Transcriptomics Reveal an Integrated Gene Regulation Network of Early Flowering Development in an Oil Sunflower Mutant Induced by Heavy Ion Beam
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Sample Collection
2.2. Histological Analysis Using Paraffin Section
2.3. RNA Extraction and Sequencing
2.4. Transcriptome Analysis
2.5. Quantitative Real-Time PCR
2.6. Statistical Analysis
3. Results
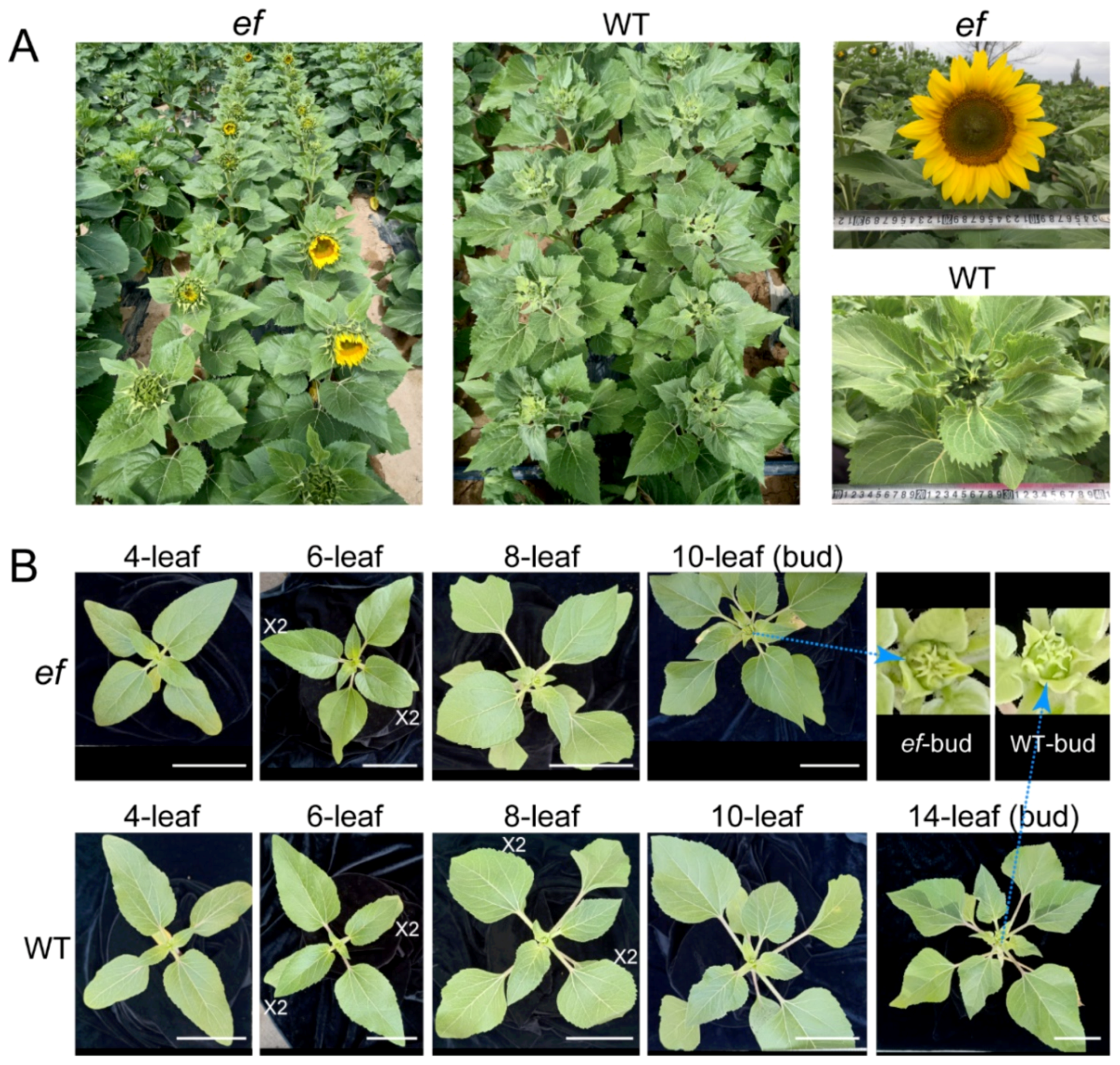
3.1. The ef Mutant Is Early-Flowering
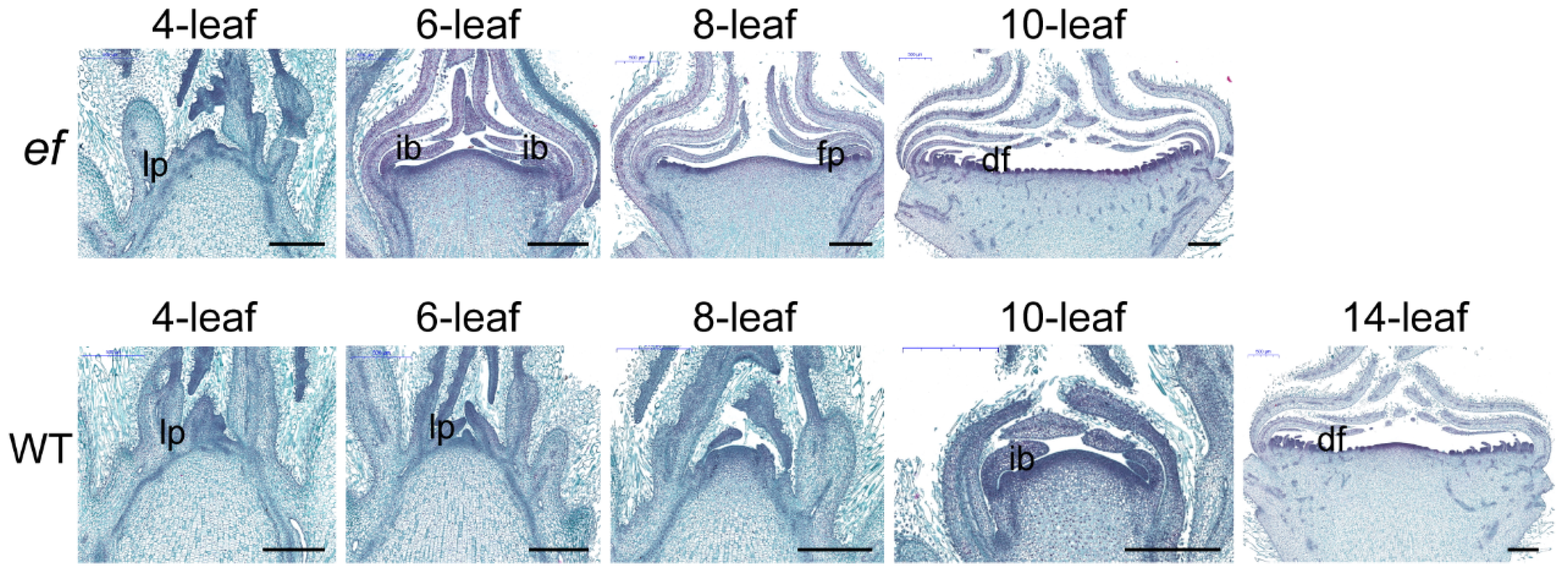
3.2. Histological Analysis of Developmental Status for ef Mutant and WT
3.3. The Gene Expression of the Shoot Apical Meristem in ef Mutant Was Different from WT
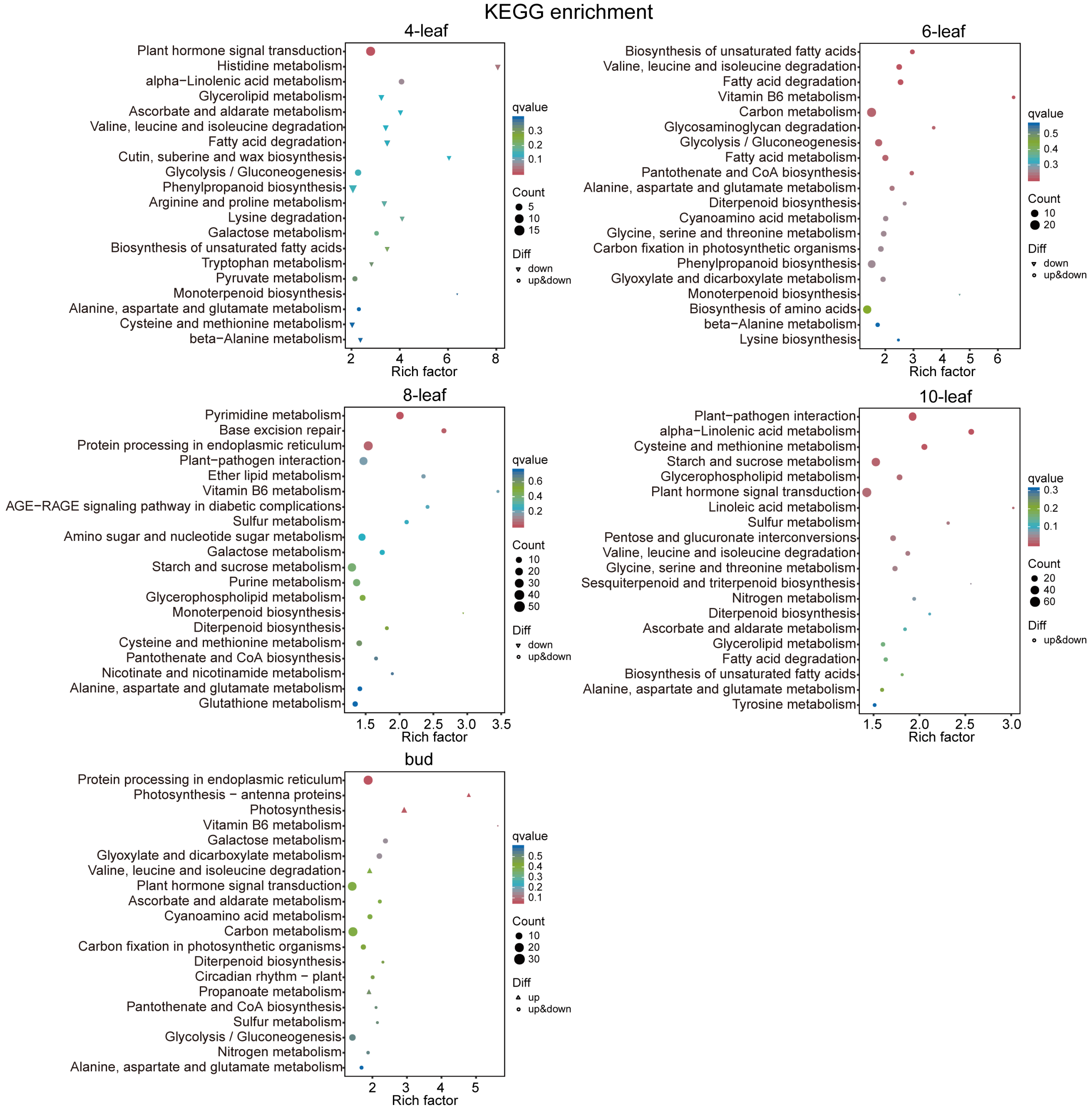
3.4. GO and KEGG Analyses of Differentially Expressed Genes at Different Periods
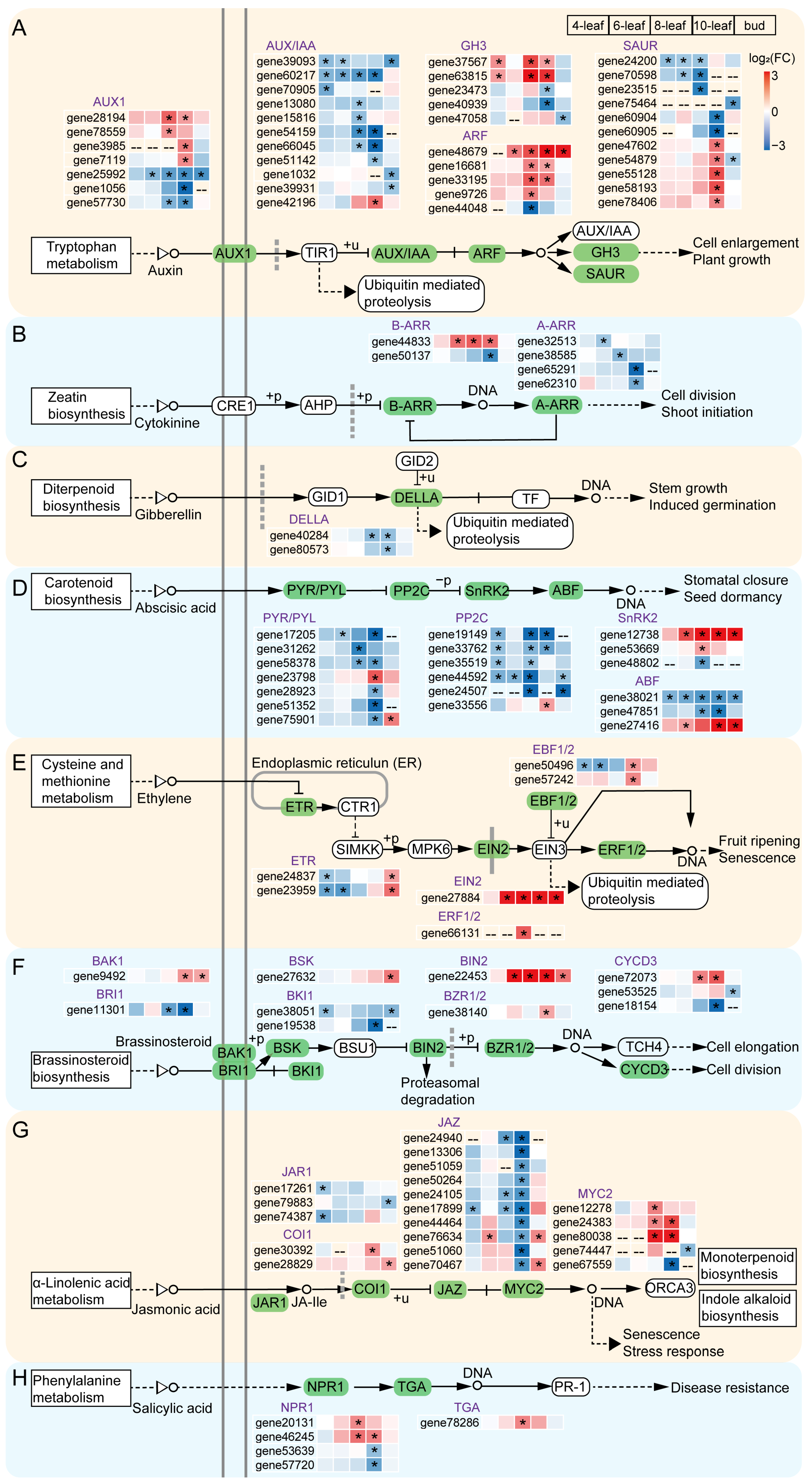
3.5. Plant Hormone Signal Transduction Is Important in Early Flowering Regulation
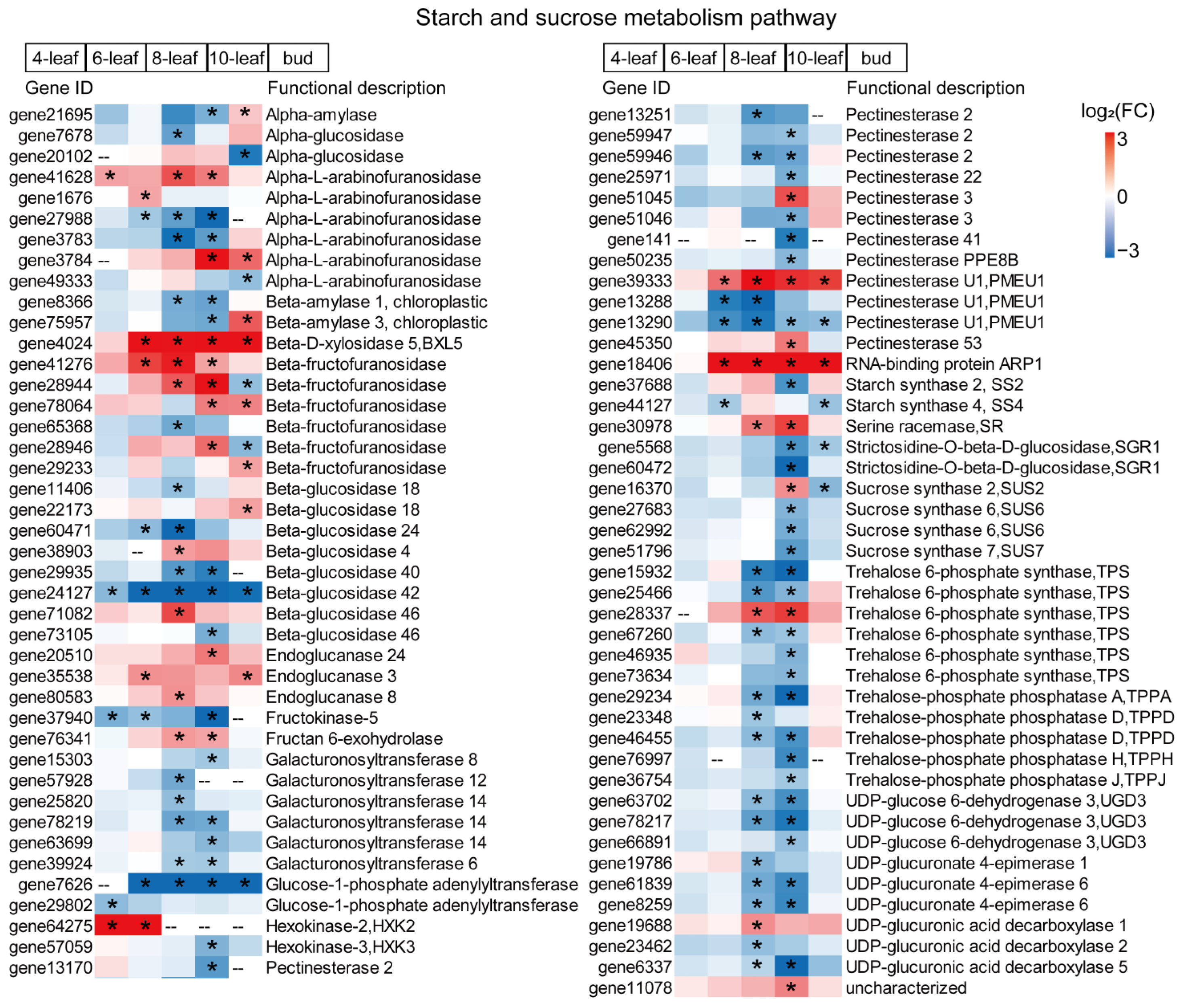
3.6. Starch and Sucrose Metabolism Pathways Have Differential Expression in the ef Mutant
3.7. Expression Profile of Flowering Genes in the ef Mutant and WT
4. Discussion
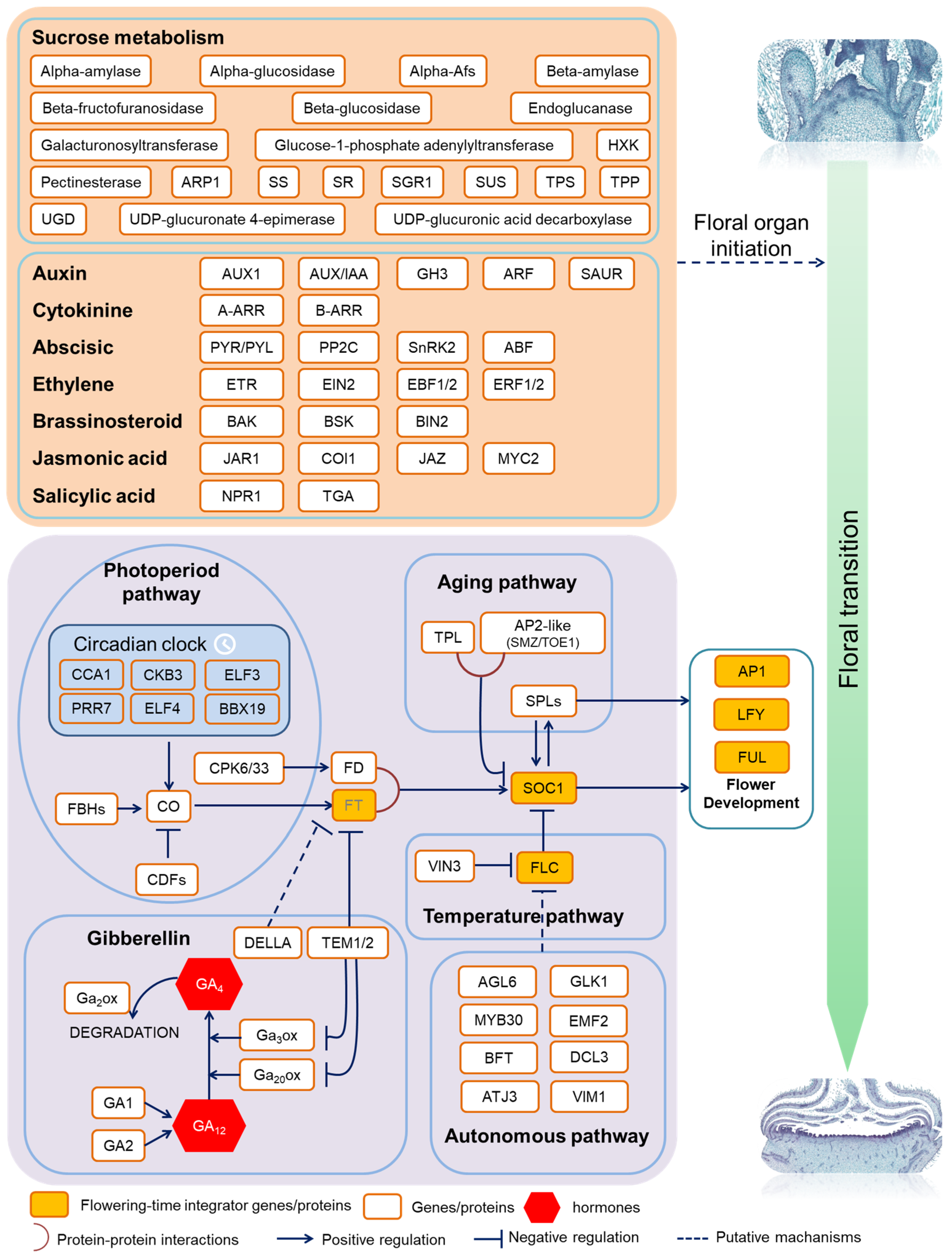
4.1. Plant Hormones Synergistically Promote Early Flowering
4.2. Sucrose Signaling in Early Flowering Mutant
4.3. Several Flowering Pathways Regulated Early Flowering
4.4. Cross-Talk between Flowering Genes, Hormones and Sucrose Signaling
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Pilorgé, É. Sunflower in the global vegetable oil system: Situation, specificities and perspectives. OCL 2020, 27, 34. [Google Scholar] [CrossRef]
- Seiler, G.J.; Qi, L.L.; Marek, L.F. Utilization of sunflower crop wild relatives for cultivated sunflower improvement. Crop Sci. 2017, 57, 1083–1101. [Google Scholar] [CrossRef]
- Prado, D.M.; Almeida, A.B.; Filho, J.G.; Alves, C.C.; Egea, M.B.; Lemes, A.C. Extraction of bioactive proteins from seeds (corn, sorghum, and sunflower) and sunflower byproduct: Enzymatic hydrolysis and antioxidant properties. Curr. Nutr Food Sci. 2020, 16, 310–320. [Google Scholar] [CrossRef]
- Wanjari, N.; Waghmare, J.S. Phenolic and antioxidant potential of sunflower meal. Adv Appl Sci Res. 2015, 6, 221–229. [Google Scholar]
- de Oliveira Filho, J.G.; Egea, M.B. Sunflower seed byproduct and its fractions for food application: An attempt to improve the sustainability of the oil process. J Food Sci. 2021, 86, 1497–1510. [Google Scholar] [CrossRef]
- Zhao, S.; Zhao, Y.; Liang, H.; Su, Y. Formaldehyde removal in the air by six plant systems with or without rhizosphere microorganisms. Int. J. Phytoremediation 2019, 21, 1296–1304. [Google Scholar] [CrossRef]
- Bonnafous, F.; Fievet, G.; Blanchet, N.; Boniface, M.C.; Carrère, S.; Gouzy, J.; Legrand, L.; Marage, G.; Bret-Mestries, E.; Munos, S.; et al. Comparison of GWAS models to identify non-additive genetic control of flowering time in sunflower hybrids. Theor. Appl Genet. 2018, 131, 319–332. [Google Scholar] [CrossRef]
- Greenup, A.; Peacock, W.J.; Dennis, E.S.; Trevaskis, B. The molecular biology of seasonal flowering-responses in Arabidopsis and the cereals. Ann. Bot. 2009, 103, 1165–1172. [Google Scholar] [CrossRef]
- Srikanth, A.; Schmid, M. Regulation of flowering time: All roads lead to Rome. Cell. Mol. Life Sci. CMLS 2011, 68, 2013–2037. [Google Scholar] [CrossRef]
- Basu, U.; Hegde, V.S.; Daware, A.; Jha, U.C.; Parida, S.K. Transcriptome landscape of early inflorescence developmental stages identifies key flowering time regulators in chickpea. Plant Mol. Biol. 2022, 108, 565–583. [Google Scholar] [CrossRef]
- Amasino, R.M.; Cheung, A.Y.; Dresselhaus, T.; Kuhlemeier, C. Focus on flowering and reproduction. Plant Physiol. 2017, 173, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Bouché, F.; Lobet, G.; Tocquin, P.; Périlleux, C. FLOR-ID: An interactive database of flowering-time gene networks in Arabidopsis thaliana. Nucleic Acids Res. 2016, 44, D1167–D1171. [Google Scholar] [CrossRef]
- Corbesier, L.; Vincent, C.; Jang, S.; Fornara, F.; Fan, Q.; Searle, I.; Giakountis, A.; Farrona, S.; Gissot, L.; Turnbull, C.; et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007, 316, 1030–1033. [Google Scholar] [CrossRef] [PubMed]
- Nagel, D.H.; Doherty, C.J.; Pruneda-Paz, J.L.; Schmitz, R.J.; Ecker, J.R.; Kay, S.A. Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc. Natl. Acad. Sci. USA 2015, 112, E4802–E4810. [Google Scholar] [CrossRef] [PubMed]
- Campos-Rivero, G.; Osorio-Montalvo, P.; Sánchez-Borges, R.; Us-Camas, R.; Duarte-Aké, F.; De-la-Peña, C. Plant hormone signaling in flowering: An epigenetic point of view. J. Plant Physiol. 2017, 214, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Martínez, C.; Pons, E.; Prats, G.; León, J. Salicylic acid regulates flowering time and links defence responses and reproductive development. Plant J. Cell Mol. Biol. 2004, 37, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, S.; Böhlenius, H.; Moritz, T.; Nilsson, O. GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell 2006, 18, 2172–2181. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Lee, I. Regulation and function of SOC1, a flowering pathway integrator. J. Exp. Bot. 2010, 61, 2247–2254. [Google Scholar] [CrossRef]
- Wang, J.W.; Czech, B.; Weigel, D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 2009, 138, 738–749. [Google Scholar] [CrossRef]
- Chowdhury, Z.; Mohanty, D.; Giri, M.K.; Venables, B.J.; Chaturvedi, R.; Chao, A.; Petros, R.A.; Shah, J. Dehydroabietinal promotes flowering time and plant defense in Arabidopsis via the autonomous pathway genes FLOWERING LOCUS D, FVE, and RELATIVE OF EARLY FLOWERING 6. J. Exp. Bot. 2020, 71, 4903–4913. [Google Scholar] [CrossRef]
- Lu, Q.; Zhao, L.; Li, D.; Hao, D.; Zhan, Y.; Li, W. A GmRAV ortholog is involved in photoperiod and sucrose control of flowering time in soybean. PLoS ONE 2014, 9, e89145. [Google Scholar] [CrossRef]
- Gaur, P.M.; Samineni, S.; Thudi, M.; Tripathi, S.; Sajja, S.B.; Jayalakshmi, V.; Mannur, D.M.; Vijayakumar, A.G.; Ganga Rao, N.V.P.R.; Ojiewo, C.; et al. Integrated breeding approaches for improving drought and heat adaptation in chickpea (Cicer arietinum L.). Plant Breed. 2019, 138, 389–400. [Google Scholar] [CrossRef]
- Digel, B.; Pankin, A.; von Korff, M. Global transcriptome profiling of developing leaf and shoot apices reveals distinct genetic and environmental control of floral transition and inflorescence development in barley. Plant Cell 2015, 27, 2318–2334. [Google Scholar] [CrossRef] [PubMed]
- Feng, N.; Song, G.; Guan, J.; Chen, K.; Jia, M.; Huang, D.; Wu, J.; Zhang, L.; Kong, X.; Geng, S.; et al. Transcriptome profiling of wheat inflorescence development from spikelet initiation to floral patterning identified stage-specific regulatory genes. Plant Physiol. 2017, 174, 1779–1794. [Google Scholar] [CrossRef] [PubMed]
- Parvathaneni, R.K.; Bertolini, E.; Shamimuzzaman, M.; Vera, D.L.; Lung, P.Y.; Rice, B.R.; Zhang, J.; Brown, P.J.; Lipka, A.E.; Bass, H.W.; et al. The regulatory landscape of early maize inflorescence development. Genome Biol. 2020, 21, 165. [Google Scholar] [CrossRef] [PubMed]
- Osnato, M.; Lacchini, E.; Pilatone, A.; Dreni, L.; Grioni, A.; Chiara, M.; Horner, D.; Pelaz, S.; Kater, M.M. Transcriptome analysis reveals rice MADS13 as an important repressor of the carpel development pathway in ovules. J. Exp. Bot. 2021, 72, 398–414. [Google Scholar] [CrossRef] [PubMed]
- Badouin, H.; Gouzy, J.; Grassa, C.J.; Murat, F.; Staton, S.E.; Cottret, L.; Lelandais-Brière, C.; Owens, G.L.; Carrère, S.; Mayjonade, B.; et al. The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 2017, 546, 148–152. [Google Scholar] [CrossRef] [PubMed]
- Blackman, B.K.; Michaels, S.D.; Rieseberg, L.H. Connecting the sun to flowering in sunflower adaptation. Mol. Ecol. 2011, 20, 3503–3512. [Google Scholar] [CrossRef] [PubMed]
- Blackman, B.K.; Rasmussen, D.A.; Strasburg, J.L.; Raduski, A.R.; Burke, J.M.; Knapp, S.J.; Michaels, S.D.; Rieseberg, L.H. Contributions of flowering time genes to sunflower domestication and improvement. Genetics 2011, 187, 271–287. [Google Scholar] [CrossRef]
- Tanaka, A.; Shikazono, N.; Hase, Y. Studies on biological effects of ion beams on lethality, molecular nature of mutation, mutation rate, and spectrum of mutation phenotype for mutation breeding in higher plants. J. Radiat. Res. 2010, 51, 223–233. [Google Scholar] [CrossRef]
- Fambrini, M.; Cionini, G.; Bertini, D.; Michelotti, V.; Conti, A.; Pugliesi, C. Missing flowers gene controls axillary meristems initiation in sunflower. Genesis. 2003, 36, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Mariotti, L.; Fambrini, M.; Scartazza, A.; Picciarelli, P.; Pugliesi, C. Characterization of lingering hope, a new brachytic mutant in sunflower (Helianthus annuus L.) with altered salicylic acid metabolism. J. Plant Physiol. 2018, 231, 402–414. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Florea, L.; Song, L.; Salzberg, S.L. Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues. F1000Research 2013, 2, 188. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C(T)) Method. Methods. 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Fambrini, M.; Salvini, M.; Pugliesi, C. Molecular cloning, phylogenetic analysis, and expression patterns of LATERAL SUPPRESSOR-LIKE and REGULATOR OF AXILLARY MERISTEM FORMATION-LIKE genes in sunflower (Helianthus annuus L.). Dev. Genes Evol. 2017, 227, 159–170. [Google Scholar] [CrossRef]
- Marín-González, E.; Matías-Hernández, L.; Aguilar-Jaramillo, A.E.; Lee, J.H.; Ahn, J.H.; Suárez-López, P.; Pelaz, S. SHORT VEGETATIVE PHASE Up-Regulates TEMPRANILLO2 Floral Repressor at Low Ambient Temperatures. Plant Physiol. 2015, 169, 1214–1224. [Google Scholar] [CrossRef]
- Oh, E.; Yamaguchi, S.; Hu, J.; Yusuke, J.; Jung, B.; Paik, I.; Lee, H.S.; Sun, T.P.; Kamiya, Y.; Choi, G. PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 2007, 19, 1192–1208. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Poethig, R.S. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development. 2006, 133, 3539–3547. [Google Scholar] [CrossRef]
- Xu, S.; Xiao, J.; Yin, F.; Guo, X.; Xing, L.; Xu, Y.; Chong, K. The protein modifications of O-GlcNAcylation and phosphorylation mediate vernalization response for flowering in winter wheat. Plant Physiol. 2019, 180, 1436–1449. [Google Scholar] [CrossRef]
- Arefian, M.; Bhagya, N.; Prasad, T.S.K. Phosphorylation-mediated signalling in flowering: Prospects and retrospects of phosphoproteomics in crops. Biol. Rev. Camb. Philos. Soc. 2021, 96, 2164–2191. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, A.; Chaudhury, R.; Dutta, S.; Basak, M.; Dey, S.; Schäffner, A.R.; Das, M. Role of metabolites in flower development and discovery of compounds controlling flowering time. Plant Physiol. Biochem. PPB 2022, 190, 109–118. [Google Scholar] [CrossRef]
- Borghi, M.; Fernie, A.R. Outstanding questions in flower metabolism. Plant J. Cell Mol. Biol. 2020, 103, 1275–1288. [Google Scholar] [CrossRef] [PubMed]
- Cucinotta, M.; Cavalleri, A.; Chandler, J.W.; Colombo, L. Auxin and flower development: A blossoming field. Cold Spring Harb Perspect Biol. 2021, 13, a039974. [Google Scholar] [CrossRef]
- Su, Y.H.; Liu, Y.B.; Zhang, X.S. Auxin-cytokinin interaction regulates meristem development. Mol. Plant 2011, 4, 616–625. [Google Scholar] [CrossRef]
- Yadav, S.; Kumar, H.; Yadav, R.K. Local auxin biosynthesis promotes stem cell differentiation and organogenesis in Arabidopsis shoot apex. Development. 2023, 150, dev202014. [Google Scholar] [CrossRef]
- Martignago, D.; Siemiatkowska, B.; Lombardi, A.; Conti, L. Abscisic acid and flowering regulation: Many targets, different places. Int J Mol Sci. 2020, 21, 9700. [Google Scholar] [CrossRef]
- Park, S.Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.F.; et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009, 324, 1068–1071. [Google Scholar] [CrossRef]
- Yoshida, T.; Fujita, Y.; Maruyama, K.; Mogami, J.; Todaka, D.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 2015, 38, 35–49. [Google Scholar] [CrossRef]
- Riboni, M.; Robustelli Test, A.; Galbiati, M.; Tonelli, C.; Conti, L. ABA-dependent control of GIGANTEA signalling enables drought escape via up-regulation of FLOWERING LOCUS T in Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 6309–6322. [Google Scholar] [CrossRef]
- Siriwardana, C.L.; Gnesutta, N.; Kumimoto, R.W.; Jones, D.S.; Myers, Z.A.; Mantovani, R.; Holt, B.F., 3rd. NUCLEAR FACTOR Y, Subunit A (NF-YA) proteins positively regulate flowering and act through FLOWERING LOCUS T. PLoS Genet. 2016, 12, e1006496. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Xing, X.; Tang, Y.; Jin, J.; Ding, L.; Song, A.; Chen, S.; Chen, F.; Jiang, J.; Fang, W. An ethylene-responsive transcription factor and a flowering locus KH domain homologue jointly modulate photoperiodic flowering in chrysanthemum. Plant Cell Environ. 2022, 45, 1442–1456. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, Y.; Chen, S.; An, L. Involvement of brassinosteroid signals in the floral-induction network of Arabidopsis. J. Exp. Bot. 2010, 61, 4221–4230. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Lee, H.; Lee, J.; Kim, H.; Ryu, H. ABSCISIC ACID-INSENSITIVE 3 is involved in brassinosteroid-mediated regulation of flowering in plants. Plant Physiol. Biochem. PPB 2019, 139, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Luo, B.; Hu, M.; Fu, S.; Liu, J.; Jiang, M.; Zhao, Y.; Huang, S.; Wang, S.; Wang, X. Brassinosteroid signaling downstream suppressor BIN2 interacts with SLFRIGIDA-LIKE to induce early flowering in tomato. Int J Mol Sci. 2022, 23, 11264. [Google Scholar] [CrossRef] [PubMed]
- Domagalska, M.A.; Schomburg, F.M.; Amasino, R.M.; Vierstra, R.D.; Nagy, F.; Davis, S.J. Attenuation of brassinosteroid signaling enhances FLC expression and delays flowering. Development. 2007, 134, 2841–2850. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zou, Z.; Qian, K.; Xia, C.; He, Y.; Zeng, H.; Zhou, X.; Riemann, M.; Yin, C. Jasmonic acid deficiency leads to scattered floret opening time in cytoplasmic male sterile rice Zhenshan 97A. J. Exp. Bot. 2017, 68, 4613–4625. [Google Scholar] [CrossRef] [PubMed]
- Rastegari, S.; Naser Alavi, S.M.; Mohayeji, M. Effect of salicylic acid and pre-cold treatment on flower induction in saffron. Scientifica 2022, 2022, 6108161. [Google Scholar] [CrossRef] [PubMed]
- Kazan, K.; Manners, J.M. MYC2: The master in action. Mol. Plant 2013, 6, 686–703. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.; Cho, L.H.; Tun, W.; Jeon, J.S.; An, G. Sucrose signaling in higher plants. Plant Sci. Int. J. Exp. Plant Biol. 2021, 302, 110703. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.L. Sucrose metabolism: Gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 2014, 65, 33–67. [Google Scholar] [CrossRef] [PubMed]
- Lejeune, P.; Bernier, G.; Requier, M.-C.; Kinet, J.-M. Sucrose increase during floral induction in the phloem sap collected at the apical part of the shoot of the long-day plant Sinapis alba L. Planta 1993, 190, 71–74. [Google Scholar] [CrossRef]
- Friend, D.J.; Bodson, M.; Bernier, G. Promotion of flowering in Brassica campestris L. cv Ceres by sucrose. Plant Physiol. 1984, 75, 1085–1089. [Google Scholar] [CrossRef]
- Sozzi, G.O.; Greve, L.C.; Prody, G.A.; Labavitch, J.M. Gibberellic acid, synthetic auxins, and ethylene differentially modulate alpha-L-Arabinofuranosidase activities in antisense 1-aminocyclopropane-1-carboxylic acid synthase tomato pericarp discs. Plant Physiol. 2002, 129, 1330–1340. [Google Scholar] [CrossRef]
- Purgatto, E.; Lajolo, F.M.; do Nascimento, J.R.; Cordenunsi, B.R. Inhibition of beta-amylase activity, starch degradation and sucrose formation by indole-3-acetic acid during banana ripening. Planta 2001, 212, 823–828. [Google Scholar] [CrossRef]
- Jin, S.; Kanagaraj, A.; Verma, D.; Lange, T.; Daniell, H. Release of hormones from conjugates: Chloroplast expression of β-glucosidase results in elevated phytohormone levels associated with significant increase in biomass and protection from aphids or whiteflies conferred by sucrose esters. Plant Physiol. 2011, 155, 222–235. [Google Scholar] [CrossRef]
- Meng, L.; Zhang, X.; Wang, L.; Liu, H.; Zhao, Y.; Yi, K.; Cui, G.; Yin, X. Transcriptome profiling unveils the mechanism of phenylpropane biosynthesis in rhizome development of Caucasian clover. PLoS ONE 2021, 16, e0254669. [Google Scholar] [CrossRef] [PubMed]
- Fichtner, F.; Lunn, J.E. The role of trehalose 6-phosphate (Tre6P) in plant metabolism and development. Annu. Rev. Plant Biol. 2021, 72, 737–760. [Google Scholar] [CrossRef]
- Ó’Maoiléidigh, D.S.; Graciet, E.; Wellmer, F. Gene networks controlling Arabidopsis thaliana flower development. New Phytol. 2014, 201, 16–30. [Google Scholar] [CrossRef]
- Balanzà, V.; Martínez-Fernández, I.; Ferrándiz, C. Sequential action of FRUITFULL as a modulator of the activity of the floral regulators SVP and SOC1. J. Exp. Bot. 2014, 65, 1193–1203. [Google Scholar] [CrossRef]
- Ferrándiz, C.; Gu, Q.; Martienssen, R.; Yanofsky, M.F. Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development. 2000, 127, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Blackman, B.K.; Strasburg, J.L.; Raduski, A.R.; Michaels, S.D.; Rieseberg, L.H. The role of recently derived FT paralogs in sunflower domestication. Curr. Biol. CB 2010, 20, 629–635. [Google Scholar] [CrossRef]
- Shim, J.S.; Imaizumi, T. Circadian clock and photoperiodic response in Arabidopsis: From seasonal flowering to redox homeostasis. Biochemistry 2015, 54, 157–170. [Google Scholar] [CrossRef]
- Kawamoto, N.; Sasabe, M.; Endo, M.; Machida, Y.; Araki, T. Calcium-dependent protein kinases responsible for the phosphorylation of a bZIP transcription factor FD crucial for the florigen complex formation. Sci. Rep. 2015, 5, 8341. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.; Shen, L.; Wu, Y.; Chen, H.; Robertson, M.; Helliwell, C.A.; Ito, T.; Meyerowitz, E.; Yu, H. A repressor complex governs the integration of flowering signals in Arabidopsis. Dev. Cell 2008, 15, 110–120. [Google Scholar] [CrossRef]
- Mateos, J.L.; Madrigal, P.; Tsuda, K.; Rawat, V.; Richter, R.; Romera-Branchat, M.; Fornara, F.; Schneeberger, K.; Krajewski, P.; Coupland, G. Combinatorial activities of SHORT VEGETATIVE PHASE and FLOWERING LOCUS C define distinct modes of flowering regulation in Arabidopsis. Genome Biol. 2015, 16, 31. [Google Scholar] [CrossRef]
- Micallef, B.J.; Haskins, K.A.; Vanderveer, P.J.; Roh, K.-S.; Shewmaker, C.K.; Sharkey, T.D. Altered photosynthesis, flowering, and fruiting in transgenic tomato plants that have an increased capacity for sucrose synthesis. Planta 1995, 196, 327–334. [Google Scholar] [CrossRef]
- Roldán, M.; Gómez-Mena, C.; Ruiz-García, L.; Salinas, J.; Martínez-Zapater, J.M. Sucrose availability on the aerial part of the plant promotes morphogenesis and flowering of Arabidopsis in the dark. Plant J. Cell Mol. Biol. 1999, 20, 581–590. [Google Scholar] [CrossRef]
- Lugassi, N.; Stein, O.; Egbaria, A.; Belausov, E.; Zemach, H.; Arad, T.; Granot, D.; Carmi, N. Sucrose synthase and fructokinase are required for proper meristematic and vascular development. Plants (Basel). 2022, 11, 1035. [Google Scholar] [CrossRef]
- Wang, M.; Le Gourrierec, J.; Jiao, F.; Demotes-Mainard, S.; Perez-Garcia, M.D.; Ogé, L.; Hamama, L.; Crespel, L.; Bertheloot, J.; Chen, J.; et al. Convergence and divergence of sugar and cytokinin signaling in plant development. Int J Mol Sci. 2021, 22, 1282. [Google Scholar] [CrossRef]
- Bonhomme, F.; Kurz, B.; Melzer, S.; Bernier, G.; Jacqmard, A. Cytokinin and gibberellin activate SaMADS A, a gene apparently involved in regulation of the floral transition in Sinapis alba. Plant J. Cell Mol. Biol. 2000, 24, 103–111. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.H.; Hsu, W.H.; Mao, W.T.; Yang, C.H. The oncidium ethylene synthesis gene oncidium 1-aminocyclopropane-1 carboxylic acid synthase 12 and ethylene receptor gene oncidium ETR1 affect GA-DELLA and jasmonic acid signaling in regulating flowering time, anther dehiscence, and flower senescence in Arabidopsis. Front. Plant Sci. 2022, 13, 785441. [Google Scholar] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Mao, X.; Chen, J.; Du, Y.; Jin, W.; Liu, R.; Zhou, L.; Qu, Y. Transcriptomics Reveal an Integrated Gene Regulation Network of Early Flowering Development in an Oil Sunflower Mutant Induced by Heavy Ion Beam. Agriculture 2024, 14, 449. https://doi.org/10.3390/agriculture14030449
Liu X, Mao X, Chen J, Du Y, Jin W, Liu R, Zhou L, Qu Y. Transcriptomics Reveal an Integrated Gene Regulation Network of Early Flowering Development in an Oil Sunflower Mutant Induced by Heavy Ion Beam. Agriculture. 2024; 14(3):449. https://doi.org/10.3390/agriculture14030449
Chicago/Turabian StyleLiu, Xiao, Xuhui Mao, Jihong Chen, Yan Du, Wenjie Jin, Ruiyuan Liu, Libin Zhou, and Ying Qu. 2024. "Transcriptomics Reveal an Integrated Gene Regulation Network of Early Flowering Development in an Oil Sunflower Mutant Induced by Heavy Ion Beam" Agriculture 14, no. 3: 449. https://doi.org/10.3390/agriculture14030449
APA StyleLiu, X., Mao, X., Chen, J., Du, Y., Jin, W., Liu, R., Zhou, L., & Qu, Y. (2024). Transcriptomics Reveal an Integrated Gene Regulation Network of Early Flowering Development in an Oil Sunflower Mutant Induced by Heavy Ion Beam. Agriculture, 14(3), 449. https://doi.org/10.3390/agriculture14030449





