Abstract
Global warming poses a serious threat to food security because of its impacts on thermosensitive food crop production. Rice is of paramount significance due to the world’s three-billion-population dependence on it as a staple food. It is well established that the high temperatures at day or night times during the grain-filling period can reduce rice grain yield, although the intriguing impact of high temperatures on head rice yields (HRY) is poorly discussed. This is because high and stable HRY is vital to meet the demand for rice grain, which is a staple food for many developing and developed nations. Hence, identifying the novel heat-tolerant rice germplasm with higher head rice yields may help mitigate a critical problem threatening global food security resulting from climate change. This review addresses the key factors, including pre-and-post-harvest scenarios related to overall reductions in the HRY and how grain molecular composition can play a significant role in determining head rice yields. Moreover, the underlying genetics of head rice is discussed as and possible mechanism to breach the complexity of HRY before identifying the key alleles and genomic regions related to the reduction in the HRY. Future research should focus on understanding the mechanisms of tolerating heat stress in rice by combining modern statistical, physiological, and molecular techniques to increase HRY. This may include high-throughput phenotyping techniques, mapping quantitative loci affecting HRY loss processes and genomic prediction using a broad wild and cultivated rice germplasm.
1. Introduction
Freshly harvested rice (Oryza sativa L.) grains, known as “paddy” or “rough” rice, are usually milled before human consumption. Paddy typically comprises 20% husk, 11% bran (aleurone layer) and 69% starchy endosperm [1,2,3]. Milling involves de-hulling the paddy and polishing, where the bran layer and part of the embryo are removed from brown rice to produce broken and un-broken (whole grain) starchy endosperm kernels, known as polished rice [1]. Polishing is important because brown rice, produced after the removal of the indigestible husk tissue, contains lipids and fatty acids in the bran layer, which is problematic for long-term grain storage. They hydrolyze and oxidize to form aldehydes and ketones, making the rice grain rancid over long-term storage periods [4,5].
In the context of the global rice production pattern, head rice yields derive from the paddy (Table 1). Head rice yield is typically affected by the milling operations (dehulling and polishing) and could result in grain breakage. The whole grain portion of the polished white rice—the head rice yield—is typically around 60–65% of the paddy yield [6,7]. Head rice is used for human consumption, while broken kernels are used for animal feed, rice flour, and the brewing industry [8]. The degree to which the aleurone layer (bran) and embryo are removed in the polishing process has implications for the quality of the milled product and depends on the milling intensity, length of time the rice is milled and grain shape. For these reasons, milling time and intensity are generally optimized for individual rice cultivars [9] to maximize head rice yield while meeting market requirements [10,11,12].

Table 1.
Production patterns of head rice yields (million metric tons) across the globe in major rice-producing regions Source: [13].
Other factors include grain moisture during harvesting and drying [14], and methods used to dry down the grain/paddy (hot air or infrared drying) [15,16,17] have a critical impact on head rice yield [14]. In addition, seasonal conditions have been linked to higher grain breakage during milling [18,19,20]. High day/night temperatures, especially during gametogenesis and early anthesis, result in substantial grain yield losses [9]. Moreover, heat stress during early and late grain filling periods affects the grain yields and milling outturn [14]. Grain breakage during milling is higher in immature grains [21], and it is therefore likely that seasonal conditions affect the uniformity of grain-filling or reduce the number of immature grains at harvest, increasing head rice yields. Further, some lines of evidence suggest that rice grain molecular components (starch, seed storage proteins, starch lipids) may be linked to grain breakage during milling [22,23,24,25]. Therefore, in addition to reviewing known factors that influence head rice yields, we review the key molecular components of rice grains, rice grain chalkiness and the impact of seasonal conditions (including temperature) on grain molecular composition, chalkiness, and the uniformity of grain-filling, with the view that these factors may ultimately have consequences on head rice yield.
This review discusses the potential impact of high temperature on HRY in respect to pre-and-post harvest scenarios. This review further resolves the effect of heat stress on losses during milling operations associated with the reductions of HRY and elaborates on genetic variation for the response of HRY and component traits, including grain molecular composition, which could be exploited to map quantitative trait loci (QTLs) or single-nucleotide polymorphisms (SNPs).
2. Factors Affecting Head Rice Yields
2.1. Rice Grain Size and Shape
There is wide variation in rice grain size and shape, which has necessitated the construction of a system to classify the rice grains by size and shape. Rice grain is generally grouped into one of the four grain length categories—short, medium, long and extra-long/slender grain (Figure 1), where short grains are ≤5.50 mm in length, medium grains are 5.51–6.60 mm, long grains are 6.61–7.5 mm and extra-long/slender grains are >7.5 mm in length [26].
Rice grain size and shape are largely determined by genetics and a few quantitative trait loci (QTLs). Rice grain size and shape are identified in Table 2. For example, the QTL GS3 (Table 2) plays a major effect on QTL for rice grain length and weight and a minor effect on QTL for rice grain width and thickness [27,28]. GL3.1 regulates the grain length and weight, while qgl3 has a favorable effect on grain length, filling and weight [29]. GW2 increases spikelet hull width and accelerates the grain-filling rate, grain width and weight [30]. However, none of the QTLs for grain length and width (Table 2) have been directly linked to head rice yield.

Table 2.
Quantitative traits loci of rice grain size and shape.
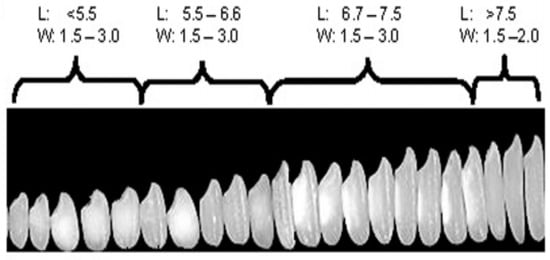
Figure 1.
Rice grain diversity in size (thickness) and shape (length, width and length-to-width ratio). Source: [31].
Figure 1.
Rice grain diversity in size (thickness) and shape (length, width and length-to-width ratio). Source: [31].
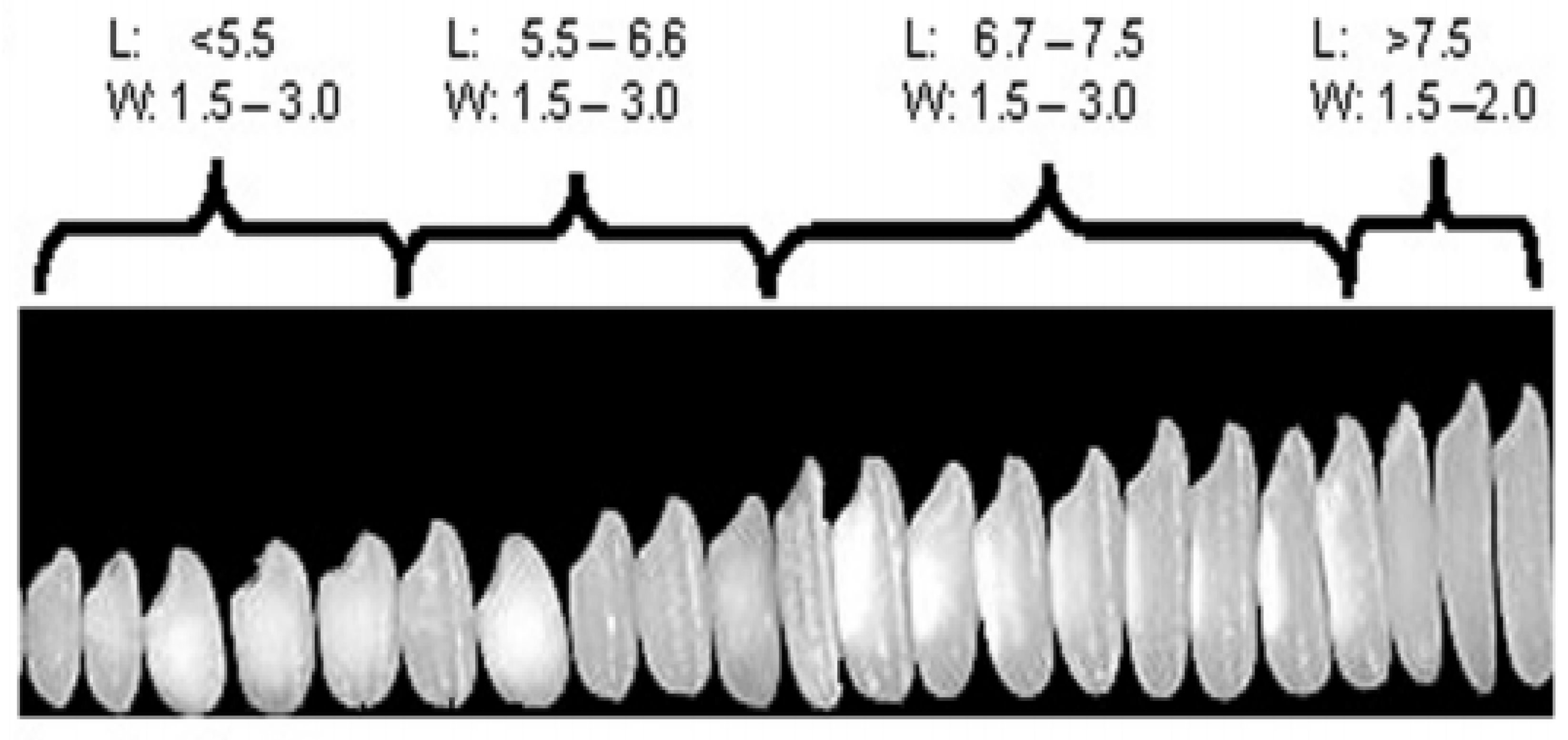
Head rice yield varies with the shape and size of the rice grain, but contradictory findings have been reported. For example, [56,57] observed higher head rice yield in short grains than in medium and long grains. In contrast, [58,59] found higher head rice yields in medium-grain cultivars than in short- and long-grain cultivars. The contradictory findings could be due to the seasonal environmental conditions where rice cultivars were grown and/or factors beyond physiological maturity or may be due to genetics. However, ultimately, the reason for the discrepancy still needs to be solved.
2.2. Rice Grain Molecular Composition
Rice grain is mainly composed of starch (89–92%), seed storage proteins (6–8%) and lipids (1–2%) [60,61], and these components largely determine the rice grain functional properties, including gelatinization temperature, gelatinization index, flour viscosity and RVA (rapid viscos analyzer) pasting properties [62,63]. The roles of starch, lipids and seed storage proteins have been widely studied from a cooking and eating perspective. However, the role of these grain components in determining the head rice yield still needs to be determined despite several studies demonstrating a potential association between one or more of these components and grain breakage, as reviewed below.
2.2.1. Starch
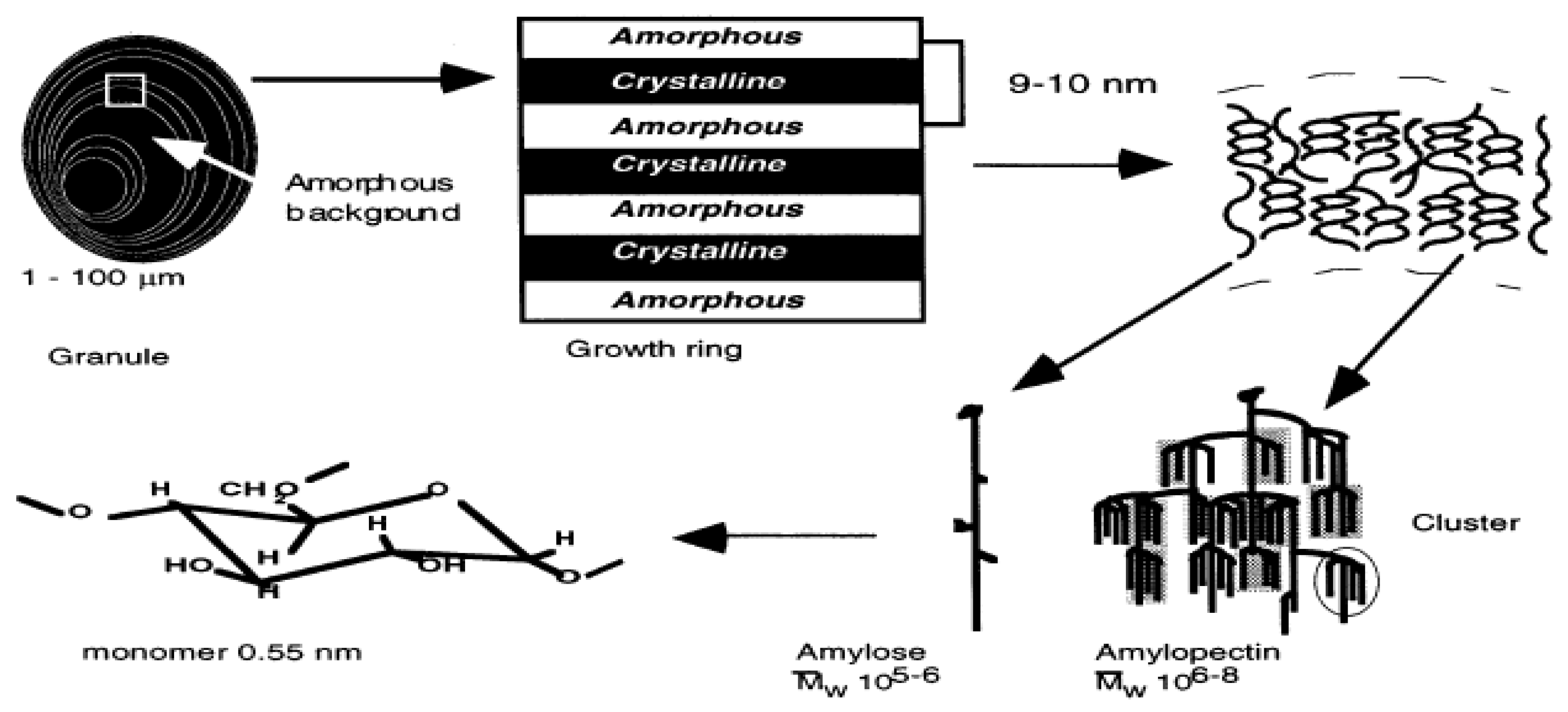
Starch, the dominant component of the rice grain endosperm, is a polymer composed of glucose monomers and is found in one of two types: amylose (0–30%) or amylopectin (75–80%) [64]. Amylose is primarily a linear molecule joined by α1-4 links while amylopectin has α1-6 linkages in addition to α1-4 links [65]. The α1-6 linkage creates a branched structure, and these structural differences affect physio-chemical properties such as solubility: amylopectin is water soluble, while amylose is not soluble in water. Structural analysis of starch granules [66] shows that amylose molecules reside between the amylopectin molecules (Figure 2), and their relative concentration is important for starch stability.
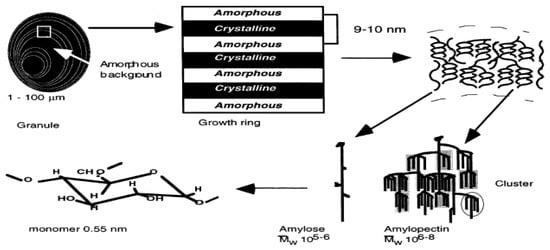
Figure 2.
Schematic representation of different structural levels of the starch granule and the involvement of amylose and amylopectin in developing starch molecules [64].
Amylose is synthesized by a granule-bound starch synthase (GBSSI) coded by the waxy gene (Waxy/wx) [67] while amylopectin is synthesized by (Sbe) starch synthase branching and debranching enzymes [68,69]. DNA sequence differences within the rice starch synthesis structural genes generate allelic diversity, which results in rice starch composition differences. However, any of the changes caused by Wx and Sbe genes have not been linked to head rice yield.
The importance of starch in determining head rice yield is somewhat unclear (Table 3). A temperature stress study [70] reported that head rice yields declined with increasing night-time temperature as amylopectin carbon chain lengths increased. In contrast, another study [63] found no association between amylose:amylopectin ratios and head rice yield across a range of rice cultivars. Similar conflicting results have been reported for the relationship between amylose and grain chalkiness, a trait linked to reduced head rice yields, discussed in Section 2.2.4 below.

Table 3.
Quantitative traits loci for endosperm starch and their importance in rice grain quality.
2.2.2. Seed Storage Proteins
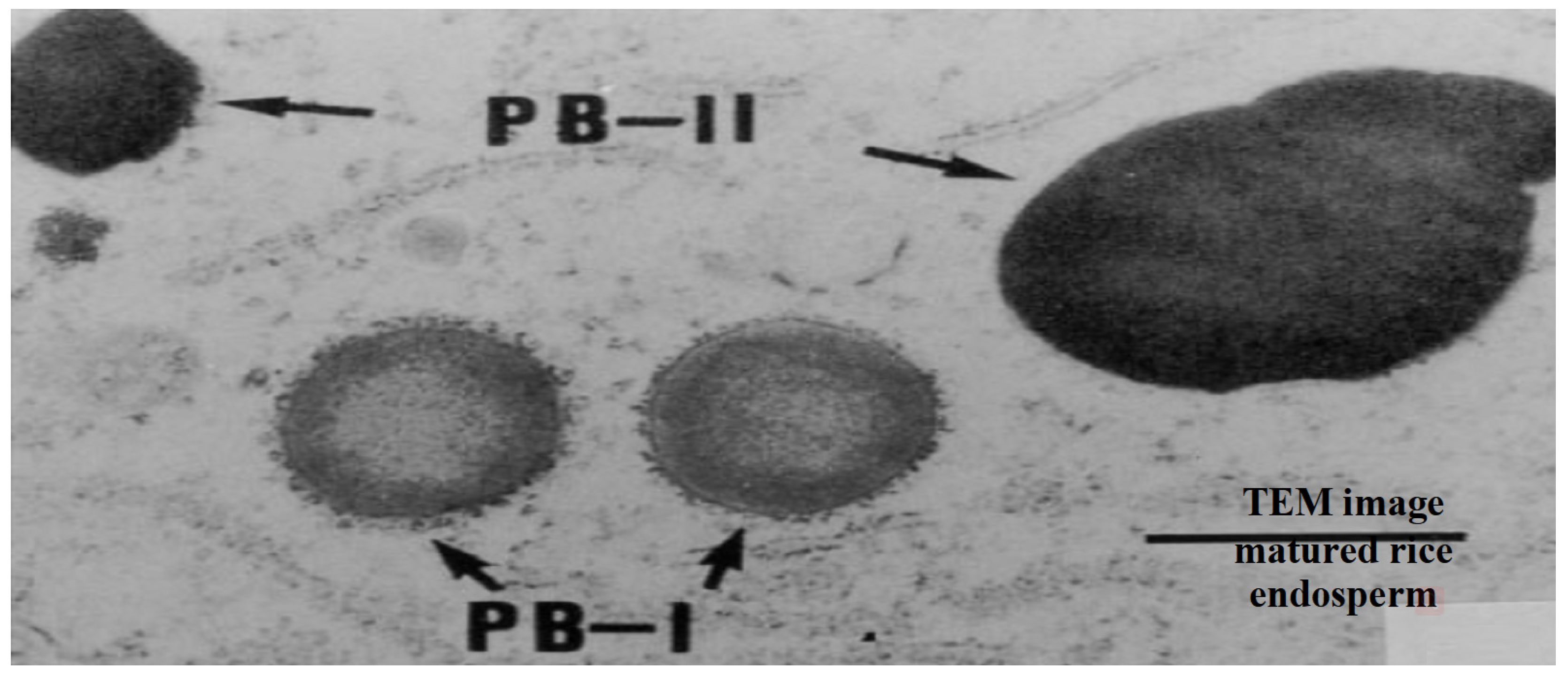
Seed storage proteins in rice endosperm are composed of glutelins (60–80%), prolamins (20–30%) and globulins (5–6%) [60]. Seed storage proteins are traditionally defined by their solubility, where glutelins are soluble in dilute acid or base, prolamins in aqueous alcohol solutions, and globulins in salt solutions. These seed storage proteins, referred to as protein body I (PB-I) and protein body II (PB-II) (Figure 3), are embedded between the starch granules, helping them to bind [75]. Prolamins are found within PB-I and have molecular masses of 10, 13 and 16 kDa [60,61,75]. The 13 kDa prolamins are the dominant component of PB-I and are further classified based on Cys residues [76]. The Cys-poor prolamins have one cysteine residue per molecule, while the Cys-rich have five to nine cysteine residues per molecule. Prolamins accumulate within the smooth endoplasmic reticulum [77], which then mature and form PB-I.
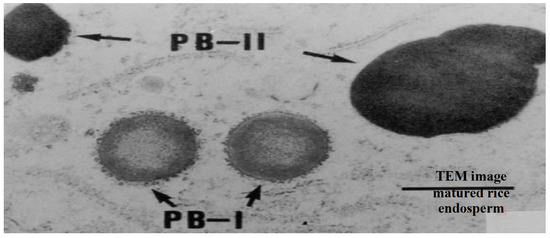
Figure 3.
Transmission electron microscope (TEM) image of ‘Protein body I (PB-I; where prolamins accumulate within the smooth endoplasmic reticulum)’ and Protein body-II (PB-II, where globulins and glutelins are accumulated into protein storage vacuoles)’ in matured rice endosperm. Source [75].
Globulins and glutelins are deposited into protein storage vacuoles, which then mature and form PB-II. Approximately 15 glutelin coding genes are divided based on DNA sequence into four gene families: GluA, GluB, GluC and GluD [78,79,80]. Globulin is dominated by a protein derived from a single-copy α-globulin encoding gene [81]. Several studies have reported the QTLs for seed storage proteins, but none for rice grain seed storage proteins (Table 4) have been linked to head rice yield.

Table 4.
Quantitative traits loci for rice grain proteins.
2.2.3. Starch Lipids
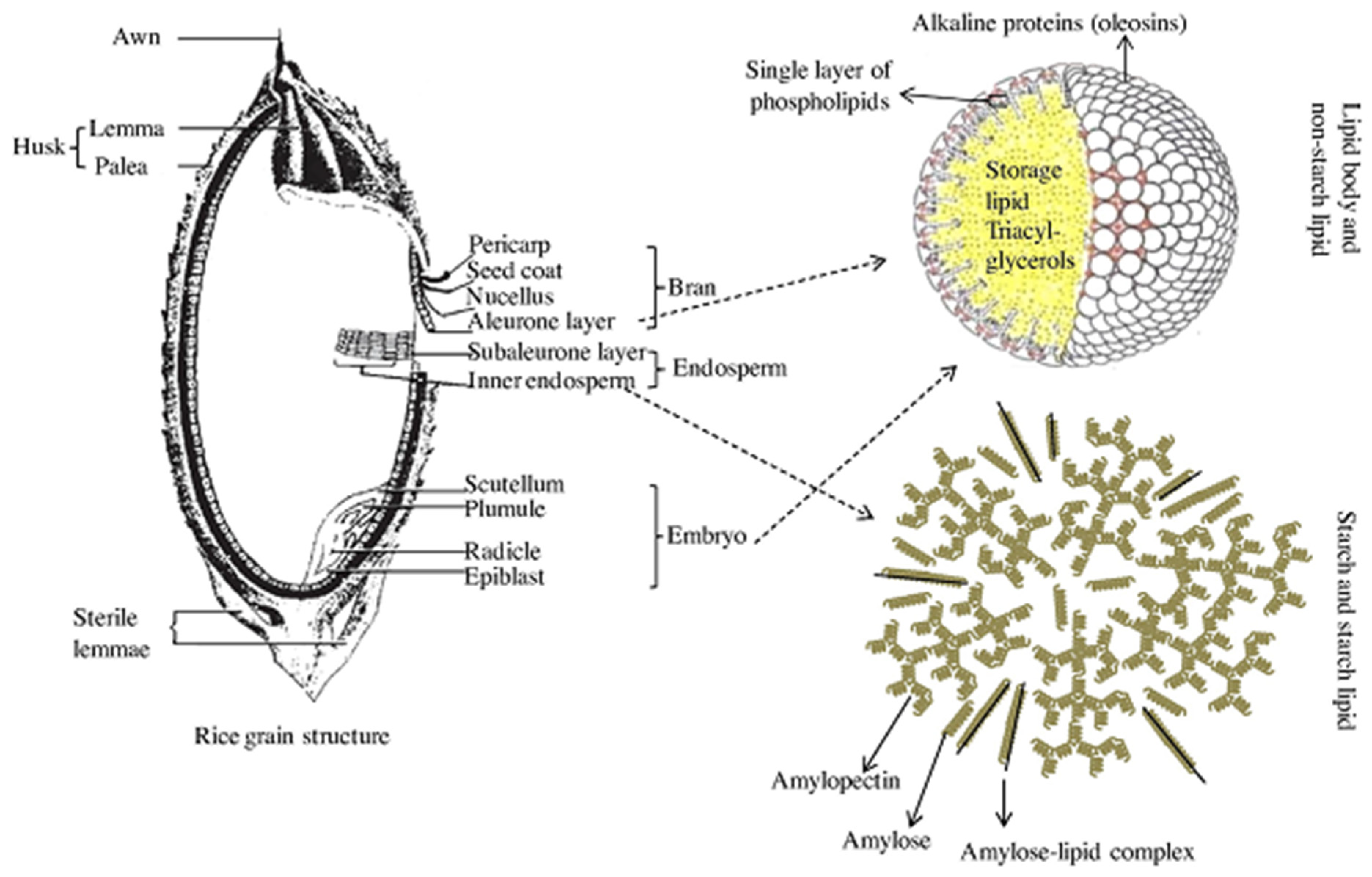
Typically, brown rice is composed of 1–2% lipids [89,90,91], which are generally classified as non-starch lipids and starch lipids [92,93,94], as shown in Figure 4. The non-starch lipids are found in the bran and embryo that are removed during milling, while starch lipids are located within the starchy endosperm portion of the rice grain. The starch lipids are chiefly composed of surface lipids, lysophosphatidyl choline, free fatty acids and lysophosphatidyl ethanolamine [95]. The surface lipids contain surface acylglycerols that are hydrolyzed through lipase action into free fatty acids [4,96] and are considered important for grain cracking during milling. For example, [22] observed high amounts of surface lipids in polished broken kernels compared to head rice (whole-grain polished kernels). Puroindoline proteins in wheat are lipid-binding proteins that play a key role in determining wheat hardness and flour milling yield [97], a trait that is analogous to grain breakage in rice. Several studies in rice have reported the QTLs for lipid contents (Table 5), but none of these have been linked definitively to head rice yields.
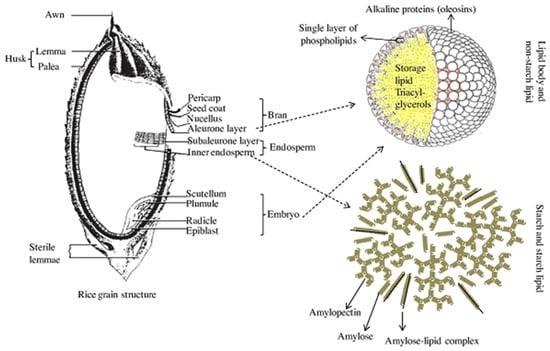
Figure 4.
Distribution and illustration of starch and non-starch lipids. Source [94].

Table 5.
Quantitative traits loci for grain lipids in rice.
2.2.4. Chalkiness
Chalkiness refers to an opaque area of the rice endosperm instead of a translucent endosperm in non-chalky rice. Chalkiness is considered a highly undesirable trait because the chalky appearance lowers the marketability of rice and causes significant milling losses [108,109,110,111,112,113,114,115].
Chalkiness is a highly undesirable trait that favors grain cracking during milling operations (dehusking and polishing), which ultimately reduces head rice yields [113]. The reason for the development of chalky grains has not been fully resolved yet [116]. Therefore, the following points need to be considered regarding the effect of rice crop physiology coupled with high-temperature stress for chalkiness.
- (1)
- High-temperature stress during grain filling [69,113,117] was considered a major contributing factor to the development of chalky grains.
- (2)
- However, other seasonal conditions such as wetting and drying cycles, are also associated with the development of chalky grains [118,119].
- (3)
- Previous literature also suggested that loosely packed starch molecules cause chalkiness, possibly due to variations in amylose content [108,120,121].
- (4)
- Other studies suggested that amylose and chalkiness are negatively correlated [113,114,122,123].
- (5)
- Environmental factors such as temperature, transpiration rate of canopy and evaporation rate contribute to developing chalkiness. Therefore, future studies are required to investigate which factors, environmental and genetic (in the context of Table 6), contribute to developing the chalky grains and finding the possible link between the rice grain molecular components and chalkiness.
 Table 6. Quantitative traits loci for chalkiness in rice grain.
Table 6. Quantitative traits loci for chalkiness in rice grain.
2.3. Rice Plant Morphology and Grain Breakage
During milling operations, immature and partially filled grains are easily broken, leading to reduced head rice yields [9,14,57,58]. The extent to which grains are filled with assimilates during grain filling is driven by source–sink relationships and plant morphological traits. These morphological traits include synchrony in tillering, panicle architecture and the annual/perennial nature of a rice cultivar, which alters the source–sink relationships [21]. For example, the GIF1 (grain incomplete filling 1) gene is associated with perennial nature in rice and downregulates the cell wall invertase that restricts carbon partitioning to grains during the early stages of grain filling, leading to immature or partially filled grains [14,130]. This gene appears to have arisen in cultivated rice (O. sativa) through introgression from the perennial wild rice O. rufipogon [130]. Panicle architecture also influences the rate and uniformity of grain filling within and between the panicles. For example, panicle length greater than 22% of plant height is associated with higher uniformity of grain filling within panicles [131]. The shorter panicle length to plant phytomass ratio could increase grain filling and development uniformity within and between the panicles, which could potentially increase head rice yield [57]. If genes controlling panicle architectural traits that are associated with more uniform grain filling could be identified, a pyramiding of these genes with genes associated with reduced perennial nature and more synchronous tillering into elite rice lines may help to lower the number of immature or partially filled grains, leading to higher head rice yields.
2.4. Pre-Harvest Factors That Affect Head Rice Yield
2.4.1. Temperature
Rice is highly sensitive to high-temperature stress (day/night), especially during gametogenesis and early anthesis (booting up to 50% flowering) stages, and heat stress during these stages results in substantial grain yield losses [6,19,132,133,134,135,136]. Heat stress during early grain filling downregulates the starch synthase and prolamin genes, which affects rice grain quality and yield [137]. The development of chalky grain, a main factor that has been reported that reduces head rice yields, is a result of high-temperature stress during grain-filling periods [18,19,69,118,138,139,140,141,142,143,144]. Other than this, rice grain functional properties such as setback viscosity, porosity, gelatinization temperature, pasting value and alkali spreading values are affected by high day and night-time temperatures [19], but whether any changes to these parameters are associated with the increased grain cracking during milling is not known.
2.4.2. Soil Moisture during Grain Filling
Soil moisture showed a considerable effect in altering the grain yield across the cereal crops following grain-filling stages in rice [145,146] and wheat [147]. Soil moisture level might affect the source-to-sink relationships where the remobilization of pre-stored carbon and nitrogen assimilates either increased or decreased from the source (vegetative parts) to the sink (grain) under soil moisture stress. However, agronomic practices, such as post-anthesis (50% days after flowering) soil drying, have the potential to increase the amount of carbon partitioning from the flag leaf to grains by regulating key plant hormones such as abscisic acid, cytokinins in leaves [148,149,150,151,152,153] and trans-zeatin-type cytokinins in the roots [154]. Grain-filling period proved to be the dominant factor that influences head rice yields [14]; therefore, soil drying practice during mid- and late grain-filling periods upregulated the starch branching and debranching enzymes (SuSase, AGPase, SSS, GBSS, SBE) and genes (SuS2, SuS3, SuS4, AGPS2b, AGPL1, AGPL2, SSSIIb, SSSIIc, GBSSI, GBSSII, SBEI, and SBEIIb) [69]. There is a possibility to investigate the effect of post-anthesis soil drying during the grain-filling periods (mid-and-late grain filling) that influence head rice yields by altering carbon partitioning from the flag leaf to developing grains.
2.4.3. Nitrogen
Nitrogen plays a key role in determining grain yield and protein contents [24]. Nitrogen fertilizer application typically leads to increasing grain protein content, albeit this is not always the case, and substantial genotype × environment interactions were observed [155,156,157,158,159,160,161,162]. The importance of nitrogen fertilizer application across various growth stages of rice plants (ranging from vegetative growth phases to gametogenesis and grain filling periods) in terms of rice grain cracking has not been extensively studied. However, some field studies reported nitrogen fertilizer application before flowering increased the amount and density of seed storage proteins and reduced chalkiness, which coincided with reducing grain cracking during milling operations [163,164,165].
2.5. Factors beyond Physiological Maturity Affecting Head Rice Yield
Head rice yield may vary with relative humidity (RH) or vapour pressure deficit (VPD), but contradictory findings have been reported. For example, [118] observed a lower head rice yield under high relative humidity and a high negative correlation between RH and HRY (r = −0.693). In contrast, [23,129] found a very weak positive correlation between RH and HRY (r = 0.168) over two-crop growing seasons across multiple growing locations. Both studies measured the RH following the entire period, grain ripening, and maturity stages (from heading/50% flowering to harvesting). The contradictory findings could be due to variation in grain moisture content at harvesting that has a confounding impact on HRY.
Paddy harvesting is normally recommended at moisture contents of about 20–25%, as a slow drying process from these levels down to around 14% (used for milling) helps to reduce fissuring and grain breakage during milling operations [16,17]. Generally, paddy drying involves the use of flat beds in hot air and/or infrared radiation chambers to reduce the grain moisture content to around 14% for dehusking and polishing [166,167]. The temperature of hot air and infrared chambers is recommended at 55 °C or less, selected based on the rice grain glass transition temperature, which is about 60 °C [14,16]. Glass transition temperature is defined as the state when polymers of starchy rice endosperm shift from soft rubbery material to hard glassy material and vice versa. Therefore, the paddy drying at or below the temperature of 55 °C makes the starchy endosperm soft and rubbery, which assists in reducing the fissuring and grain cracking during milling operations [168,169].
During milling operations, the degree of milling strongly affects head rice yield [11,12]. The degree of milling reflects how many passes are required for polishing [168]. Generally, a lower degree of milling achieves higher head rice yields and favours the retention of key nutrients in the polished grain [10]. The optimum degree of milling is generally different for short-, medium- and long-grain cultivars [169]; therefore, each grain type is recommended to be milled separately.
2.6. Genetics of Head Rice Yield
As discussed above, a wide range of rice plant morphological traits, grain size and shape, and grain molecular composition components such as chalkiness and seed storage proteins have been associated with head rice yield. Genetic variation exists for many of these traits, and the associated genes and QTLs that contribute to these traits have been identified (Table 7). However, it needs to be clarified if any of these QTL/genes have led to quantifiable improvements in breeding programs and thus to enhanced head rice yields. Given the range of environmental (temperature) and nutritional (nitrogen) factors that are associated with head rice yield, many of the mapped QTLs may be specific to the given environment (Table 7) under which the phenotyping was conducted and may not be robust across environments.

Table 7.
Quantitative traits loci for percentage head rice yield (the amount of whole grain polished white rice of the paddy from which it is derived and then multiplied by 100).
2.6.1. QTL Mapping for Head Rice Yield
The genetics of head rice yield has not been fully resolved, and the prediction power of QTL mapping requires trait variation decomposition to determine whether it is due to the main effect QTL (QTL × QTL, QTL × epistatic) and/or environmental (QTL × environment, epistatic × environment) effects. In the case of head rice yield, the environmental effects are not only confined to field variation but are also linked to variation associated with storage and milling operations. Dehusking and polishing (milling operations) are two distinct processes, and grain breakage during each step could be associated with head rice yield. Various QTLs for head rice yield have been reported, with most found on chromosomes 1 and 6 (Table 7). However, none of the studies listed in Table 7 clarify whether these are linked to grain breakage during the dehusking or polishing step. Therefore, “QTL × milling environment” could be included as a separate parameter while performing QTL mapping for head rice yield.
A further consideration is that the embryo is diploid, while the endosperm is triploid, and the present QTL models are designed for a diploid inheritance, with no software available for triploid inheritance. The development of triploid models that could be used to capture the associated endosperm variation in relation to head rice yield may be beneficial.
2.6.2. GWAS (Genome-Wide Association Mapping)
Genome-wide association mapping is a recently developed tool in plant breeding that can capture the variation associated with candidate genes and/or SNPs to related traits. GWAS is based on linkage disequilibrium to identify trait–SNP association underlying complex traits. In rice breeding, the use of GWAS for head rice yield is limited. Most of the studies sampled their materials from the 3000 resequenced rice genome populations with population sizes ranging between 200 and 404 indica or japonica rice accessions [180,181,182,183,184,185,186,187]. These studies combined detected over one hundred loci associated with head rice yield and chalkiness-related traits distributed throughout the rice genome. However, none of these QTL overlapped among different studies with interval distance between any two adjacent QTL > 0.5 Mbp, except for one chalkiness QTL on the beginning of chromosome 3 that was detected in japonica [185] and indica germplasm [186]. This indicates that these studies detected environment-specific QTL.
All previous studies used a single trait mixed linear model to detect associated QTL. Therefore, to detect stable QTL across environments, future studies should adopt more advanced statistical models such as the multi-trait mixed linear model [187,188] or MetaGWAS model [189,190]. The latter would be more appropriate when raw data cannot be shared among different institutes, as it only requires sharing summary statistics. The major difficulty associated with combining the results of multiple studies is the ability to synchronize the SNPs used in all of them. However, this can be easily solved for a crop such as rice for which a large genomic stock exists in the 3000 resequenced genome projects, which facilitate the imputation from different genotyping platforms to a common high-density genotyping [191] or by imputing missing summary statistics [192], given that grain size and shape are strongly linked to head rice yields. Since milling is generally optimized for each grain shape, it will be interesting in future studies to fit these traits (or preferably the first few principal components of them to avoid their correlations) as covariates to ensure detecting loci for head rice yields that are independent of grain characters.
2.6.3. Genomic Selection
Genomic selection has the advantage of exploiting the whole genetic variation explained by the SNPs over QTL mapping and GWAS, which usually detect a small number of QTL with a large enough effect to pass the significance threshold [193]. However, the cumulative effect of these small numbers of QTL could not be enough to make informative long-term selections for complex quantitative traits. Therefore, QTL with a small effect can be captured with the genomic selection, which usually explains most of the phenotypic variation of the complex traits that are usually controlled by hundreds to thousands of genes. To the best of our knowledge, only two studies were conducted by the same group that investigated the potential of genomic selection to improve head rice yield and grain chalkiness [194,195]. Both studies worked on the same data, which involved 327 indica elite rice lines and 309/320 japonica elite rice lines. Both populations were phenotyped for multiple seasons in a single location.
The former study [194] investigated the prediction accuracy using two cross-validation schemes (CV1 and CV2) for five traits, including head rice yield and grain chalkiness, using different prediction models that exploit genotype by environment interactions. The indica and japonica populations were phenotyped in three seasons in a single location. In CV1, they completely masked a subset of the studied accessions to calculate the accuracy of predicting them. This approach mimics the case of introducing new materials to the breeding program. In CV2, they masked random plots across different field trials to mimic the case where different trials have different sets of lines to predict phenotypes for all lines in all trials. The heritability for head rice yield and chalkiness was considerably high, with 0.87 and 0.63 average values across all seasons for indica and japonica populations, respectively. The prediction accuracies for the top model ranged between 0.51 and 0.66 for head rice yield and between 0.33 and 0.61 across different seasons for both populations, with average values of 0.56 for head rice yield and 0.44 for chalkiness. Given the small reference population used in the study, the prediction accuracy could be further improved if more data were used to train the prediction equation.
In a later study [195], the authors used the same previous data with two extra seasons for the japonica population (a total of five seasons versus only three seasons for the indica population) to test the advantage of using environmental covariates simultaneously with their molecular variants in predicting the performance of their population in unobserved environments. Environmental covariates were recorded from the daily weather data for each rice accession on the day it reached a specific developmental stage, e.g., flowering. They used the “one-environment-out” cross-validation scheme in which they dropped the environment from the reference to be predicted. Across different seasons and applied models, the prediction accuracy reached a maximum of 0.5 for head rice yield and 0.3 for chalkiness for the indica population, while the accuracies were much higher for the japonica population with a maximum of 0.8 for head rice yield and 0.7 for chalkiness. This is not unexpected given that the reference japonica population had four seasons while the indica reference population had only two seasons. A major drawback of their study is their assumption that the phenology data (such as the flowering date used to calculate the environmental covariates) may not be possible to use for untested environments. Future studies should investigate the effect of predicted phenology traits on the prediction accuracy of untested environments. Alternatively, untested or future climates can be predicted in a more straightforward CGM-WGP, which integrates crop growth models and genomic prediction [196].
3. Conclusions and Outlooks
Several factors potentially affect the yields of head rice, including day and night-time temperatures across various growth stages including vegetative, reproductive, and grain filling periods, nitrogen fertilizer applications at days to 50% flowering and during key grain developmental stages, harvest moisture contents, paddy storage conditions and paddy moisture contents before milling operations (including dehusking and polishing). Among these factors, the temperature is critical because of the predicted changes in day and night-time temperatures expected to arise due to climate change. However, the study of rice crop physiology needs to be accounted for, for a better understanding of the heat stress that affects rice plant yield in terms of grain yields and head rice yields. Moreover, the measure of association is required between chalkiness and several other factors including temperature, nitrogen application and milling operations across the different genetic backgrounds including japonica, temperate japonica, indica, aus, aromatic, and wild types to develop potential links between chalkiness and grain breakage. Moreover, applying advanced statistical models such as metaGWAS and CGM-WGP will be required to accelerate the genetic development of head rice yield.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Juliano, B.O. The rice caryopsis and its composition. In Rice: Chemistry and Technology; Houston, D.F., Ed.; American Association of Cereal Chemists Inc.: St. Paul, MN, USA, 1985. [Google Scholar]
- Evers, T.; Millar, S. Cereal grain structure and development: Some implications for quality. J. Cereal Sci. 2002, 36, 261–284. [Google Scholar] [CrossRef]
- Bodie, A.R.; Micciche, A.C.; Atungulu, G.G.; Rothrock, M.J., Jr.; Ricke, S.C. Current Trends of Rice Milling Byproducts for Agricultural Applications and Alternative Food Production Systems. Front. Sustain. Food Syst. 2019, 3, 47. [Google Scholar] [CrossRef]
- Ohta, H.; Aibra, S.; Yamashita, H.; Sekiyama, F.; Morita, Y. Post-harvest drying of fresh rice grain and its effects on deterioration of lipids during storage. Agric. Biol. Chem. 1990, 54, 1157–1164. [Google Scholar]
- Urban-Alandete, L. Lipid Degradation during Grain Storage: Markers, Mechanisms and Shelf-Life Extension Treatments. Ph.D. Dissertation, School of Agriculture and Food Science, University of Queensland, Brisbane, Australia, 2018. [Google Scholar]
- Lyman, N.B.; Jagadish, K.S.V.; Nalley, L.L.; Dixon, B.L.; Siebenmorgen, T. Neglecting rice milling yield and quality underestimates economic losses from high-temperature stress. PLoS ONE 2013, 8, e72157. [Google Scholar] [CrossRef]
- Chandrajith, U.; Gunathilake, D.; Bandara, B.; Swarnasiri, D. Effects of Combine Harvesting on Head Rice Yield and Chaff Content of Long and Short Grain Paddy Harvest in Sri Lanka. Procedia Food Sci. 2016, 6, 242–245. [Google Scholar] [CrossRef]
- Esa, N.M.; Ling, T.B.; Peng, L.S. By-products of Rice Processing: An Overview of Health Benefits and Applications. Rice Res. Open Access 2013, 1, 1–11. [Google Scholar] [CrossRef]
- Ali, F.; Waters, D.; Ovenden, B.; Bundock, P.; Raymond, C.A.; Rose, T.J. Australian rice varieties vary in grain yield response to heat stress during reproductive and grain filling stages. J. Agron. Crop Sci. 2018, 205, 179–187. [Google Scholar] [CrossRef]
- Roy, P.; Ijiri, T.; Okadome, H.; Nei, D.; Orikasa, T.; Nakamura, N.; Shiina, T. Effect of processing conditions on overall energy consumption and quality of rice (Oryza sativa L.). J. Food Eng. 2008, 89, 343–348. [Google Scholar] [CrossRef]
- Sadeghi, M.; Hoseinian, S.H.; Hemmat, A. Influence of moisture content and whitening method on degree of milling and head rice yield of three Iranian rice varieties. Aust. J. Crop Sci. 2012, 6, 1481–1485. [Google Scholar]
- Sadeghi, M.; Nasrnia, E.; Masoumi, A.; Hemmat, A. Head rice yield response to low and high drying and tempering conditions. Int. Agrophysics 2013, 27, 219–223. [Google Scholar] [CrossRef]
- Statista. World Agriculture Production. 2022; p. 28. Available online: https://apps.fas.usda.gov/psdonline/circulars/production.pdf (accessed on 2 February 2023).
- Ali, F.; Waters, D.; Ovenden, B.; Bundock, P.; Raymond, C.; Rose, T. Heat stress during grain-filling reduces head rice yield through genotype dependant increased husk biomass and grain breakage. J. Cereal Sci. 2019, 90, 102820. [Google Scholar] [CrossRef]
- Abe, T.; Afzal, T.M. Tthin-layer infrared radiation drying of rough rice. J. Agric. Eng. Res. 1997, 67, 289–297. [Google Scholar] [CrossRef]
- Cnossen, A.G.; Siebenmorgen, T.J.; Yang, W. The glass transition temperature concept in rice drying and tempering: Effect on drying rate. Trans. ASAE 2002, 45, 759–766. [Google Scholar] [CrossRef]
- Dong, R.; Lu, Z.; Liu, Z.; Koide, S.; Cao, W. Effect of drying and tempering on rice fissuring analysed by integrating intra-kernel moisture distribution. J. Food Eng. 2010, 97, 161–167. [Google Scholar] [CrossRef]
- Peng, S.; Huang, J.; Sheehy, J.E.; Laza, R.C.; Visperas, R.M.; Zhong, X.; Centeno, G.S.; Khush, G.S.; Cassman, K.G. Rice yields decline with higher night temperature from global warming. Proc. Natl. Acad. Sci. USA 2004, 101, 9971–9975. [Google Scholar] [CrossRef]
- Lanning, S.B.; Siebenmorgen, T.J.; Ambardekar, A.A.; Counce, P.A.; Bryant, R.J. Effects of Nighttime Air Temperature During Kernel Development of Field-Grown Rice on Physicochemical and Functional Properties. Cereal Chem. 2012, 89, 168–175. [Google Scholar] [CrossRef]
- Nalley, L.; Dixon, B.; Tack, J.; Barkley, A.; Jagadish, K. Optimal harvest moisture content for maximizing mid-south rice milling yields and returns. Agron. J. 2016, 108, 701–712. [Google Scholar] [CrossRef]
- Jongkaewwattana, S.; Geng, S.; Hill, J.E.; Miller, B.C. Within-Panicle Variability of grain filling in Rice Cultivars with Different Maturities. J. Agron. Crop Sci. 1993, 171, 236–242. [Google Scholar] [CrossRef]
- Monsoor, M.A.; Proctor, A.; Siebenmorgen, T.J. Surface lipid and free fatty acids (FFA) content of head and broken rice produced by milling after different drying treatments. Cereal Chem. 2004, 81, 705–709. [Google Scholar] [CrossRef]
- Li, X.; Wu, L.; Geng, X.; Xia, X.; Wang, X.; Xu, Z.; Xu, Q. Deciphering the Environmental Impacts on Rice Quality for Different Rice Cultivated Areas. Rice 2018, 11, 7. [Google Scholar] [CrossRef]
- Grigg, B.C.; Siebenmorgen, T.J.; Norman, R.J. Effects of Nitrogen Rate and Harvest Moisture Content on Physicochemical Properties and Milling Yields of Rice. Cereal Chem. 2015, 93, 172–181. [Google Scholar] [CrossRef]
- Gao, Y.; Liu, C.; Li, Y.; Zhang, A.; Dong, G.; Xie, L.; Bin Zhang, B.; Ruan, B.; Hong, K.; Xue, D.; et al. QTL analysis for chalkiness of rice and fine mapping of a candidate gene for qACE9. Rice 2016, 9, 41. [Google Scholar] [CrossRef]
- Jennings, P.R.; Coffman, W.R.; Kauffman, H.E. Rice Improvement; IRRI: Los Banos, Laguna, Philippines, 1979; pp. 101–120. [Google Scholar]
- Fan, C.; Xing, Y.; Mao, H.; Lu, T.; Han, B.; Xu, C.; Li, X.; Zhang, Q. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 2006, 112, 1164–1171. [Google Scholar] [CrossRef] [PubMed]
- Takano-Kai, N.; Jiang, H.; Kubo, T.; Sweeney, M.; Matsumoto, T.; Kanamori, H.; Padhukasahasram, B.; Bustamante, C.; Yoshimura, A.; Doi, K.; et al. Evolutionary History of GS3, a Gene Conferring Grain Length in Rice. Genetics 2009, 182, 1323–1334. [Google Scholar] [CrossRef] [PubMed]
- Qi, P.; Lin, Y.S.; Song, X.J.; Shen, J.B.; Huang, W.; Shan, J.X.; Zhu, M.Z.; Jiang, L.W.; Gao, J.P.; Lin, H.X. The novel quantitative trait locus GL3.1 control rice grain size and yield by regulating Cyclin-T1;3. Cell Res. 2012, 22, 1666–1680. [Google Scholar] [CrossRef] [PubMed]
- Song, X.J.; Huang, W.; Shi, M.; Zhu, M.Z.; Lin, H.X. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 2007, 39, 623–630. [Google Scholar] [CrossRef]
- Calingacion, M.; Laborte, A.; Nelson, A.; Resurreccion, A.; Concepcion, J.C.; Daygon, V.D.; Mumm, R.; Reinke, R.; Dipti, S.; Bassinello, P.Z.; et al. Diversity of Global Rice Markets and the Science Required for Consumer-Targeted Rice Breeding. PLoS ONE 2014, 9, e85106. [Google Scholar] [CrossRef]
- Kubo, T.; Takano-Kai, N.; Yoshimura, A. RFLP mapping of genes for long kernel and awn on chromosome 3 in rice. Rice Genet. Newsl. 2001, 18, 26–28. [Google Scholar]
- Thomson, M.; Tai, T.H.; McClung, A.M.; Lai, X.-H.; Hinga, M.E.; Lobos, K.B.; Xu, Y.; Martinez, C.P.; McCouch, S.R. Mapping quantitative trait loci for yield, yield components and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar Jefferson. Theor. Appl. Genet. 2003, 107, 479–493. [Google Scholar] [CrossRef]
- Li, J.; Thomson, M.; McCouch, S.R. Fine Mapping of a Grain-Weight Quantitative Trait Locus in the Pericentromeric Region of Rice Chromosome 3. Genetics 2004, 168, 2187–2195. [Google Scholar] [CrossRef]
- Zhou, L.Q.; Wang, Y.P.; Li, S.G. Genetic analysis and physical mapping of lk-4(t), a major gene controlling grain length in rice, with a BC2F2 population. Acta Genet. Sin. 2006, 33, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Weng, J.; Gu, S.; Wan, X.; Gao, H.; Guo, T.; Su, N.; Lei, C.; Zhang, X.; Cheng, Z.; Guo, X.; et al. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 2008, 18, 1199–1209. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Qian, Q.; Liu, Z.; Sun, H.; He, S.; Luo, D.; Xia, G.; Chu, C.; Li, J.; Fu, X. Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 2009, 41, 494–497. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Luo, L.; Yan, W.; Kovi, M.R.; Zhan, W.; Xing, Y. Genetic dissection of rice grain shape using a recombinant inbred line population derived from two contrasting parents and fine mapping a pleiotropic quantitative trait locus qGL7. BMC Genet. 2010, 11, 16. [Google Scholar] [CrossRef]
- Kato, T.; Segami, S.; Toriyama, M.; Kono, I.; Ando, T.; Yano, M.; Kitano, H.; Miura, K.; Iwasaki, Y. Detection of QTLs for grain length from large grain rice (Oryza sativa L.). Breed. Sci. 2011, 61, 269–274. [Google Scholar] [CrossRef]
- Qiu, X.; Gong, R.; Tan, Y.; Yu, S. Mapping and characterization of the major quantitative trait locus qSS7 associated with increased length and decreased width of rice seeds. Theor. Appl. Genet. 2012, 125, 1717–1726. [Google Scholar] [CrossRef]
- Ishimaru, K.; Hirotsu, N.; Madoka, Y.; Murakami, N.; Hara, N.; Onodera, H.; Kashiwagi, T.; Ujiie, K.; Shimizu, B.-I.; Onishi, A.; et al. Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat. Genet. 2013, 45, 707–711. [Google Scholar] [CrossRef]
- Zhao, D.; Li, P.; Wang, L.; Sun, L.; Xia, D.; Luo, L.; Gao, G.; Zhang, Q.; He, Y. Genetic dissection of large grain shape in rice cultivar ‘Nanyangzhan’ and validation of a grain thickness QTL (qGT3.1) and a grain length QTL (qGL3.4). Mol. Breed. 2017, 37, 42. [Google Scholar] [CrossRef]
- Segami, S.; Takehara, K.; Yamamoto, T.; Kido, S.; Kondo, S.; Iwasaki, Y.; Miura, K. Overexpression of SRS5 improves grain size of brassinosteroid-related dwarf mutants in rice (Oryza sativa L.). Breed. Sci. 2017, 67, 393–397. [Google Scholar] [CrossRef]
- Hu, J.; Wang, Y.; Fang, Y.; Zeng, L.; Xu, J.; Yu, H.; Shi, Z.; Pan, J.; Zhang, D.; Kang, S.; et al. A rare allele of gs2 enhances grain size and grain yield in rice. Mol. Plant 2015, 8, 1455–1465. [Google Scholar] [CrossRef]
- Xu, C.; Liu, Y.; Li, Y.; Xu, X.; Xu, C.; Li, X.; Xiao, J.; Zhang, Q. Differential expression of GS5 regulates grain size in rice. J. Exp. Bot. 2015, 66, 2611–2623. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Tong, H.; Xiao, Y.; Che, R.; Xu, F.; Hu, B.; Liang, C.; Chu, J.; Li, J.; Chu, C. Activation of Big Grain1 significantly improves grain size by regulating auxin transport in rice. Proc. Natl. Acad. Sci. USA 2015, 112, 11102–11107. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xiong, G.; Hu, J.; Jiang, L.; Yu, H.; Xu, J.; Fang, Y.; Zeng, L.; Xu, E.; Xu, J.; et al. Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 2015, 47, 944–948. [Google Scholar] [CrossRef] [PubMed]
- Shao, G.; Wei, X.; Chen, M.; Tang, S.; Luo, J.; Jiao, G.; Xie, L.; Hu, P. Allelic variation for a candidate gene for GS7, responsible for grain shape in rice. Theor. Appl. Genet. 2012, 125, 1303–1312. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, J.; Huang, J.; Lan, H.; Wang, C.; Yin, C.; Wu, Y.; Tang, H.; Qian, Q.; Li, J.; et al. Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc. Natl. Acad. Sci. USA 2012, 109, 21534–21539. [Google Scholar] [CrossRef]
- Segami, S.; Kono, I.; Ando, T.; Yano, M.; Kitano, H.; Miura, K.; Iwasaki, Y. Small and round seed 5 gene encodes alpha-tubulin regulating seed cell elongation in rice. Rice 2012, 5, 4. [Google Scholar] [CrossRef]
- Si, L.; Chen, J.; Huang, X.; Gong, H.; Luo, J.; Hou, Q.; Zhou, T.; Lu, T.; Zhu, J.; Shangguan, Y.; et al. OsSPL13 controls grain size in cultivated rice. Nat. Genet. 2016, 48, 447–456. [Google Scholar] [CrossRef]
- Zhou, Y.; Hou, J.; Li, P. Genetic dissection and validation of QTLs for grain shape and weight in rice and fine mapping of qGL1.3, a major QTL for grain length and weight. Mol. Breed. 2019, 39, 170. [Google Scholar] [CrossRef]
- Zhang, T.; Wang, S.; Sun, S.; Zhang, Y.; Li, J.; You, J.; Su, T.; Chen, W.; Ling, Y.; He, G.; et al. Analysis of QTL for grain size in a rice chromosome segment substitution line Z1392 with long grains and fine mapping of qGL-6. Rice 2020, 13, 40. [Google Scholar] [CrossRef]
- Li, X.; Wei, Y.; Li, J.; Yang, F.; Chen, Y.; Chen, Y.; Guo, S.; Sha, A. Identification of QTL TGW12 responsible for grain weight in rice based on recombinant inbred line population crossed by wild rice (Oryza minuta) introgression line K1561 and indica rice G1025. BMC Genet. 2020, 21, 10. [Google Scholar] [CrossRef]
- Anilkumar, C.; Sah, R.P.; Azharudheen, T.P.M.; Behera, S.; Singh, N.; Prakash, N.R.; Sunitha, N.C.; Devanna, B.N.; Marndi, B.C.; Patra, B.C.; et al. Understanding complex genetic architecture of rice grain weight through QTL-meta analysis and candidate gene identification. Sci. Rep. 2022, 12, 13832. [Google Scholar] [CrossRef] [PubMed]
- Koutroubas, S.D.; Mazzini, F.; Pons, B.; Ntanos, D.A. Grain quality variation and relationships with morpho-physiological traits in rice (Oryza sativa L.) genetic resources in Europe. Field Crops Res. 2004, 86, 115–130. [Google Scholar] [CrossRef]
- Xie, L.; Tang, S.; Chen, N.; Luo, J.; Jiao, G.; Shao, G.; Wei, X.; Hu, P. Rice Grain Morphological Characteristics Correlate with Grain Weight and Milling Quality. Cereal Chem. 2013, 90, 587–593. [Google Scholar] [CrossRef]
- Jongkaewwattana, S.; Geng, S. Inter-relationships amongst grain characteristics, grain-filling parameters and rice (Oryza sativa L.) milling quality. J. Agron. Crop. Sci. 2001, 187, 223–229. [Google Scholar] [CrossRef]
- Jongkaewwattana, S.; Geng, S. Non-Uniformity of Grain Characteristics and Milling Quality of California Rice (Oryza sativa L.) of Different Maturities. J. Agron. Crop Sci. 2002, 188, 161–167. [Google Scholar] [CrossRef]
- Shewry, P.R.; Napier, J.A.; Tatham, A.S. Seed storage proteins: Structures and biosynthesis. Plant Cell 1995, 7, 945–956. [Google Scholar]
- Shewry, P.R.; Halford, N.G. Cereal seed storage proteins: Structures, properties and role in grain utilization. J. Exp. Bot. 2002, 53, 947–958. [Google Scholar] [CrossRef]
- Zhou, Z.K.; Robards, K.; Helliwell, S.; Blanchard, C. Composition and functional properties of rice. Int. J. Food Sci. Technol. 2002, 37, 849–868. [Google Scholar] [CrossRef]
- Wang, Y.J.; Wang, L.F.; Shephard, D.; Wang, F.D.; Patindol, J. Properties and structures of flours and starches from whole, broken, and yellowed rice kernels in a model study. Cereal Chem. 2002, 79, 383–386. [Google Scholar] [CrossRef]
- Buléon, A.; Colonna, P.; Planchot, V.; Ball, S. Starch granules: Structure and biosynthesis. Int. J. Biol. Macromol. 1998, 23, 85–112. [Google Scholar] [CrossRef]
- Berg, J.M.; Tymoczko, J.L.; Stryer, L. Complex Carbohydrates Are Formed by Linkage of Monosaccharides. Biochemistry, 5th ed.; W H Freeman: New York, NY, USA, 2002. [Google Scholar]
- Bemiller, J.; Whistler, R. Starch: Chemistry and Technology, 3rd ed.; Academy Press: Cambridge, UK, 2009; pp. 310–315. [Google Scholar]
- Sano, Y. Differential regulation of waxy gene expression in rice endosperm. Theor. Appl. Genet. 1984, 68, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Xu, M.; Liu, X.; Yan, C.; Korban, S.S.; Chen, X.; Gu, M. Genes coding for starch branching enzymes are major contributors to starch viscosity characteristics in waxy rice (Oryza sativa L.). Plant Sci. 2004, 166, 357–364. [Google Scholar] [CrossRef]
- Cuevas, R.P.; Daygon, V.D.; Morell, M.K.; Gilbert, R.G.; Fitzgerald, M.A. Using chain-length distributions to diagnose genetic diversity in starch biosynthesis. Carbohydr. Polym. 2010, 81, 120–127. [Google Scholar] [CrossRef]
- Counce, P.A.; Bryant, R.J.; Bergman, C.J.; Bautista, R.C.; Wang, Y.-J.; Siebenmorgen, T.J.; Moldenhauer, K.A.K.; Meullenet, J.-F.C. Rice milling quality, grain dimensions, and starch branching as affected by high night temperatures. Cereal Chem. 2005, 82, 645–648. [Google Scholar] [CrossRef]
- Kawasaki, T.; Mizuno, K.; Shimada, H.; Satoh, H.; Kishimoto, N.; Okumura, S.; Ichikawa, N.; Baba, T. Coordinated regulation of the genes participating in starch biosynthesis by the rice Floury-2 locus. Plant Physiol. 1996, 110, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Baba, T.; Nishihara, M.; Mizuno, K.; Kawasaki, T.; Shimada, H.; Kobayashi, E.; Ohnishi, S.; Tanaka, K.; Arai, Y. Identification, cdna cloning, and gene-expression of soluble starch synthase in rice (Oryza sativa L) immature seeds. Plant Physiol. 1993, 103, 565–573. [Google Scholar] [CrossRef]
- She, K.-C.; Kusano, H.; Koizumi, K.; Yamakawa, H.; Hakata, M.; Imamura, T.; Fukuda, M.; Naito, N.; Tsurumaki, Y.; Yaeshima, M.; et al. A Novel factor Floury Endosperm2 is involved in regulation of rice grain size and starch quality. Plant Cell 2010, 22, 3280–3294. [Google Scholar] [CrossRef]
- Zhao, L.; Zhao, C.; Zhou, L.; Zhao, Q.; Zhu, Z.; Chen, T.; Yao, S.; Zhang, Y.; Wang, C. QTL mapping for starch paste viscosity of rice (Oryza sativa L.) using chromosome segment substitution lines derived from two sequenced cultivars with the same Wx allele. BMC Genom. 2021, 22, 596. [Google Scholar] [CrossRef]
- Yamagata, H.; Tanaka, K. The Site of Synthesis and Accumulation of Rice Storage Proteins. Plant Cell Physiol. 1986, 27, 135–145. [Google Scholar] [CrossRef]
- Miflin, B.J.; Field, J.M.; Shewry, P.R. Cereal storage proteins and their effects on technological properties. In Seed Proteins; Daussant, J., Mosse, J., Vaughan, J., Eds.; Academic Press: London, UK, 1983; pp. 255–319. [Google Scholar]
- Muench, D.G.; Ogawa, M.; Okita, T.W. The Prolamins of Rice. In Seed Proteins; Shewry, P.R., Casey, R., Eds.; Springer: Dordrecht, The Netherlands, 1999. [Google Scholar]
- Xu, J.H.; Messing, J. Amplification of prolamin storage protein genes in different subfamilies of the Poaceae. Theor. Appl. Genet. 2009, 119, 1397–1412. [Google Scholar] [CrossRef]
- Kawakatsu, T.; Yamamoto, M.P.; Hirose, S.; Yano, M.; Takaiwa, F. Characterization of a new rice glutelin gene GluD-1 expressed in the starchy endosperm. J. Exp. Bot. 2008, 59, 4233–4245. [Google Scholar] [CrossRef] [PubMed]
- Kawakatsu, T.; Hirose, S.; Yasuda, H.; Takaiwa, F. Reducing Rice Seed Storage Protein Accumulation Leads to Changes in Nutrient Quality and Storage Organelle Formation. Plant Physiol. 2010, 154, 1842–1854. [Google Scholar] [CrossRef] [PubMed]
- Pedrazzini, E.; Mainieri, D.; Marrano, C.A.; Vitale, A. Where do protein bodies of cereal seeds come from? Front. Plant Sci. 2016, 7, 1139. [Google Scholar] [CrossRef] [PubMed]
- Nakase, M.; Hotta, H.; Adachi, T.; Aoki, N.; Nakamura, R.; Masumura, T.; Tanaka, K.; Matsuda, T. Cloning of the rice seed α-globulin-encoding gene: Sequence similarity of the 5′-flanking region to those of the genes encoding wheat high-molecular-weight glutenin and barley D hordein. Gene 1996, 170, 223–226, ISSN 0378-1119. [Google Scholar] [CrossRef]
- Tan, F.Y.; Sun, M.; Xing, Z.Y.; Hua, P.J.; Sun, L.X.; Zhang, F.Q.; Corke, H. Mapping quantitative trait loci for milling quality, protein content and colour characteristics of rice using a recombinant inbred line population derived from an elite rice hybrid. Theor. Appl. Genet. 2001, 103, 1037–1045. [Google Scholar] [CrossRef]
- Aluko, G.; Martinez, C.; Tohme, J.; Castano, C.; Bergman, C.; Oard, J.H. QTL mapping of grain quality traits from the interspecific cross Oryza sativa × O. glaberrima. Theor. Appl. Genet. 2004, 109, 630–639. [Google Scholar] [CrossRef]
- Hu, Z.-L.; Li, P.; Zhou, M.-Q.; Zhang, Z.-H.; Wang, L.-X.; Zhu, L.-H.; Zhu, Y.-G. Mapping of quantitative trait loci (QTLs) for rice protein and fat content using doubled haploid lines. Euphytica 2004, 135, 47–54. [Google Scholar] [CrossRef]
- Peng, B.; Kong, H.; Li, Y.; Wang, L.; Zhong, M.; Sun, L.; Gao, G.; Zhang, Q.; Luo, L.; Wang, G.; et al. OsAAP6 functions as an important regulator of grain protein content and nutritional quality in rice. Nat. Commun. 2014, 5, 4847. [Google Scholar] [CrossRef]
- Chattopadhyay, K.; Behera, L.; Bagchi, T.B.; Sardar, S.S.; Moharana, N.; Patra, N.R.; Chakraborti, M.; Das, A.; Marndi, B.C.; Sarkar, A.; et al. Detection of stable QTLs for grain protein content in rice (Oryza sativa L.) employing high throughput phenotyping and genotyping platforms. Sci. Rep. 2019, 9, 3196. [Google Scholar] [CrossRef]
- Lee, S.; Park, J.; Lee, J.; Shin, D.; Marmagne, A.; Lim, P.O.; Masclaux-Daubresse, C.; An, G.; Gil Nam, H. OsASN1 Overexpression in Rice Increases Grain Protein Content and Yield under Nitrogen-Limiting Conditions. Plant Cell Physiol. 2020, 61, 1309–1320. [Google Scholar] [CrossRef]
- Fiaz, S.; Sheng, Z.; Zeb, A.; Barman, H.N.; Shar, T.; Ali, U.; Tang, S. Analysis of genomic regions for crude protein and fractions of protein using a recombinant inbred population in Rice (Oryza sativa L.). J. Taibah Univ. Sci. 2021, 15, 579–588. [Google Scholar] [CrossRef]
- Choudhury, N.H.; Juliano, B.O. Lipids in developing and mature rice grain. Phytochemistry 1980, 19, 1063–1069. [Google Scholar] [CrossRef]
- Taira, H.; Itani, T. Lipid content and fatty acid composition of brown rice of cultivars of the United States. J. Agric. Food Chem. 1988, 36, 460–462. [Google Scholar] [CrossRef]
- Mano, Y.; Kawaminami, K.; Kojima, M.; Ohnishi, M.; Ito, S. Comparative Composition of Brown Rice Lipids (Lipid Fractions) of Indica and Japonica Rices. Biosci. Biotechnol. Biochem. 1999, 63, 619–626. [Google Scholar] [CrossRef] [PubMed]
- Whattam, J.; Cornell, H.J. Distribution and Composition of the Lipids in Starch Fractions from Wheat Flour. Starch Stärke 1991, 43, 152–156. [Google Scholar] [CrossRef]
- Liu, L.; Waters, D.L.; Rose, T.J.; Bao, J.; King, G.J. Phospholipids in rice: Significance in grain quality and health benefits: A review. Food Chem. 2013, 139, 1133–1145. [Google Scholar] [CrossRef] [PubMed]
- Fujino, Y. Rice lipids. Cereal Chem. 1978, 55, 559–571. [Google Scholar]
- Lam, H.S.; Proctor, A. Lipid hydrolysis and oxidation on the surface of milled rice. J. Am. Oil Chem. Soc. 2003, 80, 563–567. [Google Scholar] [CrossRef]
- Finnie, S.; Jeannotte, R.; Morris, C.; Giroux, M.; Faubion, J. Variation in polar lipids located on the surface of wheat starch. J. Cereal Sci. 2010, 51, 73–80. [Google Scholar] [CrossRef]
- Pasha, I.; Anjum, F.; Morris, C. Grain Hardness: A major determinant of wheat quality. Food Sci. Technol. Int. 2010, 16, 511–522. [Google Scholar] [CrossRef]
- Pauly, A.; Pareyt, B.; Fierens, E.; Delcour, J.A. Wheat (Triticum aestivum L. and T. turgidum L. ssp. durum) kernel hardness: Current View on the Role of Puroindolines and Polar Lipids. Compr. Rev. Food Sci. Food Saf. 2013, 12, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.L.; Zhang, W.W.; Liu, L.L.; Shen, Y.Y.; Wang, J.K.; Jiang, L.; Zhai, H.Q.; Wan, J.M. Dynamic QTL Analysis on Rice Fat Content and Fat Index Using Recombinant Inbred Lines. Cereal Chem. 2008, 85, 769–775. [Google Scholar] [CrossRef]
- Liu, W.; Zeng, J.; Jiang, G.; He, Y. QTLs identification of crude fat content in brown rice and its genetic basis analysis using DH and two backcross populations. Euphytica 2009, 169, 197–205. [Google Scholar] [CrossRef]
- Yu, Y.-H.; Li, G.; Fan, Y.-Y.; Zhang, K.-Q.; Min, J.; Zhu, Z.-W.; Zhuang, J.-Y. Genetic relationship between grain yield and the contents of protein and fat in a recombinant inbred population of rice. J. Cereal Sci. 2009, 50, 121–125. [Google Scholar] [CrossRef]
- Qin, Y.; Kim, S.-M.; Zhao, X.; Lee, H.-S.; Jia, B.; Kim, K.-M.; Eun, M.-Y.; Sohn, J.-K. QTL detection and MAS selection efficiency for lipid content in brown rice (Oryza sativa L.). Genes Genom. 2010, 32, 506–512. [Google Scholar] [CrossRef]
- Ying, J.-Z.; Shan, J.-X.; Gao, J.-P.; Zhu, M.-Z.; Shi, M.; Lin, H.-X. Identification of Quantitative Trait Loci for Lipid Metabolism in Rice Seeds. Mol. Plant 2012, 5, 865–875. [Google Scholar] [CrossRef]
- Lee, G.-H.; Yun, B.-W.; Kim, K.-M. Analysis of QTLs Associated with the Rice Quality Related Gene by Double Haploid Populations. Int. J. Genom. 2014, 2014, 781832. [Google Scholar] [CrossRef]
- Zhou, H.; Xia, D.; Li, P.; Ao, Y.; Xu, X.; Wan, S.; Li, Y.; Wu, B.; Shi, H.; Wang, K.; et al. Genetic architecture and key genes controlling the diversity of oil composition in rice grains. Mol. Plant 2020, 14, 456–469, ISSN 0378-1119. [Google Scholar] [CrossRef]
- Li, Q.; Zhai, W.; Wei, J.; Jia, Y. Rice lipid transfer protein, OsLTPL23, controls seed germination by regulating starch-sugar conversion and ABA homeostasis. Front. Genet. 2023, 14, 1111318. [Google Scholar] [CrossRef]
- Lisle, A.J.; Martin, M.; Fitzgerald, M.A. Chalky and Translucent Rice Grains Differ in Starch Composition and Structure and Cooking Properties. Cereal Chem. 2000, 77, 627–632. [Google Scholar] [CrossRef]
- Yamakawa, H.; Ebitani, T.; Terao, T. Comparison between locations of QTLs for grain chalkiness and genes responsive to high temperature during grain filling on the rice chromosome map. Breed. Sci. 2008, 58, 337–343. [Google Scholar] [CrossRef]
- Zhou, L.; Chen, L.; Jiang, L.; Zhang, W.; Liu, L.; Liu, X.; Zhao, Z.; Liu, S.; Zhang, L.; Wang, J.; et al. Fine mapping of the grain chalkiness QTL qPGWC-7 in rice (Oryza sativa L.). Theor. Appl. Genet. 2009, 118, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Fan, C.; Xing, Y.; Yun, P.; Luo, L.; Yan, B.; Peng, B.; Xie, W.; Wang, G.; Li, X.; et al. Chalk5 encodes a vacuolar H+-translocating pyrophosphatase influencing grain chalkiness in rice. Nat. Genet. 2014, 46, 657. [Google Scholar] [CrossRef]
- Xi, M.; Lin, Z.; Zhang, X.; Liu, Z.; Li, G.; Wang, Q.; Wang, S.; Ding, Y. Endosperm Structure of White-Belly and White-Core Rice Grains Shown by Scanning Electron Microscopy. Plant Prod. Sci. 2014, 17, 285–290. [Google Scholar] [CrossRef]
- Zhou, L.; Liang, S.; Ponce, K.; Marundon, S.; Ye, G.; Zhao, X. Factors affecting head rice yield and chalkiness in indica rice. Field Crop. Res. 2015, 172, 1–10. [Google Scholar] [CrossRef]
- Lin, Z.; Zheng, D.; Zhang, X.; Wang, Z.; Lei, J.; Liu, Z.; Li, G.; Wang, S.; Ding, Y. Chalky part differs in chemical composition from translucent part of japonica rice grains as revealed by a notched-belly mutant with white-belly. J. Sci. Food Agric. 2016, 96, 3937–3943. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Daygon, V.D.; McNally, K.L.; Hamilton, R.S.; Xie, F.; Reinke, R.F.; Fitzgerald, M.A. Identification of stable QTLs causing chalk in rice grains in nine environments. Theor. Appl. Genet. 2016, 129, 141–153. [Google Scholar] [CrossRef]
- Jin, S.-K.; Xu, L.-N.; Yang, Q.-Q.; Zhang, M.-Q.; Wang, S.-L.; Wang, R.-A.; Tao, T.; Hong, L.-M.; Guo, Q.-Q.; Jia, S.-W.; et al. High-resolution quantitative trait locus mapping for rice grain quality traits using genotyping by sequencing. Front. Plant Sci. 2023, 13, 1050882. [Google Scholar] [CrossRef]
- Lin, C.-J.; Li, C.-Y.; Lin, S.-K.; Yang, F.-H.; Huang, J.-J.; Liu, Y.-H.; Lur, H.-S. Influence of High Temperature during Grain Filling on the Accumulation of Storage Proteins and Grain Quality in Rice (Oryza sativa L.). J. Agric. Food Chem. 2010, 58, 10545–10552. [Google Scholar] [CrossRef]
- Zhao, X.; Fitzgerald, M. Climate Change: Implications for the Yield of Edible Rice. PLoS ONE 2013, 8, e66218. [Google Scholar] [CrossRef]
- Xu, Q.; Chen, W.; Xu, Z. Relationship between grain yield and quality in rice germplasms grown across different growing areas. Breed. Sci. 2015, 65, 226–232. [Google Scholar] [CrossRef]
- Fu, F.F.; Xue, H.W. Coexpression Analysis Identifies Rice Starch Regulator1, a Rice AP2/EREBP Family Transcription Factor, as a Novel Rice Starch Biosynthesis Regulator. Plant Physiol. 2010, 154, 927–938. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-C.; Xu, H.; Zhu, Y.; Liu, Q.-Q.; Cai, X.-L. OsbZIP58, a basic leucine zipper transcription factor, regulates starch biosynthesis in rice endosperm. J. Exp. Bot. 2013, 64, 3453–3466. [Google Scholar] [CrossRef]
- Zhou, P.; Tan, Y.; He, Y.; Xu, C.; Zhang, Q. Simultaneous improvement for four quality traits of Zhenshan 97, an elite parent of hybrid rice, by molecular marker-assisted selection. Theor. Appl. Genet. 2003, 106, 326–331. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Zhang, X.; Wang, Z.; Jiang, Y.; Liu, Z.; Alexander, D.; Li, G.; Wang, S.; Ding, Y. Metabolomic analysis of pathways related to rice grain chalkiness by a notched-belly mutant with high occurrence of white-belly grains. BMC Plant Biol. 2017, 17, 39. [Google Scholar] [CrossRef] [PubMed]
- He, P.; Li, G.S.; Qian, Q.; Ma, Q.Y.; Li, Z.J.; Wang, M.W.; Chen, Y.; Zhu, H.L. Genetic analysis of rice grain quality. Theor. Appl. Genet. 1999, 98, 502–508. [Google Scholar] [CrossRef]
- Tan, Y.F.; Xing, Y.Z.; Li, J.X.; Yu, S.B.; Xu, C.G.; Zhang, Q. Genetic bases of appearance quality of rice grains in Shanyou 63, an elite rice hybrid. Theor. Appl. Genet. 2000, 101, 823–829. [Google Scholar] [CrossRef]
- Wan, X.Y.; Wan, J.M.; Weng, J.F.; Jiang, L.; Bi, J.C.; Wang, C.M.; Zhai, H.Q. Stability of QTLs for rice grain dimension and endosperm chalkiness characteristics across eight environments. Theor. Appl. Genet. 2005, 110, 1334–1346. [Google Scholar] [CrossRef]
- Yang, W.; Hao, Q.; Liang, J.; Tan, Q.; Luan, X.; Lin, S.; Zhu, H.; Bu, S.; Liu, Z.; Liu, G.; et al. Fine Mapping of Two Major Quantitative Trait Loci for Rice Chalkiness With High Temperature-Enhanced Additive Effects. Front. Plant Sci. 2022, 13, 957863. [Google Scholar] [CrossRef]
- Shi, H.; Zhu, Y.; Yun, P.; Lou, G.; Wang, L.; Wang, Y.; Gao, G.; Zhang, Q.; Li, X.; He, Y. Fine Mapping of qWCR4, a Rice Chalkiness QTL Affecting Yield and Quality. Agronomy 2022, 12, 706. [Google Scholar] [CrossRef]
- Kumari, A.; Sharma, D.; Sharma, P.; Wang, C.; Verma, V.; Patil, A.; Imran, M.; Singh, M.P.; Kumar, K.; Paritosh, K.; et al. Meta-QTL and haplo-pheno analysis reveal superior haplotype combinations associated with low grain chalkiness under high temperature in rice. Front. Plant Sci. 2023, 14, 1133115. [Google Scholar] [CrossRef]
- Wang, E.; Wang, J.; Zhu, X.; Hao, W.; Wang, L.; Li, Q.; Zhang, L.; He, W.; Lu, B.; Lin, H.; et al. Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat. Genet. 2008, 40, 1370–1374. [Google Scholar] [CrossRef] [PubMed]
- Raju, G.N.; Chand, N.; Bhashyam, M.K.; Srinivas, T. Predictive model for grain cracking in terms of rice plant and panicle morphology derived from multivariate analysis. J. Sci. Food Agric. 1995, 68, 141–152. [Google Scholar] [CrossRef]
- Cho, J. Double fertilization in Oryza sativa L. and development of the endosperm with special reference to the aleurone layer. Bull. Natl. Inst. Agric. Sci. 1956, 6, 61–101. [Google Scholar]
- Ekanayake, I.J.; Datta, S.; Steponkus, P.L. Spikelet Sterility and Flowering Response of Rice to Water Stress at Anthesis. Ann. Bot. 1989, 63, 257–264. [Google Scholar] [CrossRef]
- Jagadish, S.; Craufurd, P.; Wheeler, T. High temperature stress and spikelet fertility in rice (Oryza sativa L.). J. Exp. Bot. 2007, 58, 1627–1635. [Google Scholar] [CrossRef]
- Jagadish, S.V.K.; Craufurd, P.Q.; Wheeler, T.R. Phenotyping Parents of Mapping Populations of Rice for Heat Tolerance during Anthesis. Crop Sci. 2008, 48, 1140. [Google Scholar] [CrossRef]
- Jagadish, S.V.K.; Murty, M.V.R.; Quick, W.P. Rice responses to rising temperatures–challenges, perspectives and future directions. Plant Cell Environ. 2015, 38, 1686–1698. [Google Scholar] [CrossRef]
- Yamakawa, H.; Hirose, T.; Kuroda, M.; Yamaguchi, T. Comprehensive Expression Profiling of Rice Grain Filling-Related Genes under High Temperature Using DNA Microarray. Plant Physiol. 2007, 144, 258–277. [Google Scholar] [CrossRef]
- Shi, P.; Tang, L.; Wang, L.; Sun, T.; Liu, L.; Cao, W.; Zhu, Y. Post-Heading Heat Stress in Rice of South China during 1981–2010. PLoS ONE 2015, 10, e0130642. [Google Scholar] [CrossRef]
- Shi, W.; Ishimaru, T.; Gannaban, R.B.; Oane, W.; Jagadish, S.V.K. Popular Rice (Oryza sativa L.) Cultivars Show Contrasting Responses to Heat Stress at Gametogenesis and Anthesis. Crop Sci. 2015, 55, 589–596. [Google Scholar] [CrossRef]
- Sreenivasulu, N.; Butardo, V.M.; Misra, G.; Cuevas, R.P.; Anacleto, R.; Kishor, P.B.K. Designing climate-resilient rice with ideal grain quality suited for high-temperature stress. J. Exp. Bot. 2015, 66, 1737–1748. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, M.A.; Resurreccion, A.P. Maintaining the yield of edible rice in a warming world. Funct. Plant Biol. 2009, 36, 1037–1045. [Google Scholar] [CrossRef]
- Ambardekar, A.A.; Siebenmorgen, T.J.; Counce, P.A.; Lanning, S.B.; Mauromoustakos, A. Impact of field-scale night-time air temperatures during kernel development on rice milling quality. Field Crops Res. 2011, 122, 179–185. [Google Scholar] [CrossRef]
- Wu, Y.C.; Chang, S.J.; Lur, H.S. Effects of field high temperature on grain yield and quality of a subtropical type japonica rice—Pon-Lai rice. Plant Prod. Sci. 2016, 19, 145–153. [Google Scholar] [CrossRef]
- Abayawickrama, A.; Reinke, R.; Fitzgerald, M.; Harper, J.; Burrows, G. Influence of high daytime temperature during the grain filling stage on fissure formation in rice. J. Cereal Sci. 2017, 74, 256–262. [Google Scholar] [CrossRef]
- Miah, M.N.H.; Yoshida, T.; Yamamoto, Y.; Nitta, Y. Characteristics of Dry Matter Production and Partitioning of Dry Matter to Panicles in High Yielding Semidwarf Indica and Japonica-Indica Hybrid Rice Varieties. Jpn. J. Crop Sci. 1996, 65, 672–685. [Google Scholar] [CrossRef][Green Version]
- Yang, J.; Peng, S.; Visperas, R.M.; Sanico, A.L.; Zhu, Q.; Gu, S. Grain filling pattern and cytokinin content in the grains and roots of rice plants. Plant Growth Regul. 2000, 30, 261–270. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, J.; Huang, Z.; Zhu, Q.; Wang, L. Remobilization of Carbon Reserves Is Improved by Controlled Soil-Drying during Grain Filling of Wheat. Crop Sci. 2000, 40, 1645–1655. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, J.; Wang, Z.; Zhu, Q.; Wang, W. Hormonal Changes in the Grains of Rice Subjected to Water Stress during Grain Filling. Plant Physiol. 2001, 127, 315–323. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, J.; Huang, Z.; Wang, Z.; Zhu, Q.; Liu, L. Correlation of Cytokinin Levels in the Endosperms and Roots with Cell Number and Cell Division Activity during Endosperm Development in Rice. Ann. Bot. 2002, 90, 369–377. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, J.; Wang, Z.; Zhu, Q. Hormones in the grains in relation to sink strength and postanthesis development of spikelets in rice. Plant Growth Regul. 2003, 41, 185–195. [Google Scholar] [CrossRef]
- Yang, W.; Jia, C.-C.; Howell, T. Relationship of Moisture Content Gradients and Glass Transition Temperatures to Head Rice Yield during Cross-Flow Drying. Biosyst. Eng. 2003, 86, 199–206. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, J. Grain filling of cereals under soil drying. New Phytol. 2006, 169, 223–236. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wang, Y. Chapter 13-Impact of climate change on rice grain quality. In Rice, 4th ed.; Bao, J., Ed.; AACC International Press: Washington, DC, USA, 2019; pp. 427–441. [Google Scholar]
- Zhang, H.; Chen, T.; Wang, Z.; Yang, J.; Zhang, J. Involvement of cytokinins in the grain filling of rice under alternate wetting and drying irrigation. J. Exp. Bot. 2010, 61, 3719–3733. [Google Scholar] [CrossRef]
- Nangju, D.; De Datta, S.K. Effect of Time of Harvest and Nitrogen Level on Yield and Grain Breakage in Transplanted Rice 1. Agron. J. 1970, 62, 468–474. [Google Scholar] [CrossRef]
- Seetanun, W.; De Datta, S.K. Grain Yield, Milling Quality, and Seed Viability of Rice as Influenced by Time of Nitrogen Application and Time of Harvest. Agron. J. 1973, 65, 390–394. [Google Scholar] [CrossRef]
- Atilio, J.B.; Causin, H.F. The central role of amino acids on nitrogen utilization and plant growth. J. Plant Physiol. 1996, 149, 358–362. [Google Scholar] [CrossRef]
- Perez, C.M.; Juliano, B.O.; Liboon, S.P.; Alcantara, J.M.; Cassman, K.G. Effects of late nitrogen fertilizer application on head rice yield, protein content, and grain quality of rice. Cereal Chem. 1996, 73, 556–560. [Google Scholar]
- Leesawatwong, M.; Jamjod, S.; Kuo, J.; Dell, B.; Rerkasem, B. Nitrogen Fertilizer Increases Seed Protein and Milling Quality of Rice. Cereal Chem. 2005, 82, 588–593. [Google Scholar] [CrossRef]
- Yang, Y.H.; Dai, L.; Xia, H.C.; Zhu, K.M.; Liu, H.J.; Chen, K.P. Protein profile of rice (Oryza sativa L.) seeds. Genet. Mol. Biol. 2013, 36, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Arnold, A.; Sajitz-Hermstein, M.; Nikoloski, Z. Effects of Varying Nitrogen Sources on Amino Acid Synthesis Costs in Arabidopsis thaliana under Different Light and Carbon-Source Conditions. PLoS ONE 2015, 10, e0116536. [Google Scholar] [CrossRef] [PubMed]
- Bryant, R.J.; Yeater, K.M.; McClung, A.M. Effect of Nitrogen Rate and the Environment on Physicochemical Properties of Selected High-Amylose Rice Cultivars. Cereal Chem. 2015, 92, 604–610. [Google Scholar] [CrossRef]
- Beeny, J.M.; Chin, S.N. Multipass Drying of Paddy (Rice) in the Humid Tropics. J. Agric. Eng. Res. 1970, 15, 364–374. [Google Scholar] [CrossRef]
- Kim, S.Y.; Lee, H. Effect of quality characteristics on brown rice produced from paddy rice with different moisture contents. J. Korean Soc. Appl. Biol. Chem. 2013, 56, 289–293. [Google Scholar] [CrossRef]
- Siebenmorgen, T.J.; Nehus, Z.T.; Archer, T.R. Milled Rice Breakage Due to Environmental Conditions. Cereal Chem. 1998, 75, 149–152. [Google Scholar] [CrossRef]
- Siebenmorgen, T.J.; Yang, W.; Sun, Z. Glass transition temperature of rice kernels determined by dynamic mechanical thermal analysis. Trans. Am. Soc. Agric. Biol. Eng. 2004, 47, 835–839. [Google Scholar] [CrossRef]
- Srikaeo, K.; Boonrod, C.; Rahman, M.S. Effect of storage temperatures on the head rice yield in relation to glass transition temperatures and un-freezable water. J. Cereal Sci. 2016, 70, 164–169. [Google Scholar] [CrossRef]
- Truong, T.; Truong, V.; Fukai, S.; Bhandari, B. Changes in Cracking Behavior and Milling Quality of Selected Australian Rice Varieties Due to Postdrying Annealing and Subsequent Storage. Dry. Technol. 2012, 30, 1831–1843. [Google Scholar] [CrossRef]
- Perdon, A.A.; Siebenmorgen, T.J.; Mauromoustakos, A.; Griffin, V.K.; Johnson, E.R. Degree of Milling Effects on Rice Pasting Properties. Cereal Chem. 2001, 78, 205–209. [Google Scholar] [CrossRef]
- Mei, H.-W.; Luo, L.-J.; Guo, L.-B.; Wang, Y.-P.; Yu, X.-Q.; Ying, C.-S.; Li, Z.-K. Molecular mapping of QTLs for rice milling yield traits. Acta Genet. Sin. 2002, 29, 791–797. [Google Scholar] [PubMed]
- Septiningsih, E.M.; Trijatmiko, K.R.; Moeljopawiro, S.; McCouch, S.R. Identification of quantitative trait loci for grain quality in an advanced backcross population derived from the Oryza sativa variety IR64 and the wild relative O. rufipogon. Theor. Appl. Genet. 2003, 107, 1433–1441. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Tsuzuki, E.; Lin, D.; Kamiunten, H.; Terao, H.; Matsuo, M.; Cheng, S. Molecular genetic mapping of quantitative trait loci for milling quality in rice (Oryza sativa L.). J. Cereal Sci. 2004, 40, 109–114, ISSN 0733-5210. [Google Scholar] [CrossRef]
- Zheng, T.Q.; Xu, J.L.; Li, Z.K.; Zhai, H.Q.; Wan, J.M. Genomic regions associated with milling quality and grain shape identified in a set of random introgression lines of rice (Oryza sativa L.). Plant Breed. 2007, 126, 158–163. [Google Scholar] [CrossRef]
- Kepiro, J.; McClung, A.; Chen, M.; Yeater, K.; Fjellstrom, R. Mapping QTLs for milling yield and grain characteristics in a tropical japonica long grain cross. J. Cereal Sci. 2008, 48, 477–485. [Google Scholar] [CrossRef]
- Jiang, G.-H.; Hong, X.-Y.; Xu, C.-G.; Li, X.-H.; He, Y.-Q. Identification of Quantitative Trait Loci for Grain Appearance and Milling Quality Using a Doubled-Haploid Rice Population. J. Integr. Plant Biol. 2005, 47, 1391–1403. [Google Scholar] [CrossRef]
- Hao, W.; Zhu, M.-Z.; Gao, J.-P.; Sun, S.-Y.; Lin, H.-X. Identification of Quantitative Trait Loci for Rice Quality in a Population of Chromosome Segment Substitution Lines. J. Integr. Plant Biol. 2009, 51, 500–512. [Google Scholar] [CrossRef]
- Nelson, J.C.; McClung, A.M.; Fjellstrom, R.G.; Moldenhauer, K.A.K.; Boza, E.; Jodari, F.; Oard, J.H.; Linscombe, S.; Scheffler, B.E.; Yeater, K.M. Mapping QTL main and interaction influences on milling quality in elite US rice germplasm. Theor. Appl. Genet. 2011, 122, 291–309. [Google Scholar] [CrossRef]
- Nelson, J.C.; Jodari, F.; Roughton, A.I.; McKenzie, K.M.; McClung, A.M.; Fjellstrom, R.G.; Scheffler, B.E. QTL mapping for milling quality in elite western U.S. rice germplasm. Crop Sci. 2012, 52, 242–252. [Google Scholar] [CrossRef]
- Wang, X.; Pang, Y.; Wang, C.; Chen, K.; Zhu, Y.; Shen, C.; Ali, J.; Xu, J.; Li, Z. New Candidate Genes Affecting Rice Grain Appearance and Milling Quality Detected by Genome-Wide and Gene-Based Association Analyses. Front. Plant Sci. 2017, 7, 1998. [Google Scholar] [CrossRef]
- Qiu, X.; Pang, Y.; Yuan, Z.; Xing, D.; Xu, J.; Dingkuhn, M.; Li, Z.; Ye, G. Genome-Wide Association Study of Grain Appearance and Milling Quality in a Worldwide Collection of Indica Rice Germplasm. PLoS ONE 2016, 10, e0145577. [Google Scholar] [CrossRef] [PubMed]
- Quero, G.; Gutiérrez, L.; Monteverde, E.; Blanco, P.; de Vida, F.P.; Rosas, J.; Fernández, S.; Garaycochea, S.; McCouch, S.; Berberian, N.; et al. Genome-Wide Association Study Using Historical Breeding Populations Discovers Genomic Regions Involved in High-Quality Rice. Plant Genome 2018, 11, 170076. [Google Scholar] [CrossRef]
- Misra, G.; Anacleto, R.; Badoni, S.; Butardo, V.; Molina, L.; Graner, A.; Demont, M.; Morell, M.K.; Sreenivasulu, N. Dissecting the genome-wide genetic variants of milling and appearance quality traits in rice. J. Exp. Bot. 2019, 70, 5115–5130. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhan, J.; Li, J.; Lu, X.; Liu, J.; Wang, Y.; Zhao, Q.; Ye, G. Genome-Wide Association Study (GWAS) for Mesocotyl Elongation in Rice (Oryza sativa L.) under Multiple Culture Conditions. Genes 2019, 11, 49. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Liu, Y.; Gong, C.; Chen, B.; Wang, T. Waxy is an important factor for grain fissure resistance and head rice yield as revealed by a genome-wide association study. J. Exp. Bot. 2022, 73, 6942–6954. [Google Scholar] [CrossRef] [PubMed]
- Xin, X.U.; Ye, J.H.; Yang, Y.Y.; Li, R.S.; Zhen, L.I.; Shan, W.A.N.G.; Yang, Y.L. Genetic diversity analysis and GWAS reveal the adaptive loci of milling and appearance quality of japonica rice (Oryza sativa L.) in Northeast China. J. Integr. Agric. 2022, 21, 1539–1550. [Google Scholar] [CrossRef]
- Yu, P.; Ye, C.; Li, L.; Yin, H.; Zhao, J.; Wang, Y.; Zhang, Z.; Li, W.; Long, Y.; Hu, X.; et al. Genome-wide association study and genomic prediction for yield and grain quality traits of hybrid rice. Mol. Breed. 2022, 42, 16. [Google Scholar] [CrossRef]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef]
- Zhou, X.; Stephens, M. Efficient multivariate linear mixed model algorithms for genome-wide association studies. Nat. Methods 2014, 11, 407–409. [Google Scholar] [CrossRef]
- Singh, R.; Saripalli, G.; Gautam, T.; Kumar, A.; Jan, I.; Batra, R.; Kumar, J.; Kumar, R.; Balyan, H.S.; Sharma, S.; et al. Meta-QTLs, ortho-MetaQTLs and candidate genes for grain Fe and Zn contents in wheat (Triticum aestivum L.). Physiol. Mol. Biol. Plants 2022, 28, 637–650. [Google Scholar] [CrossRef]
- Joukhadar, R.; Thistlethwaite, R.; Trethowan, R.; Keeble-Gagnère, G.; Hayden, M.J.; Ullah, S.; Daetwyler, H.D. Meta-analysis of genome-wide association studies reveal common loci controlling agronomic and quality traits in a wide range of normal and heat stressed environments. Theor. Appl. Genet. 2021, 134, 2113–2127. [Google Scholar] [CrossRef]
- Joukhadar, R.; Daetwyler, H.D.; Bansal, U.K.; Gendall, A.R.; Hayden, M.J. Genetic Diversity, Population Structure and Ancestral Origin of Australian Wheat. Front. Plant Sci. 2017, 8, 2115. [Google Scholar] [CrossRef] [PubMed]
- Jighly, A.; Benhajali, H.; Liu, Z.; Goddard, M.E. MetaGS: An accurate method to impute and combine SNP effects across populations using summary statistics. Genet. Sel. Evol. 2022, 54, 37. [Google Scholar] [CrossRef] [PubMed]
- Meuwissen, T.H.E.; Hayes, B.J.; Goddard, M.E. Prediction of Total Genetic Value Using Genome-Wide Dense Marker Maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef] [PubMed]
- Monteverde, E.; Rosas, J.E.; Blanco, P.; de Vida, F.P.; Bonnecarrère, V.; Quero, G.; Gutierrez, L.; McCouch, S. Multienvironment Models Increase Prediction Accuracy of Complex Traits in Advanced Breeding Lines of Rice. Crop Sci. 2018, 58, 1519–1530. [Google Scholar] [CrossRef]
- Monteverde, E.; Gutierrez, L.; Blanco, P.; Pérez de Vida, F.; Rosas, J.E.; Bonnecarrère, V.; McCouch, S. Integrating molecular markers and environmental covariates to interpret genotype by environment interaction in rice (Oryza sativa L.) grown in subtropical areas. G3 Genes Genomes Genet. 2019, 9, 1519–1531. [Google Scholar]
- Jighly, A.; Thayalakumaran, T.; O’Leary, G.J.; Kant, S.; Panozzo, J.; Aggarwal, R.; Hessel, D.; Forrest, K.L.; Technow, F.; Tibbits, J.F.G.; et al. Using genomic prediction with crop growth models enables the prediction of associated traits in wheat. J. Exp. Bot. 2022, 74, 1389–1402. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).