.-Y.M. Yolov4: Optimal speed and accuracy of object detection. arXiv 2020, arXiv:2004.10934. [Google Scholar] [CrossRef]
- Liu, M.; Cui, M.; Xu, B.; Liu, Z.; Li, Z.; Chu, Z.; Zhang, X.; Liu, G.; Xu, X.; Yan, Y. Detection of Varroa destructor Infestation of Honeybees Based on Segmentation and Object Detection Convolutional Neural Networks. AgriEngineering 2023, 5, 1644–1662. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).