Preparation of Barley AGPS2b Antibody and Its Application in Hormone Regulation Research
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.2. Methods
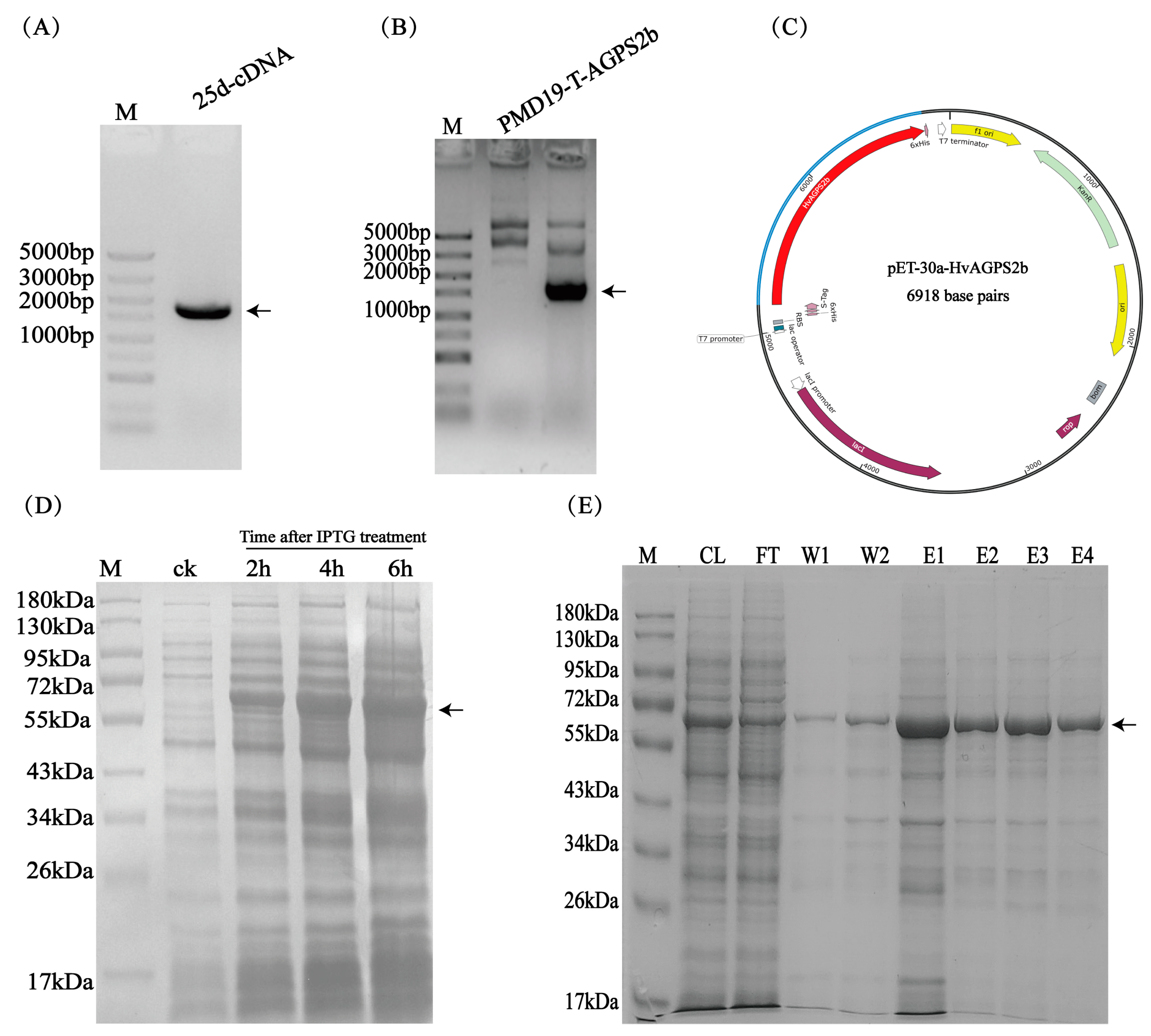
2.2.1. Cloning of HvAGPS2b Gene
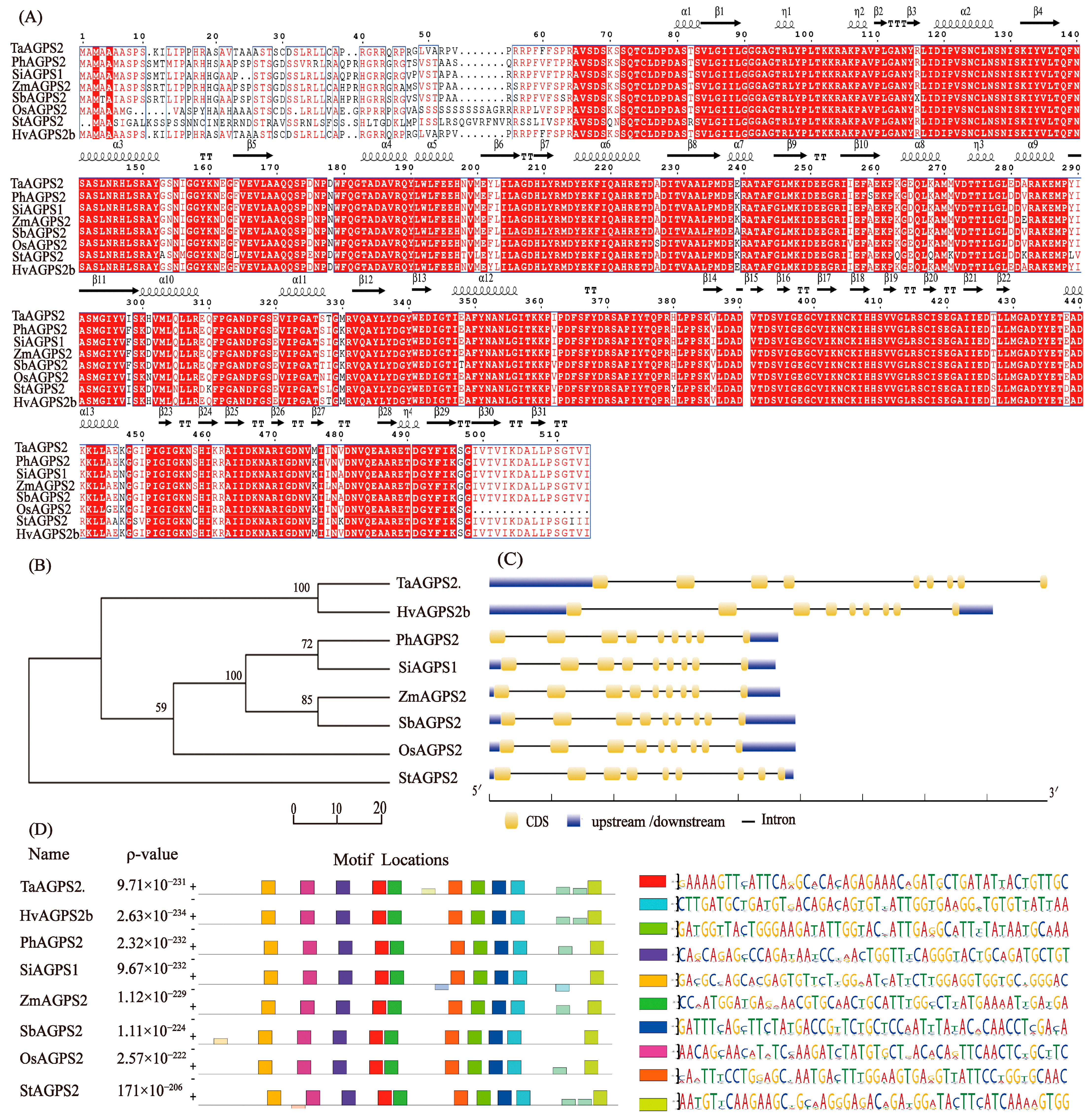
2.2.2. Bioinformatics Analysis
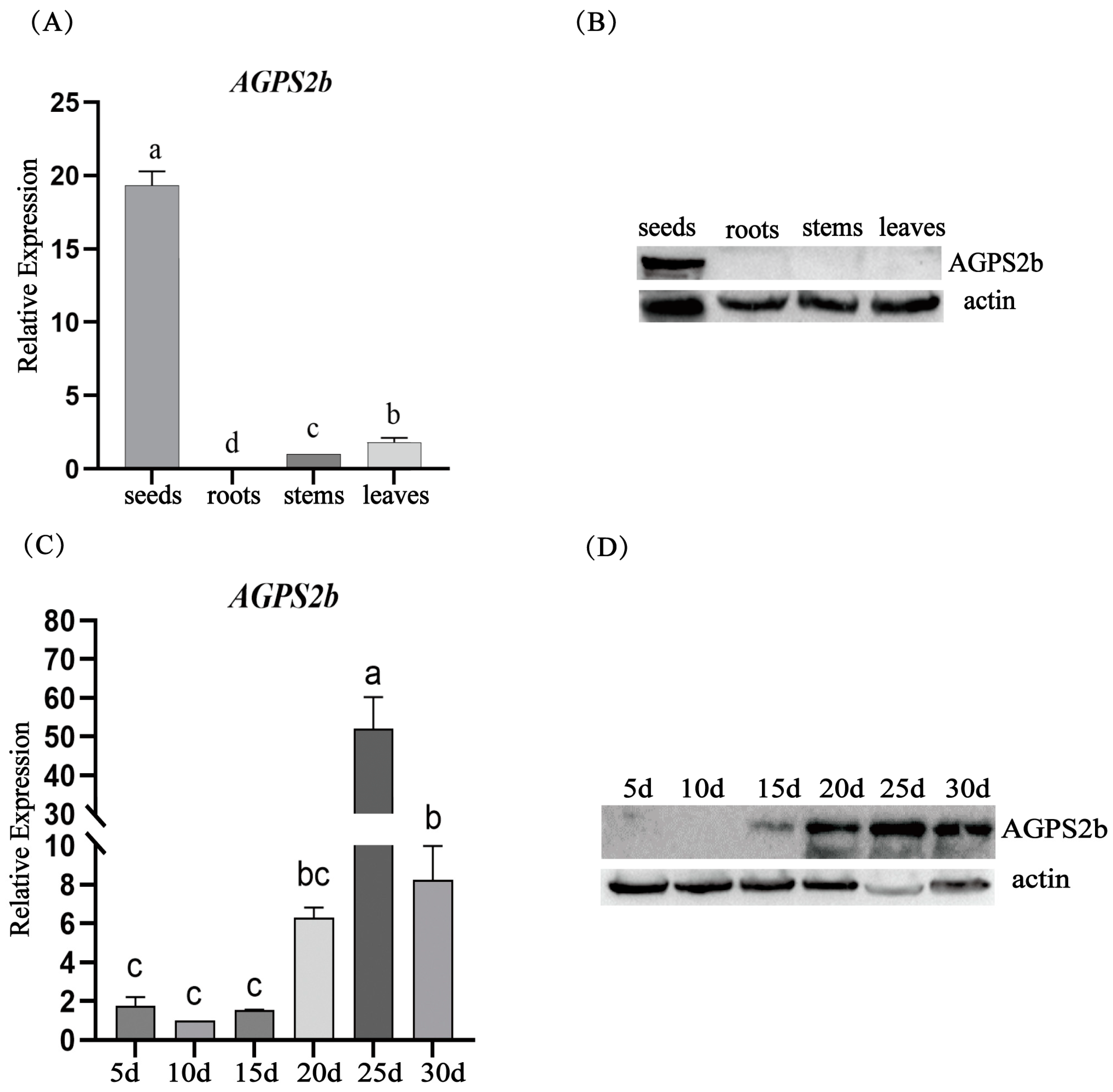
2.2.3. RNA Extraction and RT-qPCR Analysis of Tissue Expression
2.2.4. Protein Expression and Antibody Preparation
2.2.5. Western Blot
3. Results and Analysis
3.1. Biosignal Analysis of HvAGPS2b Protein
3.2. Cloning and Prokaryotic Expression of the HvAGPS2b Gene
3.3. Preparation and Titer Evaluation of HvAGPS2b Antibody
3.4. Tissue-Specific Expression Analysis
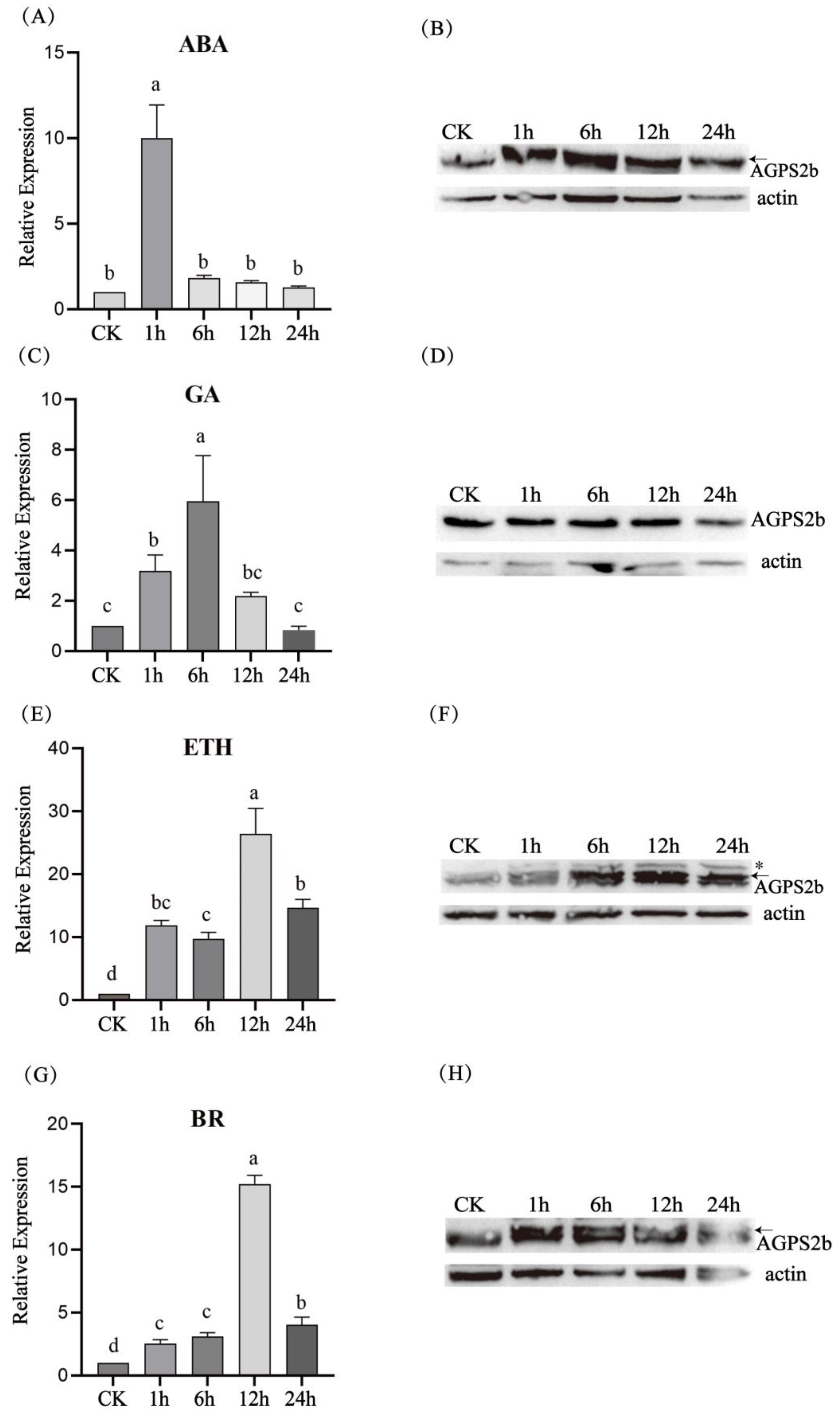
3.5. Expression Patterns of HvAGPS2b Treated with Hormones
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Keeling, P.L.; Myers, A.M. Biochemistry and genetics of starch synthesis. Annu. Rev. Food Sci. Technol. 2010, 1, 271–303. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Zhang, J.; Zhong, F.; Zhou, W.; Xu, R.; Ma, L. Comparison of the starch granule and kernel structure between feed and malt barley varieties. J. Triticeae Crops 2006, 26, 133–138. (In Chinese) [Google Scholar]
- Gao, J.; Vasanthan, T.; Hoover, R. Isolation and characterization of high-purity starch isolates from regular, waxy, and high-amylose hulless barley grains. Cereal Chem. 2009, 86, 157–163. [Google Scholar] [CrossRef]
- Gous, P.W.; Fox, G.P. Review: Amylopectin synthesis and hydrolysis—Understanding isoamylase and limit dextrinase and their impact on starch structure on barley (Hordeum vulgare) quality. Trends Food Sci. Technol. 2017, 62, 23–32. [Google Scholar] [CrossRef]
- Qu, J.; Xu, S.; Zhang, Z.; Chen, G.; Zhong, Y.; Liu, L.; Zhang, R.; Xue, J.; Guo, D. Evolutionary, structural and expression analysis of core genes involved in starch synthesis. Sci. Rep. 2018, 8, 12736. [Google Scholar] [CrossRef]
- Ohdan, T.; Francisco, P.B., Jr.; Sawada, T.; Hirose, T.; Terao, T.; Satoh, H.; Nakamura, Y. Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J. Exp. Bot. 2005, 56, 3229–3244. [Google Scholar] [CrossRef]
- Tetlow, I.J.; Emes, M.J. A review of starch-branching enzymes and their role in amylopectin biosynthesis. IUBMB Life 2014, 66, 546–558. [Google Scholar] [CrossRef] [PubMed]
- Boehlein, S.K.; Shaw, J.R.; Stewart, J.D.; Hannah, L.C. Studies of the kinetic mechanism of maize endosperm ADP-glucose pyrophosphorylase uncovered complex regulatory properties. Plant Physiol. 2010, 152, 1056–1064. [Google Scholar] [CrossRef]
- Okita, T.W.; Nakata, P.A.; Anderson, J.M.; Sowokinos, J.; Morell, M.; Preiss, J. The subunit structure of potato tuber ADPglucose pyrophosphorylase. [Solanum tuberosum L.]. Plant Physiol. 1990, 93, 785–790. [Google Scholar] [CrossRef]
- Cross, J.M.; Clancy, M.; Shaw, J.R.; Greene, T.W.; Schmidt, R.R.; Okita, T.W.; Hannah, L.C. Both subunits of ADP-glucose pyrophosphorylase are regulatory. Plant Physiol. 2004, 135, 137–144. [Google Scholar] [CrossRef]
- Kavakli, I.H.; Park, J.S.; Slattery, C.J. Analysis of allosteric effector binding sites of potato ADP-glucose pyrophosphorylase through reverse genetics. J. Biol. Chem. 2001, 276, 40834–40840. [Google Scholar] [CrossRef] [PubMed]
- Laughlin, M.J.; Payne, J.W.; Okita, T.W. Substrate binding mutants of the higher plant ADP-glucose phrophosphorylase. Phytochemistry 1998, 47, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Ballicora, M.A.; Fu, Y.; Nesbitt, N.M.; Preiss, J. ADP-Glucose pyrophosphorylase from potato tubers. Site-directed mutagenesis studies of the regulatory sites. Plant Physiol. 1998, 118, 265–274. [Google Scholar] [CrossRef][Green Version]
- Geigenberger, P. Regulation of starch biosynthesis in response to a fluctuating environment. Plant Physiol. 2011, 155, 1566–1577. [Google Scholar] [CrossRef]
- Luo, C.; Déjardin, A.; Villand, P.; Doan, D.N.; Kleczkowski, L.A. Differential processing of homologues of the small subunit of ADP-glucose pyrophosphorylase from barley (Hordeum vulgare) tissues. Z. Für Naturforschung C J. Biosci. 1997, 52, 807–811. [Google Scholar] [CrossRef]
- Siedlecka, A.; Ciereszko, I.; Mellerowicz, E.; Martz, F.; Chen, J.; Kleczkowski, L.A. The small subunit ADP-glucose pyrophosphorylase (ApS) promoter mediates okadaic acid-sensitive uidA expression in starch-synthesizing tissues and cells in Arabidopsis. Planta 2003, 217, 184–192. [Google Scholar] [CrossRef] [PubMed]
- Sikka, V.K.; Choi, S.B.; Kavakli, I.H.; Sakulsingharoj, C.; Gupta, S.; Ito, H.; Okita, T.W. Subcellular compartmentation and allosteric regulation of the rice endosperm ADPglucose pyrophosphorylase. Plant Sci. 2001, 161, 461–468. [Google Scholar] [CrossRef]
- Chen, B.Y.; Janes, H.W. Multiple forms of ADP-glucose pyrophosphorylase from tomato leaf. Physiol. Plant. 1998, 113, 235–241. [Google Scholar] [CrossRef][Green Version]
- Tiessen, A.; Hendriks, J.H.; Stitt, M.; Branscheid, A.; Gibon, Y.; Farré, E.M.; Geigenberger, P. Starch synthesis in potato tubers is regulated by post-translational redox modification of ADP-glucose pyrophosphorylase: A novel regulatory mechanism linking starch synthesis to the sucrose supply. Plant Cell Online 2002, 14, 2191–2213. [Google Scholar] [CrossRef]
- Boehlein, S.K.; Shaw, J.R.; Stewart, J.D.; Hannah, L.C. Heat stability and allosteric properties of the maize endosperm ADP-glucose pyrophosphorylase are intimately intertwined. Plant Physiol. 2008, 146, 289–299. [Google Scholar] [CrossRef][Green Version]
- Tetlow, I.J.; Morell, M.K.; Emes, M.J. Recent developments in understanding the regulation of starch metabolism in higher plants. J. Exp. Bot. 2004, 55, 2131–2145. [Google Scholar] [CrossRef]
- Hendriks, J.H.; Kolbe, A.; Gibon, Y.; Stitt, M.; Geigenberger, P. ADP-glucose pyrophosphorylase is activated by posttranslational redox-modification in response to light and to sugars in leaves of Arabidopsis and other plant species. Plant Physiol. 2003, 133, 838–849. [Google Scholar] [CrossRef]
- Tiessen, A.; Prescha, K.; Branscheid, A. Evidence that SNF1-related kinase and hexokinase are involved in separate sugar-signalling pathways modulating post-translational redox activation of ADP-glucose pyrophosphorylase in potato tubers. Plant J. 2003, 35, 490–500. [Google Scholar] [CrossRef]
- Kaur, V.; Madaan, S.; Behl, R.K. ADP-glucose pyrophosphorylase activity in relation to yield potential of wheat: Response to independent and combined high temperature and drought stress. Cereal Res. Commun. 2017, 45, 181–191. [Google Scholar] [CrossRef]
- Yu, G.; Shoaib, N.; Yang, Y.; Liu, L.; Mughal, N.; Mou, Y.; Huang, Y. Effect of phosphorylation sites mutations on the subcellular localization and activity of AGPase Bt2 subunit: Implications for improved starch biosynthesis in maize. Agronomy 2023, 13, 2119. [Google Scholar] [CrossRef]
- Hwang, S.K.; Salamone, P.R.; Okita, T.W. Allosteric regulation of the higher plant ADP-glucose pyrophosphorylase is a product of synergy between the two subunits. FEBS Lett. 2005, 579, 983–990. [Google Scholar] [CrossRef]
- Rösti, S.; Rudi, H.; Rudi, K.; Opsahl-Sorteberg, H.-G.; Fahy, B.; Denyer, K. The gene encoding the cytosolic small subunit of ADP-glucose pyrophosphorylase in barley endosperm also encodes the major plastidial small subunit in the leaves. J. Exp. Bot. 2006, 57, 3619–3626. [Google Scholar] [CrossRef][Green Version]
- Huang, B. Function of Key Enzymes in Corn Starch Synthesis: AGPase and Isoamylase; Sichuan Agricultural University: Ya’an, China, 2014. (In Chinese) [Google Scholar]
- Giroux, M.J.; Hannah, L.C. ADP-glucose pyrophosphorylase in shrunken-2 and brittle-2 mutants of maize. Mol. Gen. Genet. 1994, 243, 400–408. [Google Scholar] [CrossRef]
- Prioul, J.L.; Jeannette, E.; Reyss, A. Expression of ADP-glucose pyrophosphorylase in maize (Zea mays L.) grain and source leaf during grain filling. Plant Physiol. 1994, 104, 179–187. [Google Scholar] [CrossRef][Green Version]
- Weber, H.; Heim, U.; Wobus, B.U. Cell-type specific, coordinate expression of two ADP-glucose pyrophosphorylase genes in relation to starch biosynthesis during seed development of Vicia faba L. Planta 1995, 195, 352–361. [Google Scholar] [CrossRef] [PubMed]
- Nakata, P.A.; Greene, T.W.; Anderson, J.M. Comparison of the primary sequences of two potato tuber ADP-glucose pyrophosphorylase subunits. Plant Mol. Biol. 1991, 17, 1089–1093. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.M.; Hnilo, J.; Larson, R.; Okita, T.W.; Morell, M.; Preiss, J. The encoded primary sequence of a rice seed ADP-glucose pyrophosphorylase subunit and its homology to the bacterial enzyme. J. Biol. Chem. 1989, 264, 12238–12242. [Google Scholar] [CrossRef] [PubMed]
- Villand, P.; Olsen, O.A.; Kleczkowski, L.A. Molecular characterization of multiple cDNA clones for ADP-glucose pyrophosphorylase from Arabidopsis thaliana. Plant Mol. Biol. 1993, 23, 1279–1284. [Google Scholar] [CrossRef]
- Chen, Y.; Gao, Y. Overexpression of small subunit NtAGPase gene increases starch content and biomass in tobacco leaf. Chin. J. Biol. Eng. 2021, 37, 2845–2855. [Google Scholar] [CrossRef]
- Müller-Röber, B.T.; Kossmann, J.; Hannah, L.C. One of two different ADP-glucose pyrophosphorylase genes from potato responds strongly to elevated levels of sucrose. Mol. Gen. Genet. Mgg 1990, 224, 136–146. [Google Scholar] [CrossRef]
- Tuncel, A.; Cakir, B.; Hwang, S.K. The role of the large subunit in redox regulation of the Rice endosperm ADP-glucose pyrophosphorylase. FEBS J. 2014, 281, 4951–4963. [Google Scholar] [CrossRef]
- Lescot, M. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Hess, J.R.; Carman, J.G.; Banowetz, G.M. Hormones in wheat kernels during embryony. J. Plant Physiol. 2002, 159, 379–386. [Google Scholar] [CrossRef]
- Salamone, P.R.; Greene, T.W.; Kavakli, I.H. Isolation and characterization of a higher plant ADP-glucose pyrophosphorylase small subunit homotetramer. Febs Lett. 2000, 482, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Burton, R.A.; Johnson, P.E.; Beckles, D.M.; Fincher, G.B.; Jenner, H.L.; Naldrett, M.J.; Denyer, K. Characterization of the genes encoding the cytosolic and plastidial forms of ADP-glucose pyrophosphorylase in wheat endosperm. Plant Physiol. 2002, 130, 1464–1475. [Google Scholar] [CrossRef] [PubMed]
- Johnson, P.E.; Patron, N.J.; Bottrill, A.R.; Dinges, J.R.; Fahy, B.F.; Parker, M.L.; Waite, D.N.; Denyer, K. A low-starch barley mutant, risø 16, lacking the cytosolic small subunit of ADP-glucose pyrophosphorylase, reveals the importance of the cytosolic isoform and the identity of the plastidial small subunit. Plant Physiol. 2003, 131, 684–696. [Google Scholar] [CrossRef] [PubMed]
- Seferoglu, A.B.; Gul, S.; Dikbas, U.M.; Baris, I.; Koper, K.; Caliskan, M.; Cevahir, G.; Kavakli, I.H. Glu-370 in the large subunit influences the substrate binding, allosteric, and heat stability properties of potato ADP-glucose pyrophosphorylase. Plant Sci. 2016, 252, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Garcia, C.M.H.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2013, 217–218, 109–119. [Google Scholar] [CrossRef]
- Yu, A. Effects of Brassicin (BR) and Gibberellin (GA) on Starch Accumulation, Grain Size Distribution and Processing Characteristics of Wheat Grain; Shandong Agricultural University: Tai’an, China, 2011. [Google Scholar] [CrossRef]
- Himmelbach, A.; Iten, M.; Grill, E. Signalling of abscisic acid to regulate plant growth. Philos. Trans. R. Soc. B Biol. Sci. 1998, 353, 1439–1444. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Cai, S.; Ye, L. The effects of GA and ABA treatments on metabolite profile of germinating barley. Food Chem. 2016, 192, 928–933. [Google Scholar] [CrossRef]
- Liu, L.; Qing, Y.; Shoaib, N.; Di, R.; Liu, H.; Li, Y.; Hu, Y.; Huang, Y.; Yu, G. Preparation of polyclonal antibody against ZmBT1 protein and its application in hormone-regulated starch synthesis. Agronomy 2023, 13, 1805. [Google Scholar] [CrossRef]
- Tang, T.; Xie, H.; Wang, Y. The effect of sucrose and abscisic acid interaction on sucrose synthase and its relationship to grain filling of rice (Oryza sativa L.). J. Exp. Bot. 2009, 60, 2641–2652. [Google Scholar] [CrossRef]
- Kondhare, K.R.; Hedden, P.; Kettlewell, P.S.; Farrell, A.D. Quantifying the impact of exogenous abscisic acid and gibberellins on pre-maturity α-amylase formation in developing wheat grains. Sci. Rep. 2014, 19, 5355. [Google Scholar] [CrossRef]
- Yang, J. Activities of key enzymes in sucrose-to-starch conversion in wheat grains subjected to water deficit during grain filling. Plant Physiol. 2004, 135, 1621–1629. [Google Scholar] [CrossRef]
- Rook, F.; Corke, F.; Card, R.; Munz, G.; Smith, C. Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. Plant J. 2010, 26, 421–433. [Google Scholar] [CrossRef]
- Hauvermale, A.L.; Ariizumi, T.; Steber, C.M. Gibberellin Signaling: A theme and variations on DELLA repression. Plant Physiol. 2012, 160, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, X.; Wang, X. The influence of different plant hormones on biomass and starch accumulation of duckweed: A renewable feedstock for bioethanol production. Renew. Energy 2019, 138, 659–665. [Google Scholar] [CrossRef]


| Primer Names | Primer Sequences (5′–3′) | Size (bp) | Purpose |
|---|---|---|---|
| HvAGPS2b-F | ATGGCGATGGCCGCGGCCGCCT | 22 | gene cloning |
| HvAGPS2b-R | TCATATGACTGTTCCACTAGGG | 22 | gene cloning |
| RT-AGPS2b-F | AAAGGAGAACAGTTGAAA | 18 | RT-qPCR |
| RT-AGPS2b-R | CAGTAACCGTCGTATAGG | 18 | RT-qPCR |
| Pet-30a-AGPS2b-F | GACAGCCCAGATCTGGGTA CCATGGCGATGGCCGCGGCC | 39 | vector construction |
| Pet-30a-AGPS2b-R | TTGTCGACGGAGCTCGAATTCTCATATGACTGTTCCACTAGGGAGTAA | 48 | vector construction |
| β-Actin-F | GTTCCAATCTATGAGGGATACACGC | 25 | RT-qPCR |
| β-Actin-R | GAACCTCCACTGAGAACAACATTACC | 26 | RT-qPCR |
| Motif | Sequence | Quantities | Possible Function |
|---|---|---|---|
| TATA-box | TATA | 612 | core promoter element around -30 of transcription start |
| ARE | AAACCA | 419 | a cis-acting regulatory element essential for the anaerobic induction |
| MBS | CAACTG | 361 | MYB binding site involved in drought-inducibility |
| P-box | CCTTTTG | 1983 | gibberellin-responsive element |
| GATA-motif | GATAGGA | 1652 | part of a light-responsive element |
| LTR | CCGAAA | 1368 | cis-acting element involved in low-temperature responsiveness |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xi, B.; Zhou, Q.; Guo, Y.; Shaoib, N.; Cheng, Z.; Gao, Y.; Liu, Y.; Zhao, H.; Feng, Z.; Yu, G. Preparation of Barley AGPS2b Antibody and Its Application in Hormone Regulation Research. Agriculture 2024, 14, 1712. https://doi.org/10.3390/agriculture14101712
Xi B, Zhou Q, Guo Y, Shaoib N, Cheng Z, Gao Y, Liu Y, Zhao H, Feng Z, Yu G. Preparation of Barley AGPS2b Antibody and Its Application in Hormone Regulation Research. Agriculture. 2024; 14(10):1712. https://doi.org/10.3390/agriculture14101712
Chicago/Turabian StyleXi, Boai, Qiyan Zhou, Yang Guo, Noman Shaoib, Zhenbin Cheng, Yan Gao, Yajie Liu, Hui Zhao, Zongyun Feng, and Guowu Yu. 2024. "Preparation of Barley AGPS2b Antibody and Its Application in Hormone Regulation Research" Agriculture 14, no. 10: 1712. https://doi.org/10.3390/agriculture14101712
APA StyleXi, B., Zhou, Q., Guo, Y., Shaoib, N., Cheng, Z., Gao, Y., Liu, Y., Zhao, H., Feng, Z., & Yu, G. (2024). Preparation of Barley AGPS2b Antibody and Its Application in Hormone Regulation Research. Agriculture, 14(10), 1712. https://doi.org/10.3390/agriculture14101712






