Morphological Estimation of Primary Branch Inclination Angles in Jujube Trees Based on Improved PointNet++
Abstract
1. Introduction
2. Materials and Methods
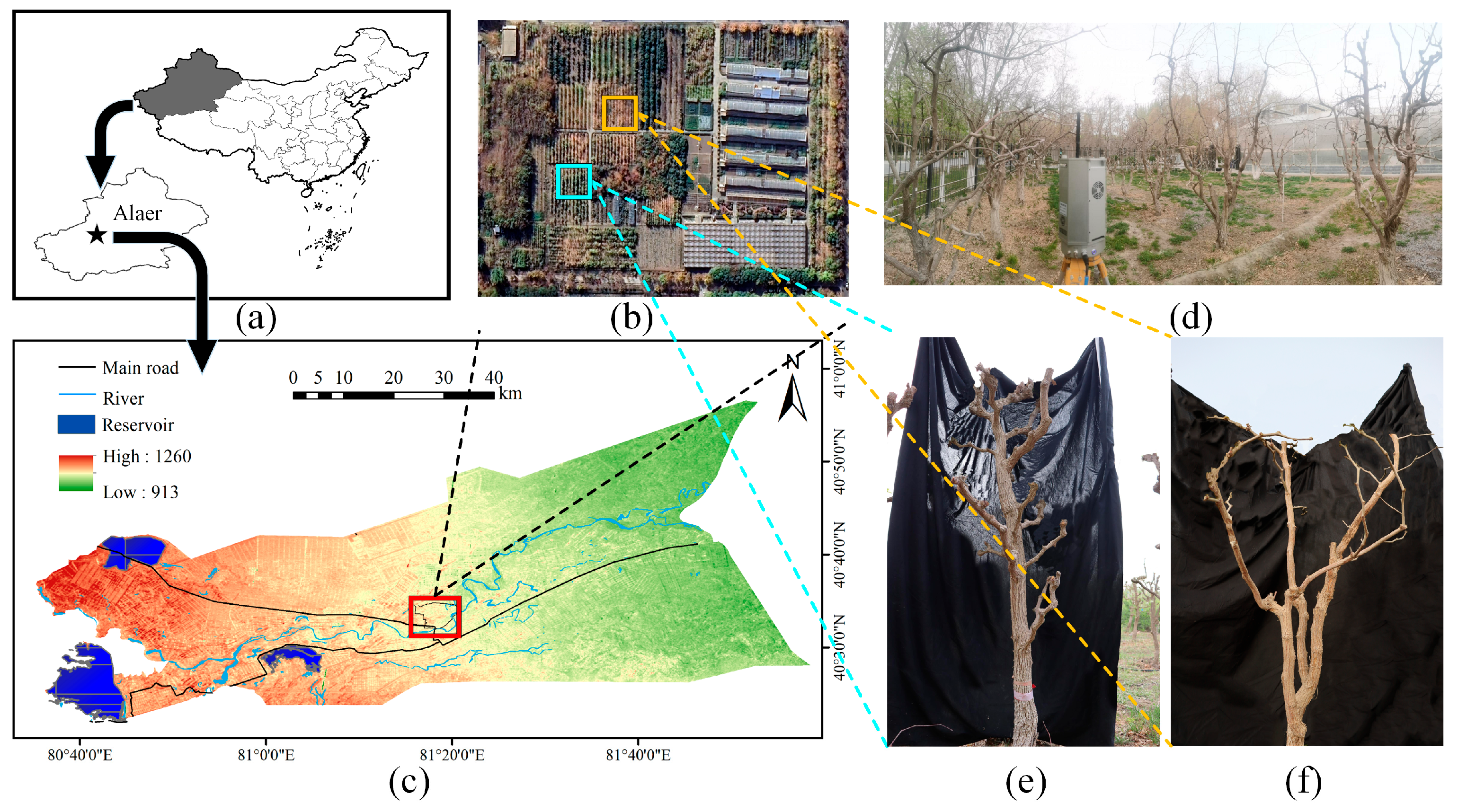
2.1. Experimental Site
2.2. Jujube Tree Dataset
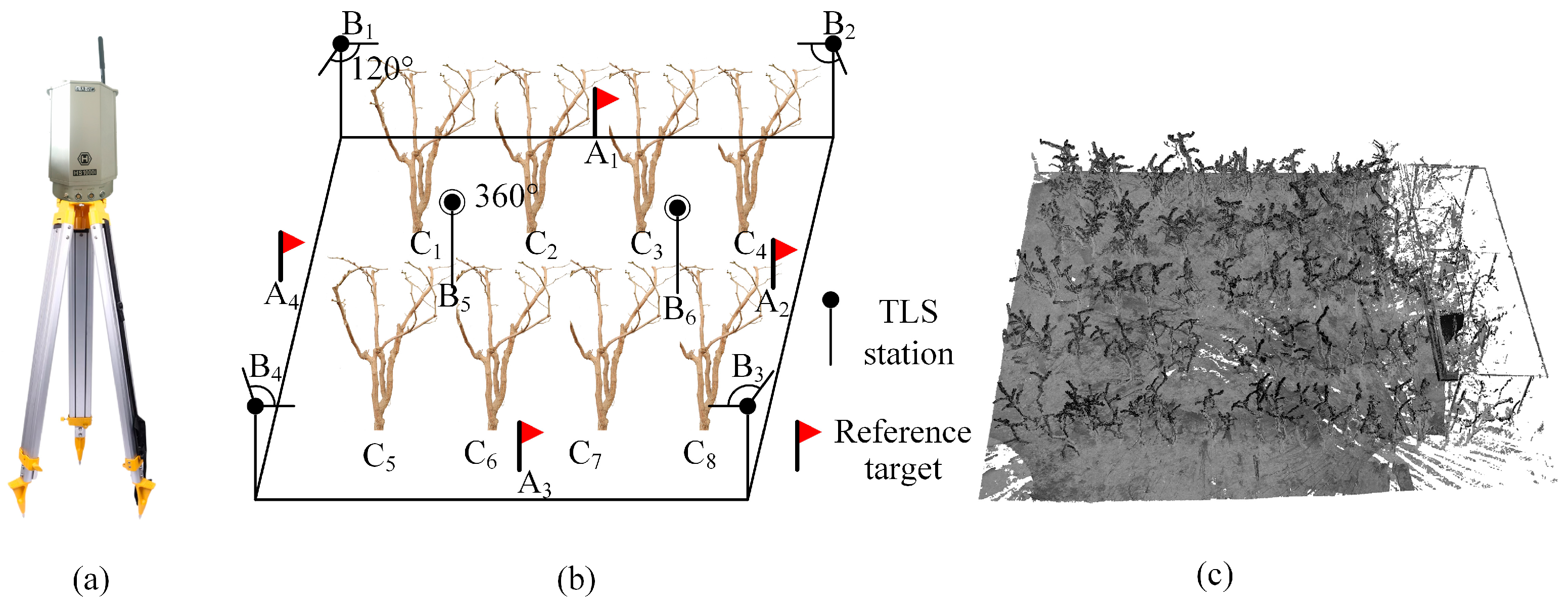
2.2.1. Jujube Tree Point Cloud Acquisition
2.2.2. Point Cloud Preprocessing
2.2.3. Jujube Tree Dataset Overview
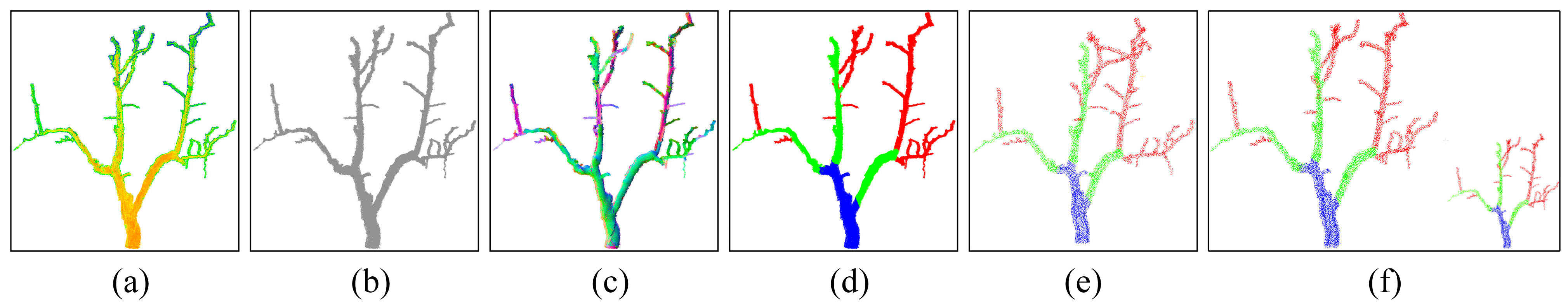
2.3. Point Cloud Segmentation
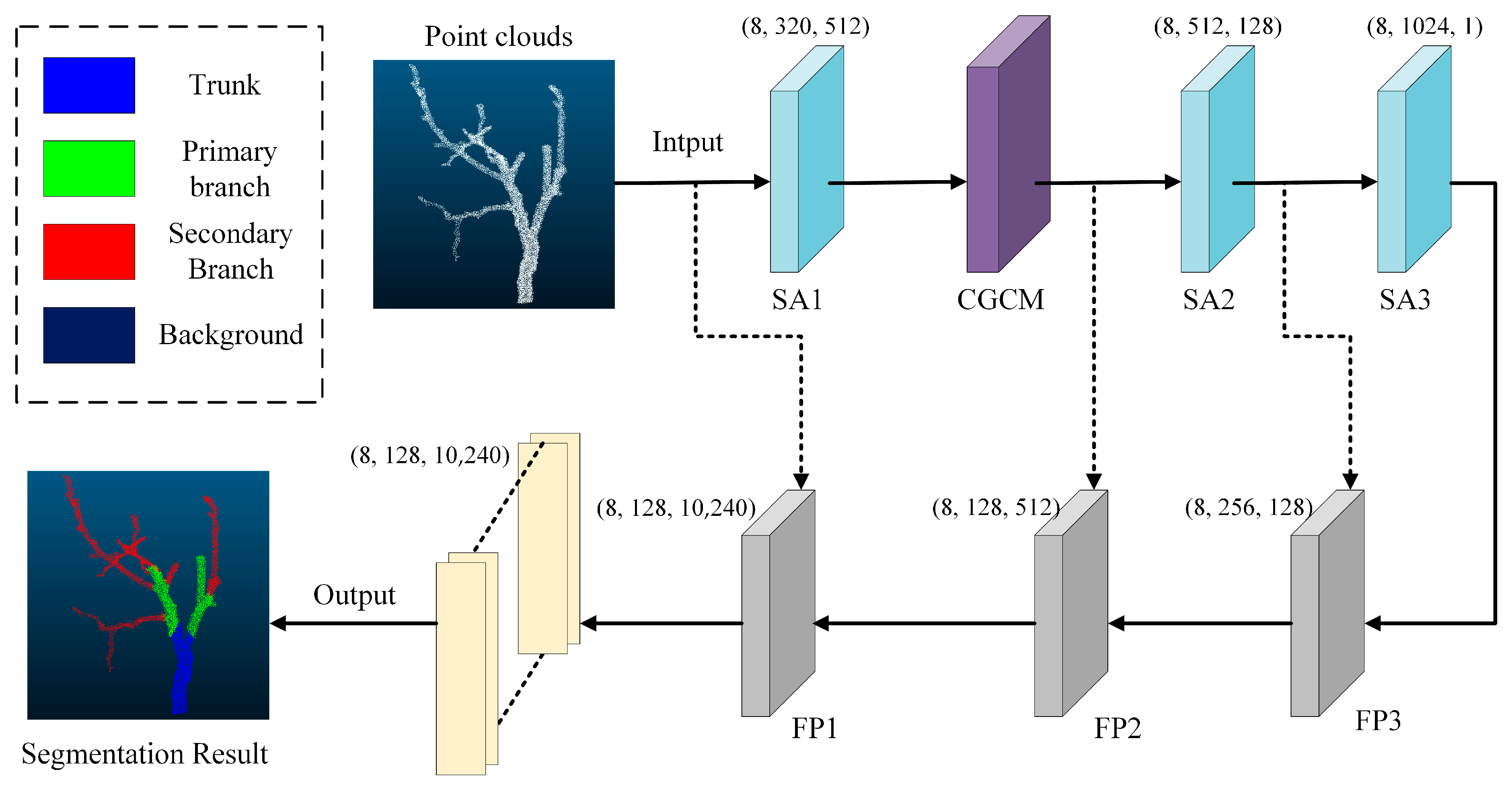
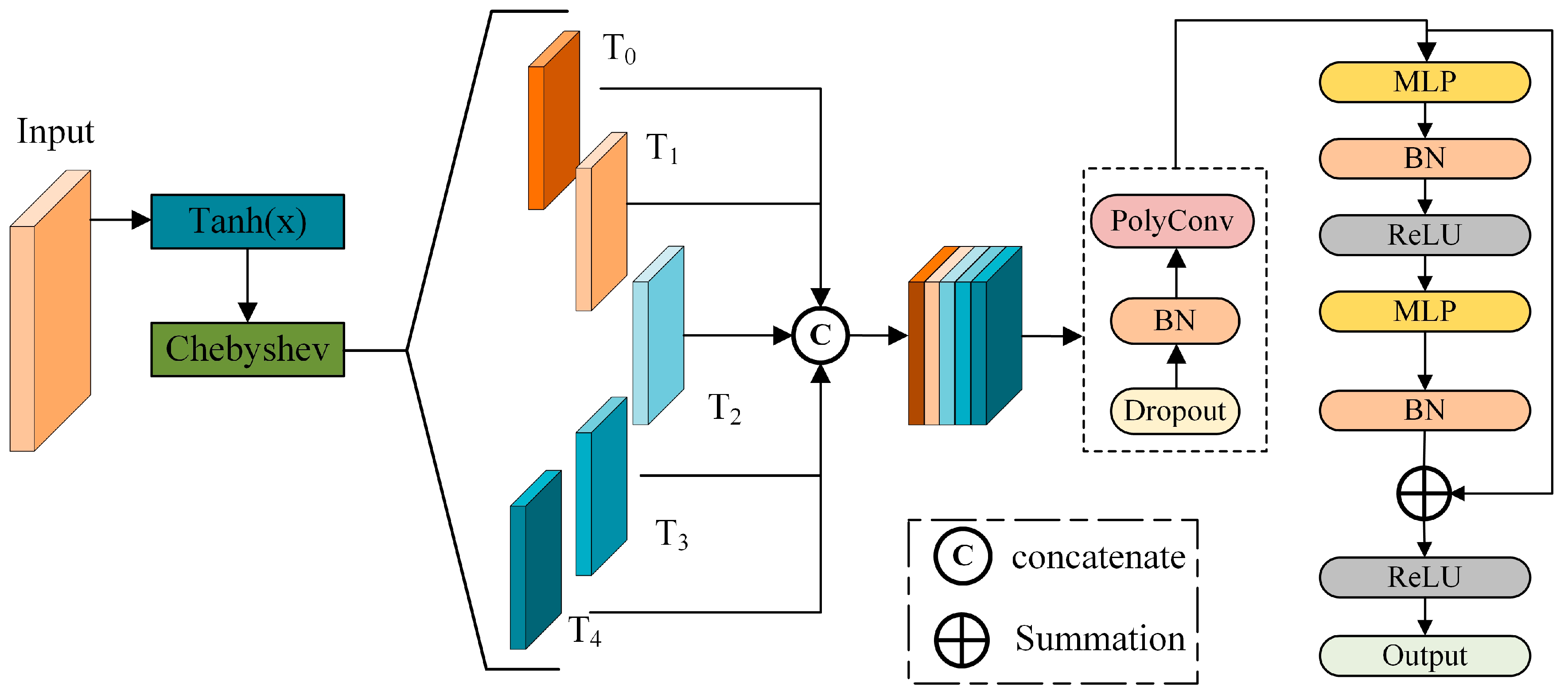
2.3.1. Improvement of the PointNet++
2.3.2. Instance Segmentation of Point Clouds
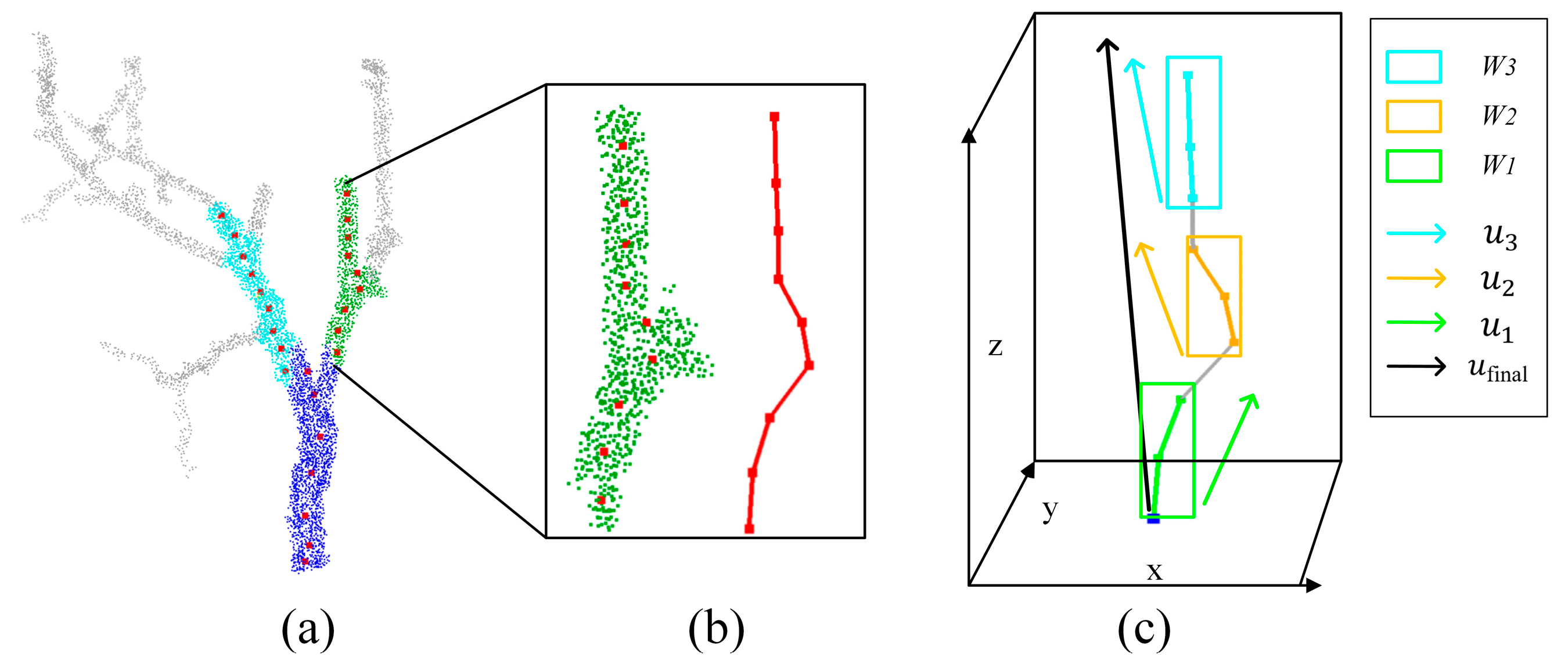
2.4. Estimation of Primary Branch Inclination Angles
2.5. Evaluation Metrics
2.5.1. Point Cloud Semantic Segmentation
2.5.2. Primary Branch Inclination Estimation
2.5.3. DBSCAN Clustering
3. Results
3.1. Semantic Segmentation of Jujube Tree Branches
3.1.1. Network Training
3.1.2. Comparison of Semantic Segmentation of Branches
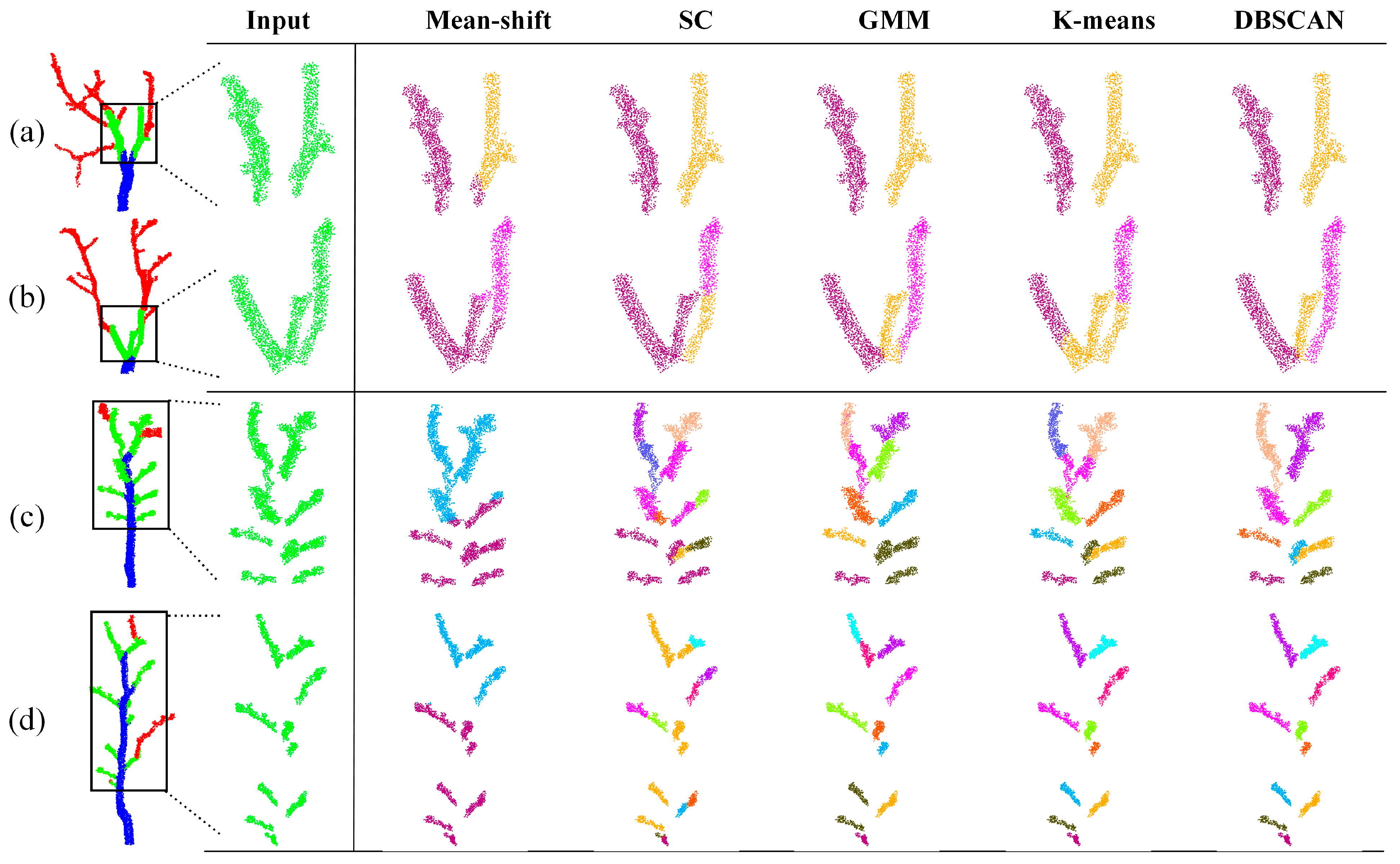
3.2. Instance Segmentation of Jujube Tree Branches
3.3. Skeleton Extraction of Jujube Tree Branches
3.4. Estimation of the Inclination Angle of Primary Branches
3.5. Ablation Experiments
4. Discussion
4.1. Analysis of Point Cloud Data Acquisition
4.2. Analysis of Point Cloud Segmentation
4.3. Analysis of Inclination Angle Estimation
4.4. Analysis of Experimental Results
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, B.; Chen, X.G.; Zhang, H.M.; Shen, C.J.; Fu, W. Design and performance test of a jujube pruning manipulator. Agriculture 2022, 12, 552. [Google Scholar] [CrossRef]
- Ma, B.J.; Du, J.; Wang, L.; Jiang, H.Y.; Zhou, M.C. Automatic branch detection of jujube trees based on 3D reconstruction for dormant pruning using the deep learning-based method. Comput. Electron. Agric. 2021, 190, 106484. [Google Scholar] [CrossRef]
- Kolmanič, S.; Strnad, D.; Kohek, Š.; Benes, B.; Hirst, P.; Žalik, B. An algorithm for automatic dormant tree pruning. Appl. Soft Comput. 2021, 99, 106931. [Google Scholar] [CrossRef]
- Xie, B.B.; Jin, Y.C.; Faheem, M.; Gao, W.J.; Liu, J.Z.; Jiang, H.K.; Cai, L.J.; Li, Y.X. Research progress of autonomous navigation technology for multi-agricultural scenes. Comput. Electron. Agric. 2023, 211, 107963. [Google Scholar] [CrossRef]
- Akbar, S.A.; Elfiky, N.M.; Kak, A. A novel framework for modeling dormant apple trees using single depth image for robotic pruning application. In Proceedings of the IEEE International Conference on Robotics and Automation, Stockholm, Sweden, 16–21 May 2016; pp. 5136–5142. [Google Scholar]
- Li, Y.L.; Zhang, Z.Y.; Wang, X.F.; Fu, W.; Li, J.B. Automatic reconstruction and modeling of dormant jujube trees using three-view image constraints for intelligent pruning applications. Comput. Electron. Agric. 2023, 212, 108149. [Google Scholar] [CrossRef]
- Qiao, Y.C.; Hu, Y.H.; Zheng, Z.Z.; Qu, Z.H.; Wang, C.; Guo, T.F.; Hou, J.C. A diameter measurement method of red jujubes trunk based on improved PSPNet. Agriculture 2022, 12, 1140. [Google Scholar] [CrossRef]
- Zahid, A.; Mahmud, M.S.; He, L.; Heinemann, P.; Choi, D.; Schupp, J. Technological advancements towards developing a robotic pruner for apple trees: A review. Comput. Electron. Agric. 2021, 189, 106383. [Google Scholar] [CrossRef]
- Lei, X.H.; Yuan, Q.C.; Xyu, T.; Qi, Y.N.; Zeng, J.; Huang, K.; Sun, Y.H.; Herbst, A.; Lyu, X.L. Technologies and Equipment of Mechanized Blossom Thinning in Orchards: A Review. Agronomy 2023, 13, 2753. [Google Scholar] [CrossRef]
- Wu, B.X.; Zheng, G.; Chen, Y.; Yu, D.S. Assessing inclination angles of tree branches from terrestrial laser scan data using a skeleton extraction method. Int. J. Appl. Earth Obs. Geoinf. 2021, 104, 102589. [Google Scholar] [CrossRef]
- Mo, J.W.; Lan, Y.B.; Yang, D.Z.; Wen, F.; Qiu, H.B.; Chen, X.; Deng, X.L. Deep learning-based instance segmentation method of litchi canopy from UAV-acquired images. Remote Sens. 2021, 13, 3919. [Google Scholar] [CrossRef]
- Pérez-Borrero, I.; Marín-Santos, D.; Gegúndez-Arias, M.E.; Cortés-Ancos, E. A fast and accurate deep learning method for strawberry instance segmentation. Comput. Electron. Agric. 2020, 178, 105736. [Google Scholar] [CrossRef]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- Ling, S.K.; Wang, N.Y.; Li, J.B.; Ding, L.P. Accurate Recognition of Jujube Tree Trunks Based on Contrast Limited Adaptive Histogram Equalization Image Enhancement and Improved YOLOv8. Forests 2024, 15, 625. [Google Scholar] [CrossRef]
- Sun, Y.Z.; Zhang, Z.X.; Sun, K.; Li, S.; Yu, J.L.; Miao, L.X.; Zhang, Z.G.; Li, Y.; Zhao, H.J.; Hu, Z.B. Soybean-MVS: Annotated three-dimensional model dataset of whole growth period soybeans for 3D plant organ segmentation. Agriculture 2023, 13, 1321. [Google Scholar] [CrossRef]
- Westling, F.; Underwood, J.; Bryson, M. A procedure for automated tree pruning suggestion using LiDAR scans of fruit trees. Comput. Electron. Agric. 2021, 187, 106274. [Google Scholar] [CrossRef]
- Gamal, A.; Wibisono, A.; Wicaksono, S.B.; Abyan, M.A.; Hamid, N.; Wisesa, H.A.; Jatmiko, W.; Ardhianto, R. Automatic LIDAR building segmentation based on DGCNN and euclidean clustering. J. Big Data 2020, 7, 102. [Google Scholar] [CrossRef]
- Suthaharan, S. Support vector machine. In Machine Learning Models and Algorithms for Big Data Classification: Thinking with Examples for Effective Learning; Springer: Boston, MA, USA, 2016; Volume 36, pp. 207–235. [Google Scholar]
- Sa, I.; Lehnert, C.; English, A.; McCool, C.; Dayoub, F.; Upcroft, B.; Perez, T. Peduncle detection of sweet pepper for autonomous crop harvesting—Combined color and 3-D information. IEEE Robot. Autom. Lett. 2017, 2, 765–772. [Google Scholar] [CrossRef]
- Wahabzada, M.; Paulus, S.; Kersting, K.; Mahlein, A.-K. Automated interpretation of 3D laserscanned point clouds for plant organ segmentation. BMC Bioinform. 2015, 16, 248. [Google Scholar] [CrossRef]
- Jin, S.C.; Su, Y.J.; Wu, F.F.; Pang, S.X.; Gao, S.; Hu, T.Y.; Liu, J.; Guo, Q.H. Stem–leaf segmentation and phenotypic trait extraction of individual maize using terrestrial LiDAR data. IEEE Trans. Geosci. Remote Sens. 2018, 57, 1336–1346. [Google Scholar] [CrossRef]
- Qiu, T.; Cheng, L.L.; Jiang, Y. 3D characterization of tree architecture for apple crop load estimation. In Proceedings of the 2022 ASABE Annual International Meeting, Houston, TX, USA, 17–20 July 2022; p. 1. [Google Scholar]
- Itakura, K.; Miyatani, S.; Hosoi, F. Estimating tree structural parameters via automatic tree segmentation from LiDAR point cloud data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2021, 15, 555–564. [Google Scholar] [CrossRef]
- Saeed, F.; Li, C. Plant organ segmentation from point clouds using Point-Voxel CNN. In Proceedings of the 2021 ASABE Annual International Virtual Meeting, Online, 12–16 July 2021; p. 1. [Google Scholar]
- Guo, R.C.; Xie, J.L.; Zhu, J.X.; Cheng, R.F.; Zhang, Y.; Zhang, X.H.; Gong, X.J.; Zhang, R.W.; Wang, H.; Meng, F.F. Improved 3D point cloud segmentation for accurate phenotypic analysis of cabbage plants using deep learning and clustering algorithms. Comput. Electron. Agric. 2023, 211, 108014. [Google Scholar] [CrossRef]
- Qi, C.R.; Su, H.; Mo, K.; Guibas, L.J. Pointnet: Deep learning on point sets for 3d classification and segmentation. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR 2017), Honolulu, HI, USA, 21–26 July 2017; pp. 652–660. [Google Scholar]
- Qi, C.R.; Yi, L.; Su, H.; Guibas, L.J. Pointnet++: Deep hierarchical feature learning on point sets in a metric space. In Proceedings of the Advances in Neural Information Processing Systems 30: Annual Conference on Neural Information Processing Systems 2017, Long Beach, CA, USA, 4–9 December 2017; pp. 1–5. [Google Scholar]
- Sun, X.M.; He, L.L.; Jiang, H.H.; Li, R.; Mao, W.L.; Zhang, D.; Majeed, Y.; Andriyanov, N.; Soloviev, V.; Fu, L.S. Morphological estimation of primary branch length of individual apple trees during the deciduous period in modern orchard based on PointNet++. Comput. Electron. Agric. 2024, 220, 108873. [Google Scholar] [CrossRef]
- Ao, Z.R.; Wu, F.F.; Hu, S.H.; Sun, Y.; Su, Y.J.; Guo, Q.H.; Xin, Q.C. Automatic segmentation of stem and leaf components and individual maize plants in field terrestrial LiDAR data using convolutional neural networks. Crop J. 2022, 10, 1239–1250. [Google Scholar] [CrossRef]
- Chang, A.X.; Funkhouser, T.; Guibas, L.; Hanrahan, P.; Huang, Q.; Li, Z.; Savarese, S.; Savva, M.; Song, S.; Su, H. Shapenet: An information-rich 3d model repository. arXiv 2015, arXiv:1512.03012. [Google Scholar]
- Defferrard, M.; Bresson, X.; Vandergheynst, P. Convolutional neural networks on graphs with fast localized spectral filtering. Adv. Neural Inf. Process. Syst. 2016, 29, 1–9. [Google Scholar]
- Ferrara, R.; Virdis, S.G.P.; Ventura, A.; Ghisu, T.; Duce, P.; Pellizzaro, G. An automated approach for wood-leaf separation from terrestrial LIDAR point clouds using the density based clustering algorithm DBSCAN. Agric. For. Meteorol. 2018, 262, 434–444. [Google Scholar] [CrossRef]
- Wang, C.; Ji, M.; Wang, J.; Wen, W.; Li, T.; Sun, Y. An improved DBSCAN method for LiDAR data segmentation with automatic Eps estimation. Sensors 2019, 19, 172. [Google Scholar] [CrossRef]
- Tagliasacchi, A.; Zhang, H.; Cohen-Or, D. Curve skeleton extraction from incomplete point cloud. ACM Trans. 2009, 28, 71. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, Y.B.; Liu, Z.W.; Sarma, S.E.; Bronstein, M.M.; Solomon, J.M. Dynamic graph cnn for learning on point clouds. ACM Trans. Graph. 2019, 38, 146. [Google Scholar] [CrossRef]
- Zheng, S.; Pan, J.Q.; Lu, C.J.; Gupta, G. Pointnorm: Dual normalization is all you need for point cloud analysis. In Proceedings of the 2023 International Joint Conference on Neural Networks (IJCNN), Gold Coast, Australia, 18–23 June 2023; pp. 1–8. [Google Scholar]
- Ma, X.; Qin, C.; You, H.X.; Ran, H.X.; Fu, Y. Rethinking network design and local geometry in point cloud: A simple residual MLP framework. arXiv 2022, arXiv:2202.07123. [Google Scholar] [CrossRef]
- Belton, D.; Moncrieff, S.; Chapman, J. Processing tree point clouds using Gaussian Mixture Models. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2013, 2, 43–48. [Google Scholar] [CrossRef]
- Miao, Y.L.; Li, S.; Wang, L.Y.; Li, H.; Qiu, R.C.; Zhang, M. A single plant segmentation method of maize point cloud based on Euclidean clustering and K-means clustering. Comput. Electron. Agric. 2023, 210, 107951. [Google Scholar] [CrossRef]
- Ma, T.; Wu, Z.Z.; Feng, L.; Luo, P.; Long, X. Point cloud segmentation through spectral clustering. In Proceedings of the 2nd International Conference on Information Science and Engineering, Hangzhou, China, 4–6 December 2010; pp. 1–4. [Google Scholar]
- Carreira-Perpiñán, M.Á. A review of mean-shift algorithms for clustering. arXiv 2015, arXiv:1503.00687. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, W.; Yang, G.; Lei, L.; Han, S.; Xu, W.; Chen, R.; Zhang, C.; Yang, H. Maize ear height and ear–plant height ratio estimation with LiDAR data and vertical leaf area profile. Remote Sens. 2023, 15, 964. [Google Scholar] [CrossRef]
- Shen, J.; Wu, T.; Zhao, J.; Wu, Z.; Huang, Y.; Gao, P.; Zhang, L. Organ segmentation and phenotypic trait extraction of cotton seedling point clouds based on a 3D lightweight network. Agronomy 2024, 14, 1083. [Google Scholar] [CrossRef]
- Liu, F.-Y.; Geng, H.; Shang, L.-Y.; Si, C.-J.; Shen, S.-Q. A cotton organ segmentation method with phenotypic measurements from a point cloud using a transformer. Plant Methods 2025, 21, 37. [Google Scholar] [CrossRef]
- Zhu, T.; Ma, X.; Guan, H.; Wu, X.; Wang, F.; Yang, C.; Jiang, Q. A method for detecting tomato canopies’ phenotypic traits based on improved skeleton extraction algorithm. Comput. Electron. Agric. 2023, 214, 108285. [Google Scholar] [CrossRef]
- Miao, T.; Zhu, C.; Xu, T.; Yang, T.; Li, N.; Zhou, Y.; Deng, H. Automatic stem-leaf segmentation of maize shoots using three-dimensional point cloud. Comput. Electron. Agric. 2021, 187, 106310. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, Q.; Yang, J.; Ren, G.; Wang, W.; Zhang, W.; Li, F. A method for tomato plant stem and leaf segmentation and phenotypic extraction based on skeleton extraction and supervoxel clustering. Agronomy 2024, 14, 198. [Google Scholar] [CrossRef]


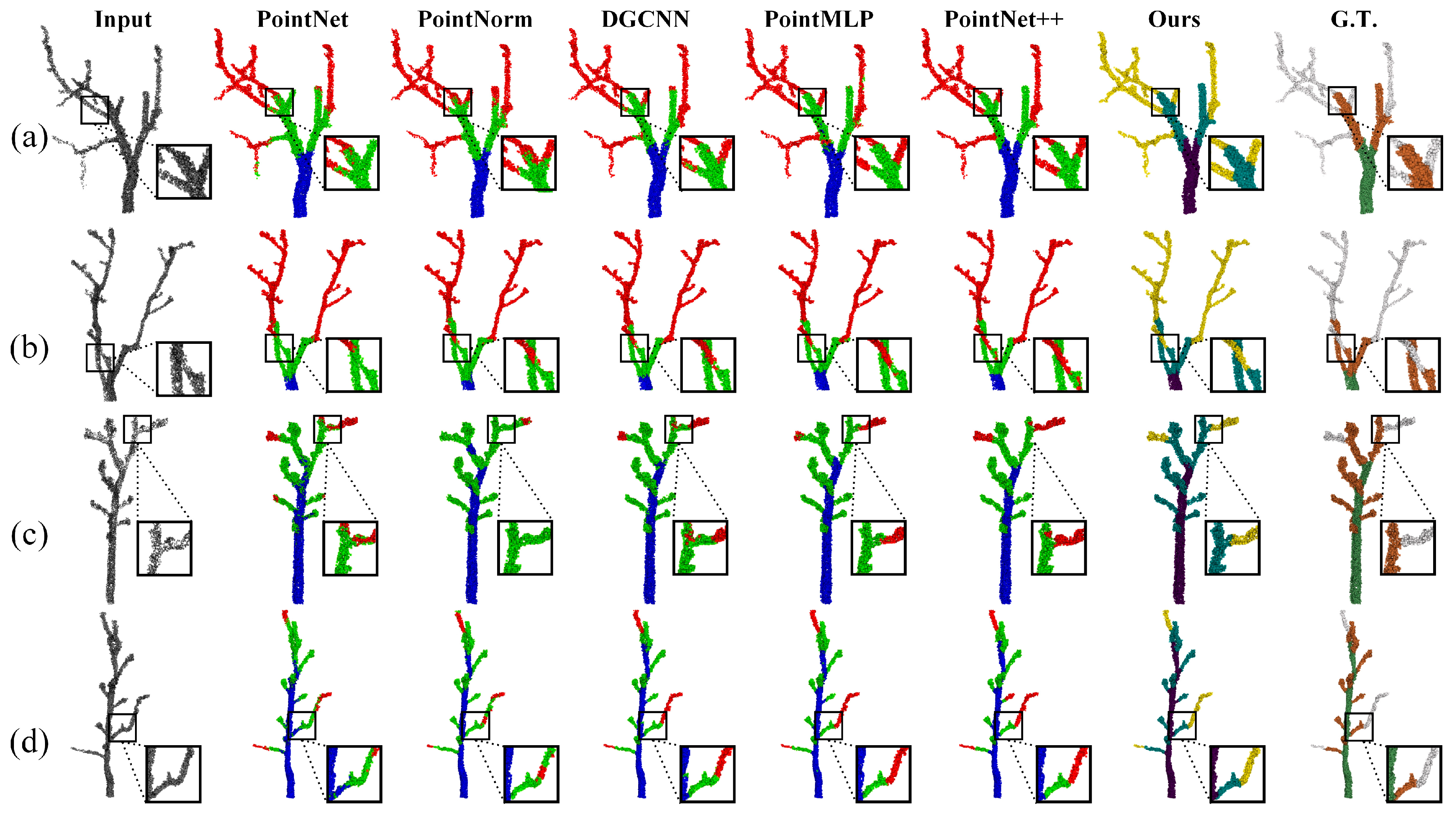


| Model | Acc | CAA | mIoU | Ocs_mIoU | Mts_mIoU |
|---|---|---|---|---|---|
| PointNet | 84.47 | 83.18 | 69.6 | 75.0 | 64.3 |
| PointNorm | 87.69 | 85.28 | 72.7 | 73.7 | 71.7 |
| DGCNN | 90.06 | 87.8 | 80.0 | 75.0 | 69.0 |
| PointMLP | 94.76 | 94.35 | 88.7 | 89.3 | 88.2 |
| PointNet++ | 95.94 | 95.79 | 90.5 | 88.1 | 93.0 |
| Ours | 97.27 | 97.26 | 93.83 | 95.2 | 92.47 |
| Method | ARI | NMI | FMI | Completeness |
|---|---|---|---|---|
| Mean-Shift | 0.459 | 0.717 | 0.584 | 0.772 |
| SC | 0.810 | 0.856 | 0.846 | 0.869 |
| GMM | 0.846 | 0.890 | 0.873 | 0.878 |
| K-Means | 0.861 | 0.910 | 0.884 | 0.898 |
| DBSCAN | 0.982 | 0.989 | 0.986 | 0.999 |
| Model | Poly | ChebPoly | ResBlock | Acc | CAA | mIoU | Ocs_mIoU | Mts_mIoU |
|---|---|---|---|---|---|---|---|---|
| PointNet++ | ✗ | ✗ | ✗ | 95.94 | 95.79 | 90.54 | 92.97 | 88.10 |
| PolyNet | ✓ | ✗ | ✗ | 96.50 | 96.40 | 92.2 | 93.02 | 91.42 |
| PolyResNet | ✓ | ✗ | ✓ | 96.78 | 96.67 | 92.78 | 93.29 | 92.26 |
| ChebPolyNet | ✓ | ✓ | ✗ | 96.93 | 96.59 | 92.52 | 93.42 | 91.62 |
| CGCM-PointNet++ | ✓ | ✓ | ✓ | 97.27 | 97.26 | 93.83 | 95.20 | 92.47 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shang, L.; Yan, F.; Teng, T.; Pan, J.; Zhou, L.; Xia, C.; Li, C.; Shi, M.; Si, C.; Niu, R. Morphological Estimation of Primary Branch Inclination Angles in Jujube Trees Based on Improved PointNet++. Agriculture 2025, 15, 1193. https://doi.org/10.3390/agriculture15111193
Shang L, Yan F, Teng T, Pan J, Zhou L, Xia C, Li C, Shi M, Si C, Niu R. Morphological Estimation of Primary Branch Inclination Angles in Jujube Trees Based on Improved PointNet++. Agriculture. 2025; 15(11):1193. https://doi.org/10.3390/agriculture15111193
Chicago/Turabian StyleShang, Linyuan, Fenfen Yan, Tianxin Teng, Junzhang Pan, Lei Zhou, Chao Xia, Chenlin Li, Mingdeng Shi, Chunjing Si, and Rong Niu. 2025. "Morphological Estimation of Primary Branch Inclination Angles in Jujube Trees Based on Improved PointNet++" Agriculture 15, no. 11: 1193. https://doi.org/10.3390/agriculture15111193
APA StyleShang, L., Yan, F., Teng, T., Pan, J., Zhou, L., Xia, C., Li, C., Shi, M., Si, C., & Niu, R. (2025). Morphological Estimation of Primary Branch Inclination Angles in Jujube Trees Based on Improved PointNet++. Agriculture, 15(11), 1193. https://doi.org/10.3390/agriculture15111193




