High Resistance and Yield: A New Cultivar ‘ZJLZS002’ of Lyophyllum decastes Suitable for Industrial Cultivation
Abstract
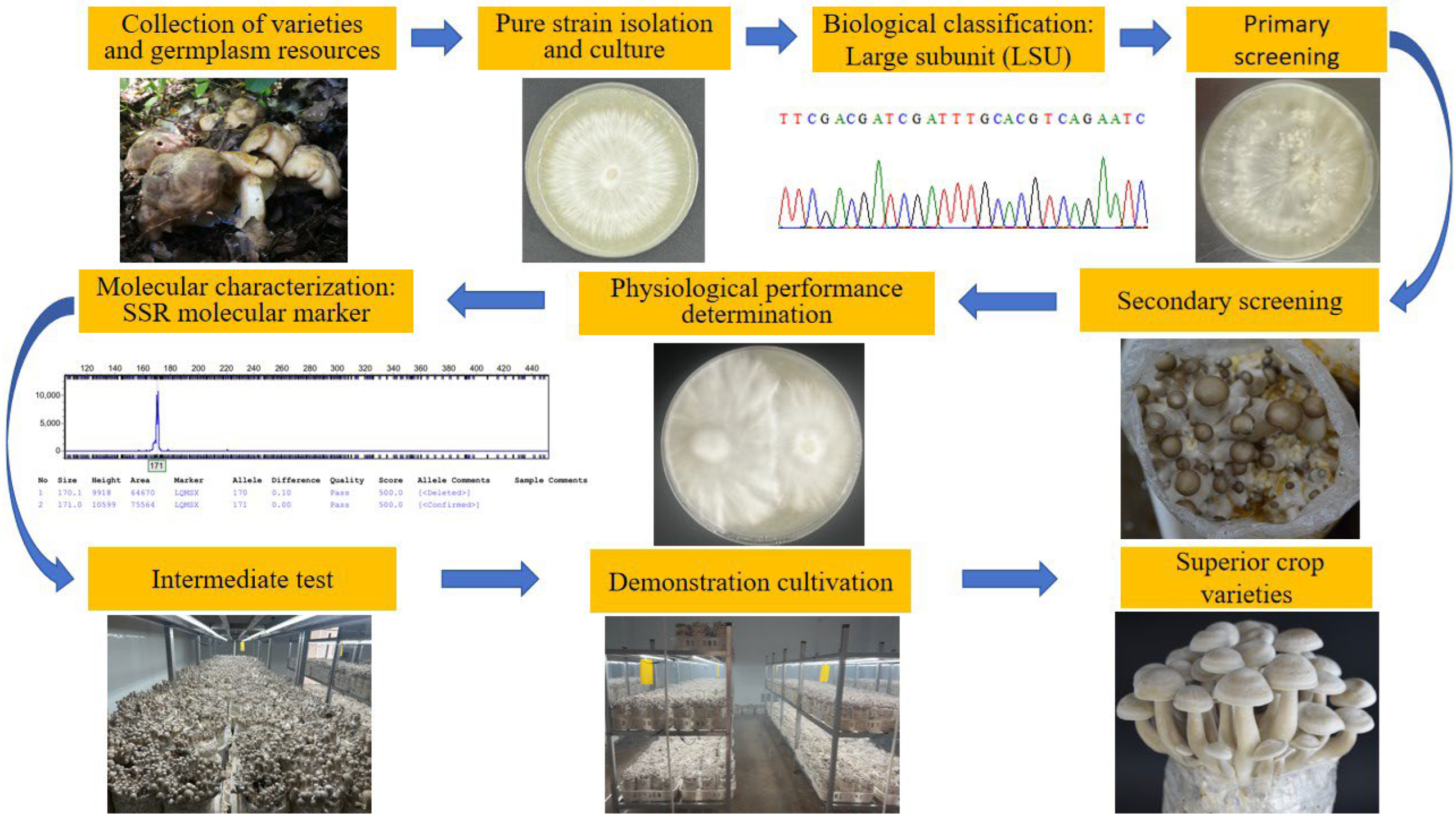
1. Introduction
2. Materials and Methods
2.1. Materials
2.1.1. Tested Strains
2.1.2. Culture Conditions
2.2. Methods
2.2.1. Tissue Isolation and Purification
2.2.2. Biological Classification
2.2.3. Primary Screening
2.2.4. Secondary Screening
2.2.5. Physiological Performance Determination of Potential Strains
Antagonistic Reaction Determination
Antibacterial TEST
2.2.6. Molecular Characterization: SSR Molecular Markers
2.2.7. Intermediate Test
2.2.8. Demonstration Cultivation
2.2.9. Determination of Nutrient Content in Fruiting Body
2.3. Analysis of Data
3. Results
3.1. Isolation of Strains
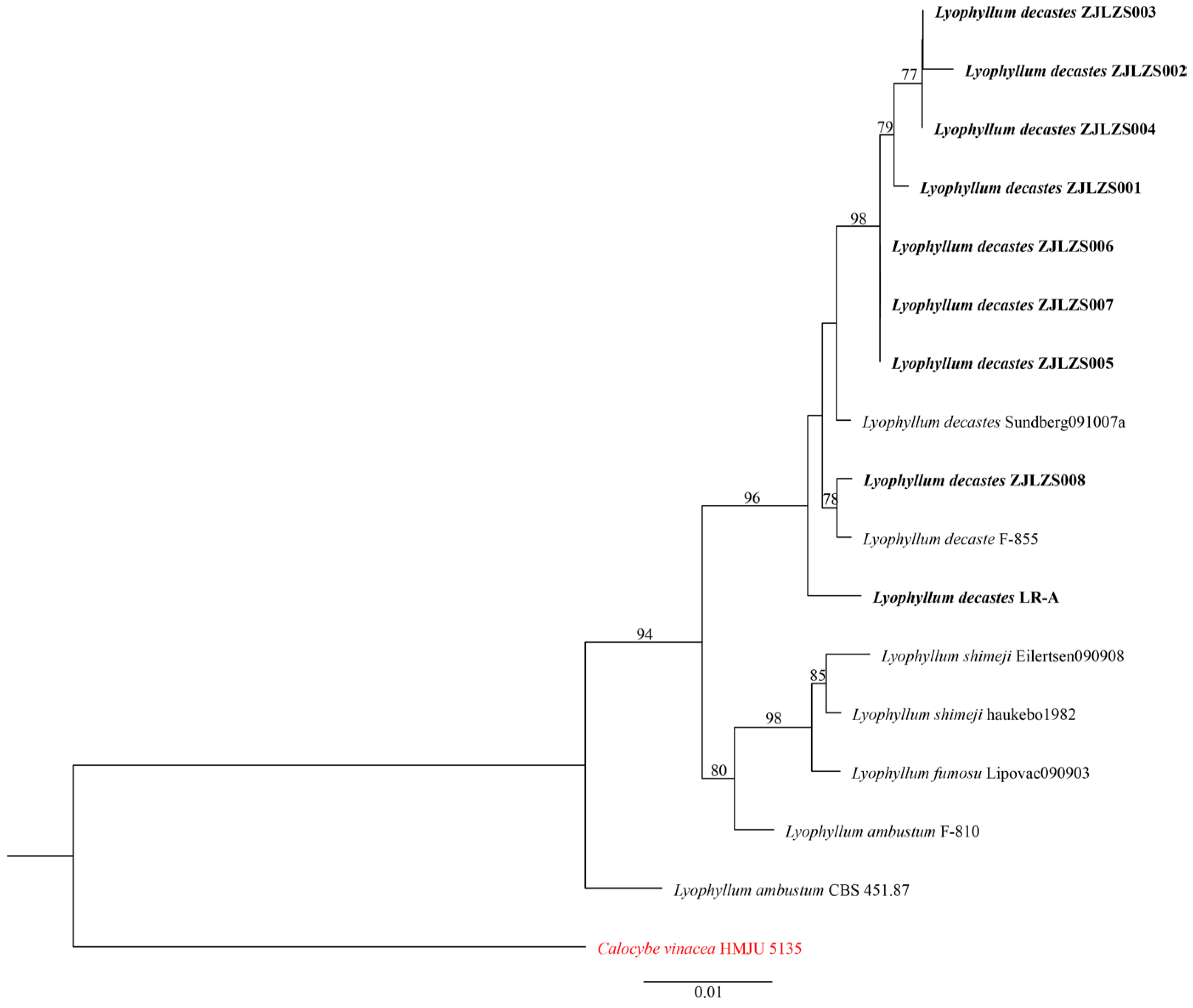
3.2. Phylogenetic Analyses
3.3. Comparison of Preliminary Screening
3.4. Comparison of Secondary Screening
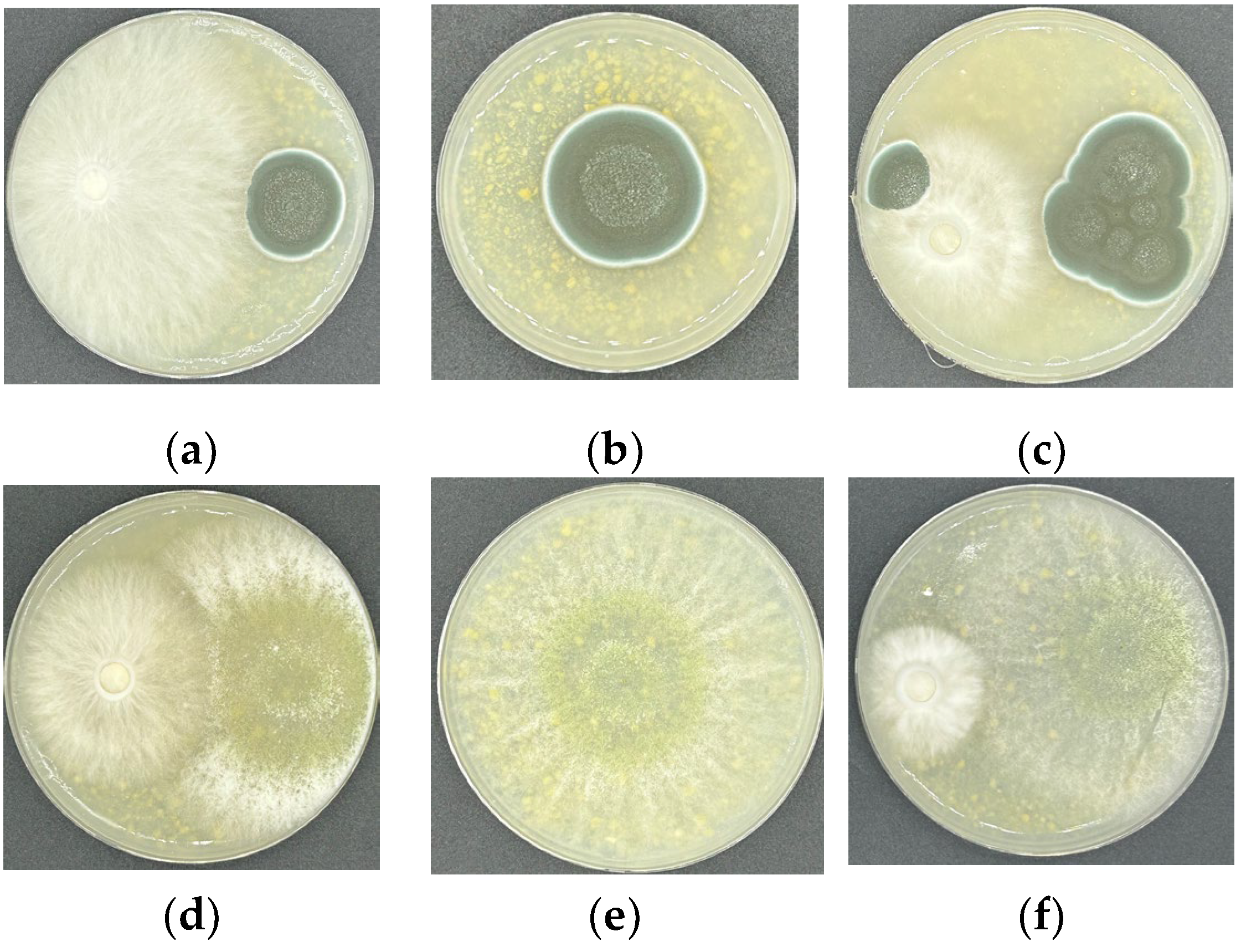
3.5. Results of Antagonistic Reaction
3.6. Results of Antibacterial Test
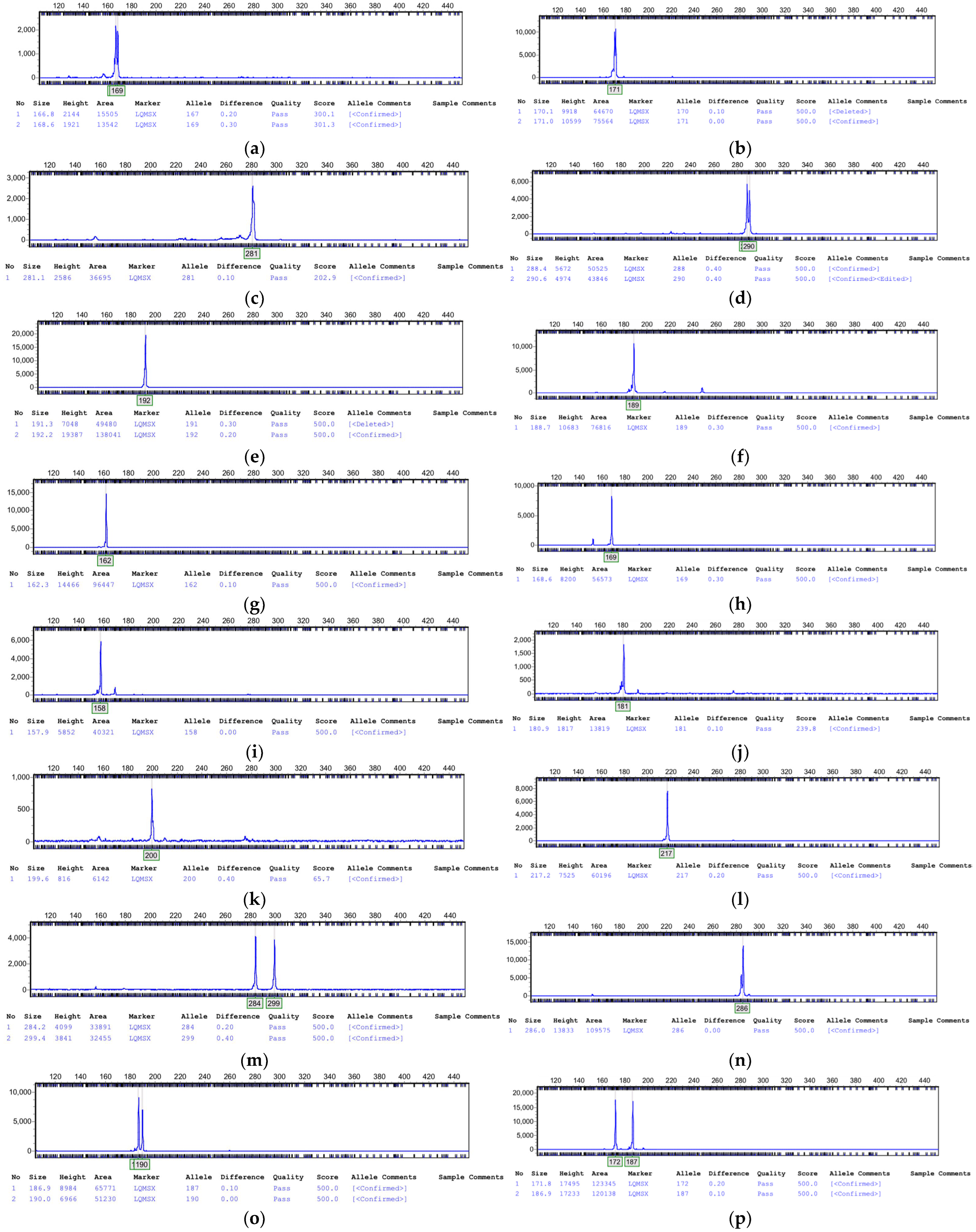
3.7. Amplification Results of SSR
3.8. Results of Intermediate Test
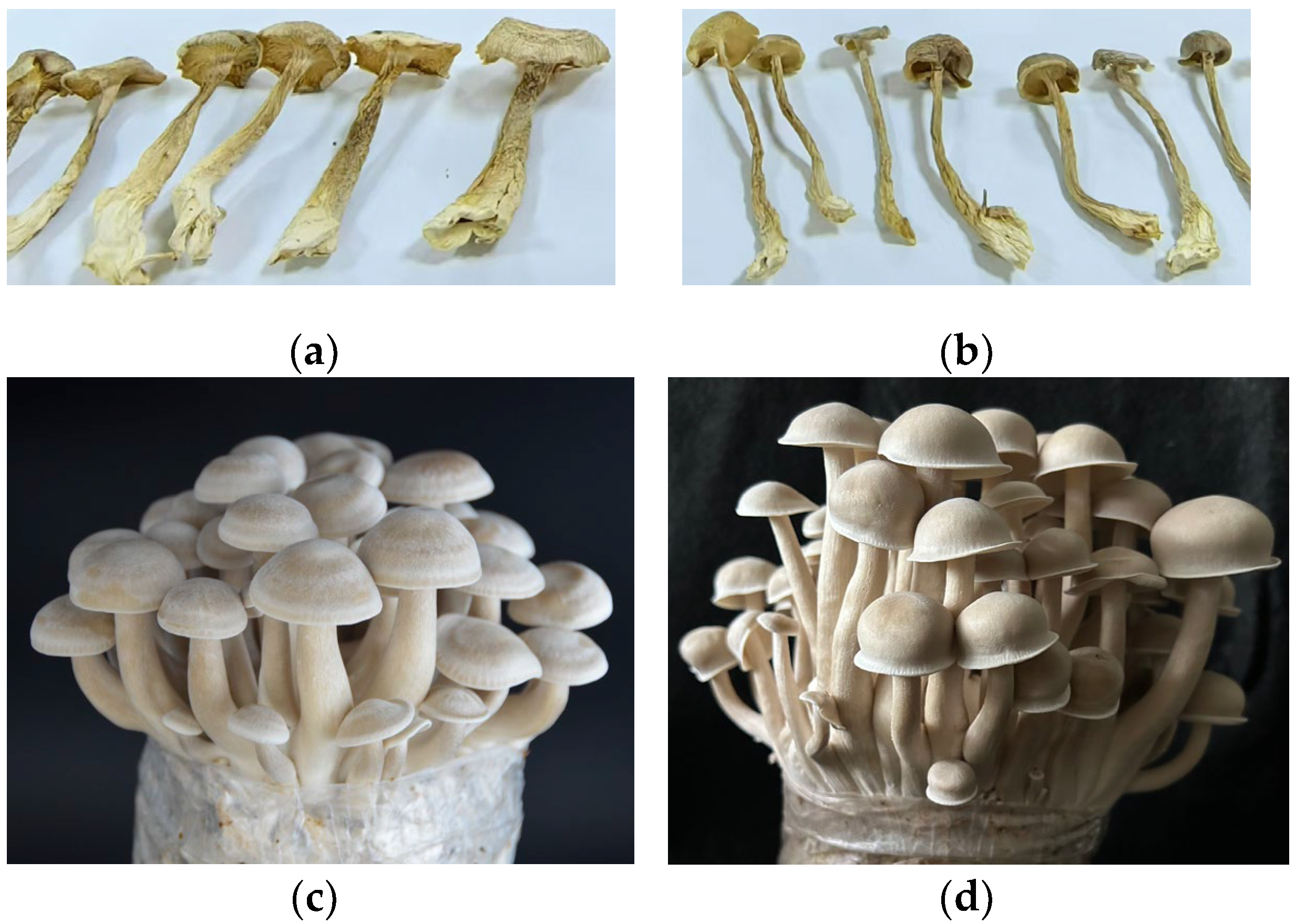
3.9. Results of Demonstration Cultivation
3.10. Nutrient Contents of ZJLZS002 Fruiting Bodies
4. Discussion
5. Conclusions
6. Patents
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, J.H.; Zheng, H.F.; Ben, W.D.; Ma, S.Z. Industrial Cultivation of Lyophyllum decastes. Acta Edulis Fungi 2008, 15, 23–25. [Google Scholar] [CrossRef]
- Li, S.H.; Tang, S.M.; He, J.; Zhou, D.Q. Two New Lyophyllum Species from Yunnan, China. Diversity 2024, 16, 210. [Google Scholar] [CrossRef]
- Tang, S.M.; Yu, F.M.; Karunarathna, S.; Luo, Z.L.; Niu, K.Y.; Li, R.Y.; Li, L.; Su, X.J.; Li, S.H. Morpho-phylogenetic analyses of two novel edible mushrooms from China and a mini review of Lyophyllum (Agaricales, Lyophyllaceae) cultivation and bioactivities. MycoKeys 2025, 112, 307–334. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, T.H.; Liang, Y.Z.; Er, T.L.G.; Chen, D.Y. Atlas of Chinese Macrofungal Resources; Central Plains Farmers: Zhengzhou, China, 2015. [Google Scholar]
- Chen, X.L.; Liu, Y.H.; Guo, W.Z.; Wang, M.F.; Zhao, J.X.; Zhang, X.; Zheng, W.G. The development and nutritional quality of Lyophyllum decastes affected by monochromatic or mixed light provided by light-emitting diode. Front. Nutr. 2024, 11, 1–13. [Google Scholar] [CrossRef]
- Huang, J.Q.; Chen, H.; Lin, Y.; Ye, J.; Wang, Y.X.; Guo, Y.X.; Ma, L.Y. Genetic Differences and Mycelial Morphology of Lyophyllum decastes Strains. Fujian J. Agric. Sci. 2023, 38, 312–321. [Google Scholar]
- Chen, J.H. Cultivation status and nutritional value of Lyophyllum decastes. Edible Med. Mushroom 2021, 29, 12–15. [Google Scholar]
- Li, X.Y.; Jiang, W.Z.; Qin, W.X.; Long, W.S. Comparative Analysis of Nutrients in Fruit Body, Mycelia and Fermentation Broth of Lyophyllum decastes. Food Sci. 2010, 31, 155–157. [Google Scholar] [CrossRef]
- Xi, Y.L.; Li, M.A.; Qin, W.X.; Hua, G.J.; Jiang, W.Z.; Long, W.S. Assessment for protein nutrition of fruit bodies, mycelia and fermentation broth of Lyophyllum decastes. Mycosystema 2010, 29, 603–607. [Google Scholar] [CrossRef]
- Zhang, H.Y.; Zhang, F.Q.; Ming, W.X.; Long, W.S. Nutrition ingredient analysis and evaluation of Lyophyllum decastes fruit body. Mycosystema 2008, 27, 696–700. [Google Scholar]
- Zhang, G.P.; Wang, Y.N.; Qin, C.Q.; Ye, S.M.; Zhang, F.M.; Linhardt, R. Structural characterization of an antioxidant polysaccharide isolated from the fruiting bodies of Lyophyllum decastes. J. Mol. Struct. 2023, 1285, 135507. [Google Scholar] [CrossRef]
- Miura, T.; Kubo, M.; Itoh, Y.; Iwamoto, N.; Kato, M.; Park, S.; Ukawa, Y.; Kita, Y.; Suzuki, I. Antidiabetic Activity of Lyophyllum decastes in Genetically Type 2 Diabetic Mice. Biol. Pharm. Bull. 2002, 25, 1234–1237. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.T.; Feng, N.; Zhan, J.S.; Han, W. Qualitative Chemical Analysis and Antitumor Activity of Lyophyllum decastes Fruit Body Extracts. J-GLOBAL. 2016, 23, 70–74. [Google Scholar]
- Ukawa, Y.; Furuichi, Y.; Kokean, Y.; Nishii, T.; Hisamatsu, M. Effect of Hatakeshimeji (Lyophyllum decastes Sing.) Mushroom on Serum Lipid Levels in Rats. J. Nutr. Sci. Vitaminol. 2002, 48, 73–76. [Google Scholar] [CrossRef]
- Xu, L.L.; Yang, W.J.; Qiu, T.M.; Gao, X.; Zhang, H.Y.; Zhang, S.L.; Cui, H.; Guo, L.Z.; Yu, H.L.; Yu, H. Complete genome sequences and comparative secretomic analysis for the industrially cultivated edible mushroom Lyophyllum decastes reveals insights on evolution and lignocellulose degradation potential. Front. Microbiol. 2023, 14, 1–14. [Google Scholar] [CrossRef]
- Tan, Q.; Song, C.Y. China Edible Fungus Seed Industry Development Report 2023; China Agriculture Press: Beijing, China, 2025; pp. 261–264. [Google Scholar]
- Woo, S.M.; Park, Y.H.; Yoo, Y.B.; Shin, P.G.; Jang, K.Y.; Lee, K.H.; Sung, J.M. Development on Artificial Cultivation method of Hatakeshimeji (Lyophyllum decastes) using fermented sawdust substrate. J. Mushroom Sci. Prod. 2009, 7, 156–162. [Google Scholar]
- Yoshihama, Y.; Kusakabe, K.; Matsui, S.; Morita, H. Biologically Pure Mushroom Culture and Method for Mushroom Cultivation. U.S. Patent 5349121, 13 December 1994. [Google Scholar]
- Wei, S.L.; Wang, Z.J.; Yu, H.P.; Chen, Y. Studies on Isolation and domestication characteristics of Lypphyllum decastes. Southwest China J. Agric. Sci. 2005, 14, 128–131. [Google Scholar]
- Qiu, C.L.; Chen, C.M.; Zhuo, Z.M.; Lu, X. A Strain of Black Brown Lyophyllum decastes and Its Application. China Patent 202310624496.9, 30 May 2023. [Google Scholar]
- Kimura, E. Factory cultivation of Lyophyllum decastes. Edible Med. Mushrooms 2019, 27, 237–240. [Google Scholar]
- Pan, H.Y.; Ning, T.Y.; Xu, Y.T.; Zhou, H.Y.; Xu, B. Breeding of high yield strains of Lyophyllum decastes. Edible Fungi China 2023, 42, 42–46. [Google Scholar]
- Liang, Q.Q.; Wei, S.L.; Ding, L.Q.; Yan, H.X.; Liang, Y.P.; Xi, Y.L. Mutational screening of Lyophyllum decastes with high Yield of Laccase and Characteristics of Laccase from the Mutant. Nat. Prod. Res. Dev. 2014, 26, 1178–1181. [Google Scholar]
- Liang, Q.Q.; Wei, S.L.; Wang, H.; Wang, X.Q.; Ding, L.Q.; Xi, Y.L. Breeding high polysaccharide producing of Lyophyllum decastes strains by protolplast mutagensis. Sci. Technol. Food Ind. 2013, 34, 178–181. [Google Scholar]
- Zhou, H.M.; Zhang, Y.Z.; Xi, Y.L.; Wei, S.L. Screening of Lyophyllum decastes highly productive cultivable strains. Mycosystema 2014, 33, 208–217. [Google Scholar] [CrossRef]
- Yin, L.L.; Yao, F.J.; Shi, C.Y.; Lu, L.X.; Wang, T.; Liu, W. Genetic diversity of wild Auricularia heimuer germplasm resources based on SSR markers. Acta Edulis Fungi 2022, 29, 1–9. [Google Scholar]
- Zhang, D.; Wu, P.; Zhang, L.J.; Tang, L.H.; Song, C.Y.; Shang, X.D.; Bao, D.P.; Tan, Q. Polymorphism of SSR markers based on the whole genime sequence of Lentinula edodes and use in strain identification. Acta Edulis Fungi 2012, 19, 7–10. [Google Scholar]
- GB/T 21125-2007; Technical Inspection for Mushroom Selecting and Breeding. National Standardization Administration: Beijing, China, 2007.
- Huang, J.; Ge, X.; Sun, M. Modified CTAB protocol using a silica matrix for isolation of plant genomic DNA. BioTechniques 2000, 28, 432–434. [Google Scholar] [CrossRef]
- Pahlich, E.; Gerlitz, C. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemistry 1980, 19, 11–13. [Google Scholar] [CrossRef]
- Vilgalys, R.; González, D. Organization of ribosomal DNA in the basidiomycete Thanatephorus praticola. Curr. Genet. 1990, 18, 277–280. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.X.; Ma, J.H.; Meng, H. The Strain ZJLZS002 of Lyophyllum decastes, and Its Application, SSR Molecular Marker Primer. China Patent 202410492240.1, 23 April 2024. [Google Scholar]
- NY/T 1845-2010; Identification of Distinctness for Edible Mushroom Cultivar by Antagonism. Ministry of Agriculture and Rural Affairs of China: Beijing, China, 2010.
- Liu, Q.M.; Hua, R.; Sun, D.F.; Zhang, J.B.; Li, J.Y.; Liu, C.L.; Luo, X.; Liu, S.X. Breeding of a New Cyclocybe chaxingu Cultivar ‘Zhongjunbaicha 1’. Acta Edulis Fungi 2023, 30, 57–66. [Google Scholar]
- GB5009.5-2016; Determination of Protein in Food of National Standard for Food Safety. China Food and Drug Administation: Beijing, China, 2016.
- GB5009.124-2016; Determination of Amino Acids in Food of National Standard for Food Safety. China Food and Drug Administation: Beijing, China, 2016.
- SN/T 4260-2015; Determination of Crude Polysaccharides in Plant Source Foods for Export-Phehol-Sulfuric Acid Colorimetry. General Administration of Quality Supervision, Inspection and Quarantine of the People’s Republic of China: Beijing, China, 2015.
- Ellen, L.; Henrik, S. Lyophyllum shimeji, a species associated with lichen pine forest in northern Fennoscandia. Mycoscience 2011, 52, 289–295. [Google Scholar] [CrossRef]
- Moncalvo, J.M.; Vilgalys, R.; Redhead, S.A.; Johnson, J.E.; James, T.Y.; Catherine Aime, M.; Hofstetter, V.; Verduin, S.J.; Larsson, E.; Baroni, T.J.; et al. One hundred and seventeen clades of euagarics. Mol. Phylogenet. Evol. 2002, 23, 357–400. [Google Scholar] [CrossRef]
- Wei, S.W.; Qi, B.; Zhang, X.Z.; Peng, Z.W.; Li, Y.; Wang, Q. Global diversity and biogeography of Lyophyllum inferred from amplicon datasets. Front. Ecol. Evol. 2024, 12, 1328569. [Google Scholar] [CrossRef]
- Zhu, X.K.; Zhang, Q.Y. Protection and development of the old skill “bark hacking method” of Lentinula edodes cultivation. Edible Med. Mushrooms 2019, 27, 96–98. [Google Scholar]
- Wang, Z.S.; Liao, J.H.; Chen, M.Y.; Cai, Z.X. Breeding and industrial development of Agaricus bisporus. Acta Edulis Fungi 2012, 19, 1–14. [Google Scholar] [CrossRef]
- Chen, S.Y. Mushroom Cultivation manual; Scientific and Technical Documents Publishing House: Beijing, China, 2003. [Google Scholar]
- Liu, Q. Fungus seed industry: A neglected new seed industry. China Dev. Obs. 2021, 14, 43–45. [Google Scholar]
- Zhao, J.; Wang, Y.F.; Sheng, C.G.; Shi, L.; Pan, C.L.; Wang, J.H.; Liu, Z.T.; Zhang, P.; Yu, H.Y.; Meng, X.H. The production status and development trend of edible fungi factory in China. For. By-Prod. Spec. China 2021, 174, 68–74. [Google Scholar]
- Zhou, X.; Zhang, Y.; Zhang, C.X.; Yu, J.J. Breeding of high polysaccharide yield strains of Lyophyllum decastes by UV induced protoplast mutagenesis. Jiangsu Agric. Sci. 2017, 45, 126–128. [Google Scholar]
- Yuan, B.; Ke, L.N.; Chen, G.X.; Lian, Y.P.; Zhang, Z.H.; Ji, P.W.; Wu, Z.Q. Agaricus bisporus germplasms in southern Fujian identified by somatic incompatibility test and SSR. Fujian J. Agric. Sci. 2020, 35, 950–956. [Google Scholar]
- Gao, L.H.; Bao, D.P.; Xu, Z.; Li, Y.; Lu, H.; Tan, Y.S.; Shang, X.D.; Chen, H.Y.; Wang, R.J.; Wu, Y.Y. Construction of core collection and DNA fingerprinting of Flammulina filiformis based on genetic diversity analysis. Mycosystema 2021, 40, 3214–3230. [Google Scholar]





| Code | Strain No. | Taxon | Type | Origin of Resource |
|---|---|---|---|---|
| 1 | ZJLZS001 | Lyophyllum decastes | Wild | Dandong City, Liaoning Province |
| 2 | ZJLZS002 | Lyophyllum decastes | Wild | Dandong City, Liaoning Province |
| 3 | ZJLZS003 | Lyophyllum decastes | Wild | Hohhot City, Inner Mongolia Autonomous Region |
| 4 | ZJLZS004 | Lyophyllum decastes | Wild | Yongping County, Yunnan Province |
| 5 | ZJLZS005 | Lyophyllum decastes | Wild | Xinjiang Uygur Autonomous Region |
| 6 | ZJLZS006 | Lyophyllum decastes | Wild | Yuxi County, Yunnan Province |
| 7 | ZJLZS007 | Lyophyllum decastes | Wild | Lijiang City, Yunnan Province |
| 8 | ZJLZS008 | Lyophyllum decastes | Wild | Diqing Tibetan Autonomous Prefecture |
| 9 | LR-A | Lyophyllum decastes | Cultivation | Jiangsu Hongsheng Biotechnology Co., Ltd. |
| Primer Name | Size of Primer Fragment |
|---|---|
| LD045 | F: CCGCACAACACCTCAACAAG; R: TCGTCGCACAAAGAGCAGTA |
| LD063 | F: TCATCGGGACAAAGCCGAAA; R: GTCAACCCAAGCGACAACAC |
| LD089 | F: CCAAACAGTGCCGTTGAGTG; R: CTCCGGCGTTGAGTGACTAG |
| LD093 | F: TGGGTGTCGTTTGGGTATGG; R: CACGAGGACAGGCACATTCT |
| LD094 | F: GGGACGGAAGGGAAGGAAAG; R: TCTTTAACGCAGCGGTCCAT |
| LD095 | F: TGATGATGAGGCTTCGACGG; R: GTCACGACAACGCACTGTTC |
| LD098 | F: TCGGTGCGTAATCGTTTGGA; R: ATCGCCGCTTTCTGCAAATG |
| LD129 | F: CGTCGTTACATCGTTGACGC; R: TGGCACATCCATGAAGCAGT |
| LD149 | F: AAACTCAAGCTGTCCCGGTC; R: TCAAAGGAAGCTCCACCGAC |
| LD157 | F: ACGAGCTATTGGACGACCAC; R: TTTCGTTTCCACGCCGACTA |
| Species | Voucher | nrLSU | Reference |
|---|---|---|---|
| Lyophyllum decastes | ZJLZS001 | PV570115 | This study |
| L. decastes | ZJLZS002 | PV570116 | This study |
| L. decastes | ZJLZS003 | PV570117 | This study |
| L. decastes | ZJLZS004 | PV570118 | This study |
| L. decastes | ZJLZS005 | PV570119 | This study |
| L. decastes | ZJLZS006 | PV570120 | This study |
| L. decastes | ZJLZS007 | PV570121 | This study |
| L. decastes | ZJLZS008 | PV570122 | This study |
| L. decastes | LR-A | PV570114 | This study |
| L. decastes | Sundberg091007a | HM572548 | [39] |
| L. decastes | F-855 | PQ652409 | Genbank |
| L. ambustum | F-810 | PQ652355 | Genbank |
| L. fumosu | Lipovac090903 | HM572538 | [39] |
| L. shimeji | Eilertsen090908 | HM572531 | [39] |
| L. shimeji | CBS 451.87 | AF223215 | [40] |
| L. shimeji | haukebo1982 | HM572529 | [39] |
| Calocybe vinacea | HMJU 5135 | NG243053 | Genbank |
| Strain | Mycelium Growth Rate (cm/d) | Growth Vigor | Fruiting |
|---|---|---|---|
| ZJLZS001 | 0.27 ± 0.01 bc | + | T |
| ZJLZS002 | 0.35 ± 0.01 a | +++ | T |
| ZJLZS003 | 0.36 ± 0.04 a | +++ | T |
| ZJLZS004 | 0.31 ± 0.01 b | ++ | N |
| ZJLZS005 | 0.3 ± 0.00 b | ++ | N |
| ZJLZS006 | 0.3 ± 0.02 b | ++ | N |
| ZJLZS007 | 0.27 ± 0.02 bc | + | T |
| ZJLZS008 | 0.3 ± 0.02 b | ++ | N |
| Strain | Growth Period /Day | Stipe Characteristics | Cap Diameter /mm | Single Fruit Mass/g | Yield /(g·bag−1) | Biological Efficiency/% | ||
|---|---|---|---|---|---|---|---|---|
| Length /cm | Upper Diameter /mm | Bottom Diameter /mm | ||||||
| ZJLZS002 | 75.7 ± 1.5 b | 9.80 ± 0.36 | 10.92 ± 0.33 a | 16.49 ± 1.00 a | 33.89 ± 0.57 | 16.00 ± 0.26 | 374.67 ± 8.50 a | 93.67 ± 2.13 a |
| ZJLZS003 | 80.7 ± 1.5 a | 10.73 ± 0.92 | 8.53 ± 0.72 b | 10.87 ± 0.11 b | 32.61 ± 1.13 | 14.03 ± 1.76 | 337.00 ± 21.0 b | 84.25 ± 6.00 b |
| Strain | P. brevicompactum | T. pleuroticola | ||||||
|---|---|---|---|---|---|---|---|---|
| R1/cm | R2/cm | R1 − R2/cm | E/% | R1/cm | R2/cm | R1 − R2/cm | E/% | |
| ZJLZS002 | 2.4 | 1.1 | 1.3 | 54.2 | 2.1 | 4.1 | 2 | 48.8 |
| LR-A | 2.4 | 1.6 | 0.8 | 33.3 | 3.2 | 4.1 | 0.9 | 21.9 |
| Primer Name | LR-A | ZJLZS002 |
|---|---|---|
| LD045 | 171 | (167, 169) |
| LD063 | (288, 290) | 281 |
| LD089 | 189 | 192 |
| LD093 | 169 | 162 |
| LD094 | 181 | 158 |
| LD095 | 217 | 200 |
| LD098 | 286 | (284, 299) |
| LD129 | (172, 187) | (187, 190) |
| LD149 | 206 | 346 |
| LD157 | 135 | (142, 145) |
| Strain | Growth Period /Day | Stipe Characters | Cap Diameter/mm | Single Fruit Mass/g | Yield /(g·bag−1) | Biological Efficiency/% | ||
|---|---|---|---|---|---|---|---|---|
| Length /cm | Upper Diameter /mm | Bottom Diameter /mm | ||||||
| ZJLZS002 | 75.6 ± 1.3 | 9.63 ± 0.49 b | 10.38 ± 0.48 a | 16.47 ± 0.53 a | 33.89 ± 0.67 | 15.77 ± 0.95 a | 380 ± 3.6 a | 95 a |
| LR-A | 77.7 ± 1.5 | 10.9 ± 0.4 a | 7.87 ± 1.27 b | 10.93 ± 1.18 b | 33.87 ± 1.72 | 11.5 ± 0.78 b | 345.7 ± 13.9 b | 86.4 b |
| Strain | Biological Efficiency of the First Group/% | Biological Efficiency of the Second Group/% | Biological Efficiency of the Third Group/% | |||
|---|---|---|---|---|---|---|
| 1 | 2 | 1 | 2 | 1 | 2 | |
| ZJLZS002 | 95.17 ± 0.38 a | 94.5 ± 1.64 a | 94.75 ± 0.66 a | 94.92 ± 0.63 a | 94.67 ± 0.58 a | 95.92 ± 1.26 a |
| LR-A | 86.5 ± 3.19 b | 86.5 ± 2.54 b | 86.67 ± 3.15 b | 86.58 ± 3.26 b | 86.42 ± 2.98 b | 87.42 ± 3.36 b |
| Item | ZJLZS002 | Units | Item | ZJLZS002 | Units |
|---|---|---|---|---|---|
| Asp | 1.27 | g/100 g | Protein | 16.9 | g/100 g |
| Thr | 0.59 | g/100 g | Crude fiber | 24.1 | % |
| Ser | 0.63 | g/100 g | Fat | 1.6 | g/100 g |
| Glu | 1.65 | g/100 g | Total reducing sugar | 55.6 | % |
| Pro | 0.45 | g/100 g | Polysaccharide | 5.59 | g/100 g |
| Gly | 0.57 | g/100 g | Mg | 1217 | mg/kg |
| Ala | 0.64 | g/100 g | Se | 0.08 | mg/kg |
| Val | 0.58 | g/100 g | Mn | 9.09 | mg/kg |
| Met | 0.13 | g/100 g | Zn | 40.3 | mg/kg |
| Ile | 0.55 | g/100 g | Fe | 93.2 | mg/kg |
| Leu | 0.77 | g/100 g | Ca | 28.7 | mg/kg |
| Tyr | 0.43 | g/100 g | K | 4209 | mg/100 g |
| Phe | 0.54 | g/100 g | Na | 21.8 | mg/100 g |
| His | 0.33 | g/100g | Cu | 6.7 | mg/kg |
| Lys | 1.46 | g/100 g | P | 471 | mg/100 g |
| Arg | 0.72 | g/100 g | VB2 | 1.37 | mg/100 g |
| Total amino acids | 11.31 | g/100 g | VB6 | 4.57 | mg/kg |
| Moisture | 7.25 | g/100 g | VB3 | 21 | mg/100 g |
| Ash content | 6.0 | g/100 g | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Q.; Liu, S.; Li, J.; Zhang, J.; Zhou, F.; Luo, X.; Ma, J.; Hua, R.; Sun, D. High Resistance and Yield: A New Cultivar ‘ZJLZS002’ of Lyophyllum decastes Suitable for Industrial Cultivation. Agriculture 2025, 15, 1045. https://doi.org/10.3390/agriculture15101045
Liu Q, Liu S, Li J, Zhang J, Zhou F, Luo X, Ma J, Hua R, Sun D. High Resistance and Yield: A New Cultivar ‘ZJLZS002’ of Lyophyllum decastes Suitable for Industrial Cultivation. Agriculture. 2025; 15(10):1045. https://doi.org/10.3390/agriculture15101045
Chicago/Turabian StyleLiu, Qimeng, Shaoxiong Liu, Jianying Li, Junbo Zhang, Fan Zhou, Xi Luo, Jianxiong Ma, Rong Hua, and Dafeng Sun. 2025. "High Resistance and Yield: A New Cultivar ‘ZJLZS002’ of Lyophyllum decastes Suitable for Industrial Cultivation" Agriculture 15, no. 10: 1045. https://doi.org/10.3390/agriculture15101045
APA StyleLiu, Q., Liu, S., Li, J., Zhang, J., Zhou, F., Luo, X., Ma, J., Hua, R., & Sun, D. (2025). High Resistance and Yield: A New Cultivar ‘ZJLZS002’ of Lyophyllum decastes Suitable for Industrial Cultivation. Agriculture, 15(10), 1045. https://doi.org/10.3390/agriculture15101045





