Abstract
The Szekler horse was a small-sized mountain horse of the Carpathian Mountains whose official stud book ceased to exist after WWII. Despite that, individual horses preserving all the characteristics of the Szekler horse remained scattered in remote areas. This study aims to evaluate the mitochondrial D-loop sequence (608 bp) of the founder population (n = 59) in 2021 of a breed reconstruction project started in 2012. D-loop showed 68 polymorphic sites. The number of haplotypes was 34, with haplotype diversity (Hd) 0.966 and nucleotide diversity (π) 0.02232. The value of Fu’s Fs statistic (−6.566) was significant (p < 0.001), which rejects a stable population status. Thirteen haplogroups (HG) were found with a nearly equal number of representatives (HG(n)—A (5), D (1), E (2), G (4), I (4), J–K (1), M (4), N (2), O’P (4), and R (1)). In contrast, the Q, L, and B HGs occurred in more horses (15, 9, and 7, respectively). Based on a large number of polymorphic sites and haplotypes, the founder stock is considered diverse. Since the HG Q is characteristic of Asian horses, the examined stock haplotype distribution reflects the eastern origin of the Hungarian horses brought from the East in 896 AD. It is complemented by the gene pool of horses from Europe (e.g., L) and the Middle East (e.g., B).
1. Introduction
One of the many reasons for preserving our endangered domestic animal breeds is their historical importance, which usually reflects particular virtues. In most cases, the maintenance of heritage breeds is well organized. However, when continuity is interrupted, as exemplified by the case of the Szekler horse (in Hungarian Székely ló), it becomes necessary to reorganize the preservation of the breed, an effort officially called breed reconstruction.
Over the centuries, the Hungarian horse breeds commanded respect and high esteem worldwide. Before WWII, the records listed a ninth horse breed beyond the currently existing eight Hungarian ones: the Szekler horse. Our detailed knowledge about the Szekler horse comes from several descriptions by writers on horse breeding in the eighteenth and nineteenth centuries [1]. These writers unanimously praised the Szekler horse for its constant power and enduring performance.
The Szekler horse was a small-sized mountain horse that lived in the Carpathian Mountains up until the middle of the last century, whose official stud book ceased to exist after WWII. The Szekler horse herd of Zsuk (a town in Transylvania) vanished, the studbooks were burned, and no further efforts were made to continue breeding. So much so, that the Szekler horse (if it was mentioned at all) was typically referred to as an extinct ancient breed. The drawing (Supplementary Material S1, [2,3]), illustrates well the fiery but undemanding and perseverant Transylvanian horse from the time when it had not yet been crossed with modern horses.
Relatives of the Szekler horse included the Békás horse and the Hucul horse, the latter still extant thanks to a cross-border preservation program.
The Szekler horse, like its relatives, was classified as a small horse. Its height at the withers was 140 cm, and the croup was slightly overgrown. In addition, it was an undemanding, hardy, sure-footed, and, therefore excellent pack animal for the mountains. In Szekler Land (Székelyföld), in the absence of a railway, these strong horses were used to transport the harvested crop and timber in rugged terrain. These powerful horses, with strong joints, performed tasks related to forestry and agriculture in mountainous areas. The 400 kg animals worked well even under saddle, whether at a trot or a gallop, with a highly set head and high foreleg action. Thanks to their small size and calm temperament, it was easy for women to take care of them.
The famous professor of animal husbandry, Béla Hankó [2], published a detailed study of the Szekler horse, with measurements and photo documentation of 80 mares and two stallions. Using his description, the Szekler horse type consists of “animals with a medium height and Arabian appearance, with good stature, relatively short legs, and long, strong trunk, quite wide breast and hip, and relatively small and dry head, set on a proportional, not too short neck. All in all, these are beautiful animals who observe the world with alert ears, big eyes, and dilating nostrils”.
Despite the backlog of stud booking work, individual horses preserving all the characteristics of the Szekler horse remained scattered among smallholder farmers in remote areas of Szekler Land. By crossing with other horse breeds (Arabian, Lipizzaner, and to a lesser extent English half-blood and small Nonius), efforts were made to increase her body size and improve her movement. However, these experiments failed, as the offspring did not succeed in the mountainous conditions [4].
Recently, a breed reconstruction program has been started to rescue the Szekler horse. Expeditions have been organized in the few Transylvanian Szekler counties to recover individual horses with a suitable appearance and major body measurements and to construct a database with photographs and information about their descent. The breed reconstruction program continued with the founding of two sister associations (the Mereklye Association in 2008 and the Szekler Horse Breeders’ Association in 2020), which was greeted with enthusiasm by many horse owners [5]. During the introductory part of the program, the following tasks are built on each other over time: establishing a stud book and breeding regulations, searching for breeding animals, creating a seed stock (which is desirable for the sake of safety), individual and in-depth conformation judgment, performance testing, organization of competitions, creating a mating plan, and last but not least, building a social base. The actual number of herd-booked horses is 85. These horses satisfy the conditions of the last census of the breed, which is collected in Hankó’s book [2], in terms of appearance and usability. The body measurements of the initial founding individuals correspond with those of the previous ones (Supplementary Material S2, [2,5,6,7]). The results proved that the reconstruction of the breed is going well, in regard to morphology.
It is essential to emphasize the regular utilization of the breed in its current state. In addition to the classic use of that endangered working horse breed (in forestry, in the chart), breeders also strive to develop new ones (equestrian tourism, rides for children, equine therapy, preservation of military traditions, horseback archery, education of youth, fashion). Through these, the utilization of the breed becomes even more diverse, it comes to play an important role in the life of society, and at the same time, the owner gets a secure income through the use of the horse.
The genetic pool of a community of animals kept in a given place is an exact imprint of the genetic influence of newer animals that have reached this place throughout history. Depending on the breeding method (e.g., drop-crossing or grading-up), the new individuals have a stronger or weaker effect on the genetic structure of the original stock, on the autosomes, mainly from the paternal side. In contrast, mitochondrial DNA preserves the genetic background of the mothers that created it. If there is no great loss among the maternal lineages during history, the bottleneck effect does not strongly affect the mtDNA pattern. Therefore, the initial diversity can be detected and maintained even in the present.
Nowadays, genetic research concerning the mitochondrion also examines the entire mitochondrial genome. An Italian research group led by Achilli [8] published the main redetermined haplogroups into which the horses could be classified. The currently known 83 haplotypes of mitochondrial DNA are classified into 18 haplogroups by horse genetic science, which are assigned letters from A to R. Most haplogroups occur in the domestication centers of Asia, and animals brought from there represent fewer haplogroups. Therefore, initially, the different haplogroups clustered geographically to some extent. For example, haplogroup L became much more common in Europe than in Asia, while haplogroup Q was and remains much more common in Central Asia than in Europe. As a result of the breeding and purchasing habits of modern breeders, these haplogroups have mixed to a significant extent. Haplogroups with exclusive geographical locations no longer exist. However, the occurrence of haplogroups within each region still shows a characteristic distribution and pattern. The conservative nature of the mitochondrial DNA pool can also be emphasized by the fact that the most common way to implement breeding is to buy highly selected stallions and use them in natural service or artificial insemination, which do not affect the maternal mitochondrial DNA. Therefore, we can use studies dealing with mitochondrial DNA to clarify the composition and probable origin of the breeds. The male-specific region of the Y chromosome (MSY) also contributes to a more complete understanding of the horse’s domestication process and explains the genetic variation that has emerged today [9].
The goal of this study is to examine the maternal background and genetic structure of the reconstructed stock, the phenotypic traits of which can be considered identical to the ancient Szekler horse. To accomplish this, we analyzed the D-loop (control region) of the mitochondrial DNA.
2. Materials and Methods
Blood samples were taken from individuals of the reconstructed population (n = 59, 39 mares and 20 stallions) to evaluate the mitochondrial D-loop sequence (608 bp) in January 2021. The samples came from the samples annually taken during routine veterinary procedures to detect equine infectious anemia. The further use of samples obtained during clinical veterinary procedures for research is not considered an animal experiment under EU Directive 63/2010 [10], so no ethical approval is required. Although stallions do not pass on the mitochondrial genome, their samples are suitable for assessing the haplotype diversity. Furthermore, this method provides the CR haplotype of the stallion’s collateral relatives on the maternal side.
DNA was isolated using the GenElute Blood Genomic DNA Kit (Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer’s instructions. The 25 μL PCR reaction mixture prepared for each sample contained 2.5 μL dNTP (10 mM), 2.5 μL 10× PCR buffer, 1.5 μL MgCl2 (25 mM), 2 μL primer (10 μM), 1 μL BSA (20 mg/mL), 0.4 μL Taq polymerase (5 U/μL) (ThermoFisher Scientific, Waltham, MA, USA), and 10 ng DNA template and PCR grade water to a final volume of 25 μL. The mitochondrial D-loop was amplified using forward EqCRF 5′-AAACCAGAAAAGGGGGAAAA-3′ and reverse EqCRR 5′-TGGCGAATAGCTTTGTTGTG-3′ oligonucleotides [11].
The following amplification protocol was used for an initial cycle of 94 °C for 20 s, followed by 34 cycles of 94 °C for 30 s, 55 °C for 30 s, 72 °C for 45 s, and a final extension step of 72 °C for 7 min. The expected PCR product size was 688 bp.
A Thermal Cycler 2720 PCR equipment (Applied Biosystem, Waltham, MA, USA) was used to amplify the DNA sequence. PCR products were purified with the SIGMA GenElute™ PCR Clean Up Kit (Sigma-Aldrich, St. Louis, MO, USA) according to the protocol.
BigDye® Terminator version 3.1 Cycle Sequencing Kit (ThermoFisher Scientific, Waltham, MA, USA) was used for sequencing reaction following the manufacturer’s recommendations. For sequence detection, an ABI Prism 3130XL Genetic Analyzer (Applied Biosystems, ThermoFisher Scientific, Waltham, MA, USA) was used, according to the manufacturer’s guidelines.
The mtDNA D-loop sequences were aligned, and haplogroups and haplotypes were determined by MEGA 11 [12].
The DnaSP 6 [13] software was used to estimate the genetic diversity indices (i.e., haplotype diversity, Hd and nucleotide diversity, π) and to conduct neutrality tests based on Tajima’s D [14] and Fu’s Fs [15] test. To complement these statistics, sequential mismatch distribution was derived from pair-wise nucleotide differences.
The haplogroup frequencies of the Szekler horse were compared to that of the literature [8] by using the one-sided difference test with a known number of observations [16].
The D-loop sequences are deposited in GenBank under accession numbers: OL445148-OL445206.
3. Results
3.1. D-loop Sequence Diversity
The total number of trimmed nucleotide sites and sequences were 608 (from 15,494 to 16,101 on the consensus NC001640 [17]) and 59, respectively. The total number of sites, excluding the sites with gaps and/or missing data, was 605. There were 537 invariable (monomorphic) sites and 68 variable (polymorphic) sites. The latter meets the total number of mutations as well.
In total, there were 25 singleton sites and 50 parsimony informative sites (see mutant sites in Supplementary Material S3). Parsimony is a polymorphic base site where two or more base variants can be present, and two of these base variants occur at least twice.
The total number of haplotypes in the study population was 34 (of the 83 total observed so far). The haplotype diversity (Hd) was 0.966, while the standard deviation of the haplotype diversity was 0.013.
The nucleotide diversity (π) was 0.02232, with a standard deviation of 0.00066. The nucleotide diversity, as demonstrated by the Jukes and Cantor method (π-JC) was 0.02271. The average number of nucleotide differences (k) was 13.503.
3.2. Prevalence of Haplogroups
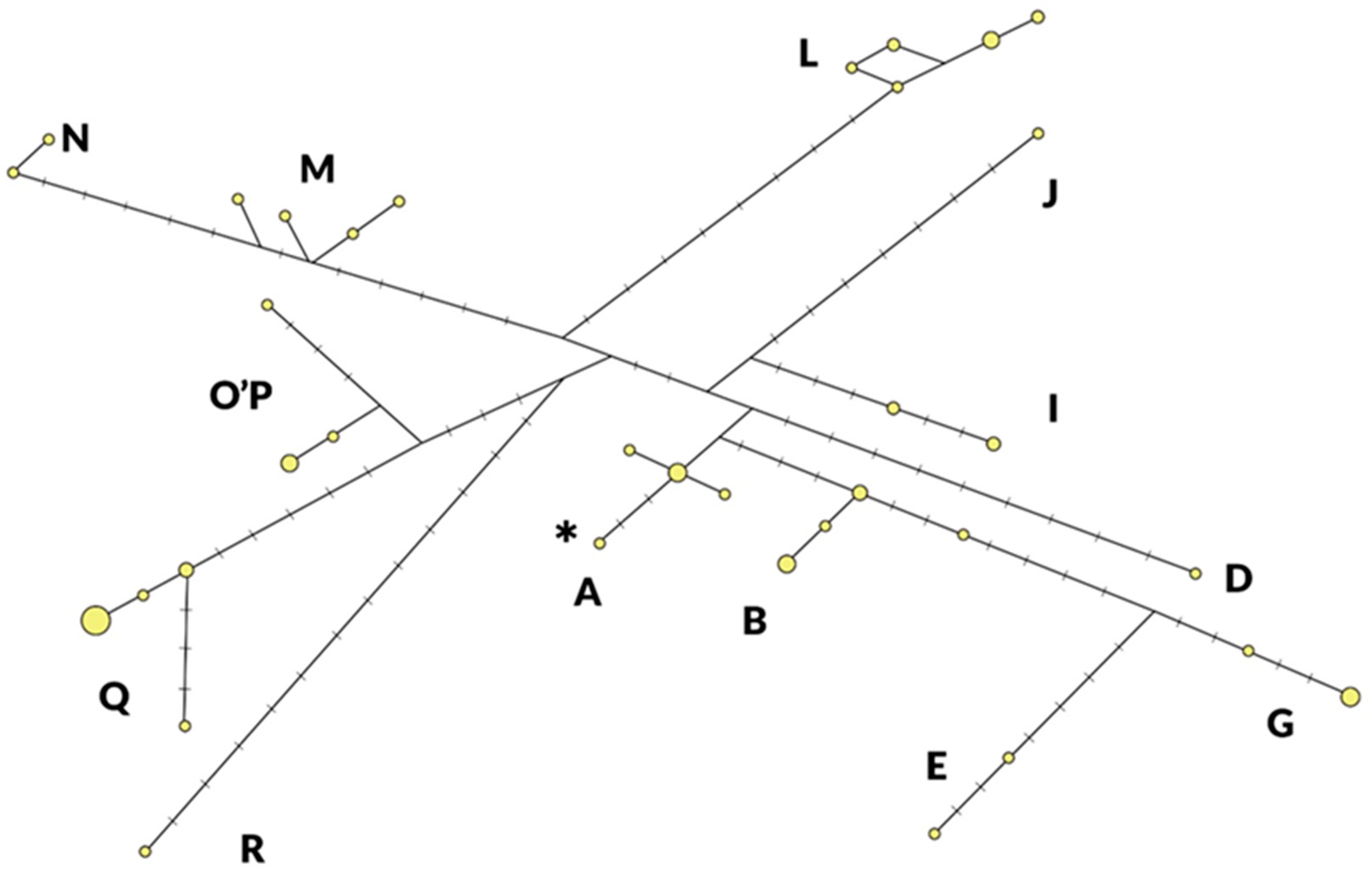
As shown in Figure 1, 13 of the known 18 haplogroups are present in the Szekler horse population with at least one representative, with an equally low number of representatives in HGs A (5), D (1), E (2), G (4), I (4), J (1), M (4), N (2), O’P (4) and R (1). As exceptions, the HGs Q, L, and B occurred with a higher number of individuals (15, 9, and 7, respectively). An individual belonging to haplogroup J has been identified, which will be listed as belonging to haplogroup J–K in the comparison. The asterisk indicates the haplotype identical to Equine Reference Sequence, NC001640, published in GenBank [17].
Figure 1.
Distribution of haplogroups (A–R) and haplotypes (yellow circles) detected in Szekler horse (n = 59). The size of the circles is in proportion to the number of individuals observed per haplotype. The lines crossed in the sections connecting the haplotypes indicate the number of mutations between them. The asterisk indicates the haplotype identical to Equine Reference Sequence, NC001640, published in GenBank.
In Table 1, the frequency values of the Szekler horse haplogroups are compared with the results of Achilli’s analysis [8]. It can be seen which horses the Szekler horse is closer to according to their geographical distribution. In the case of eight haplogroups (A, I, J–K, L, M, N, O’P, and R), the Szekler horse merged into the range of frequency values of the three geographical regions.

Table 1.
Haplogroup frequencies of Szekler horse compared with the former results.
In two cases (haplogroups D and G), the frequencies remained below the frequency values of the three geographical regions. Moreover, in three cases (haplogroup B, E, and Q), the frequencies surpassed them. Concerning the haplogroup Q, the frequency was significantly higher (p < 0.006).
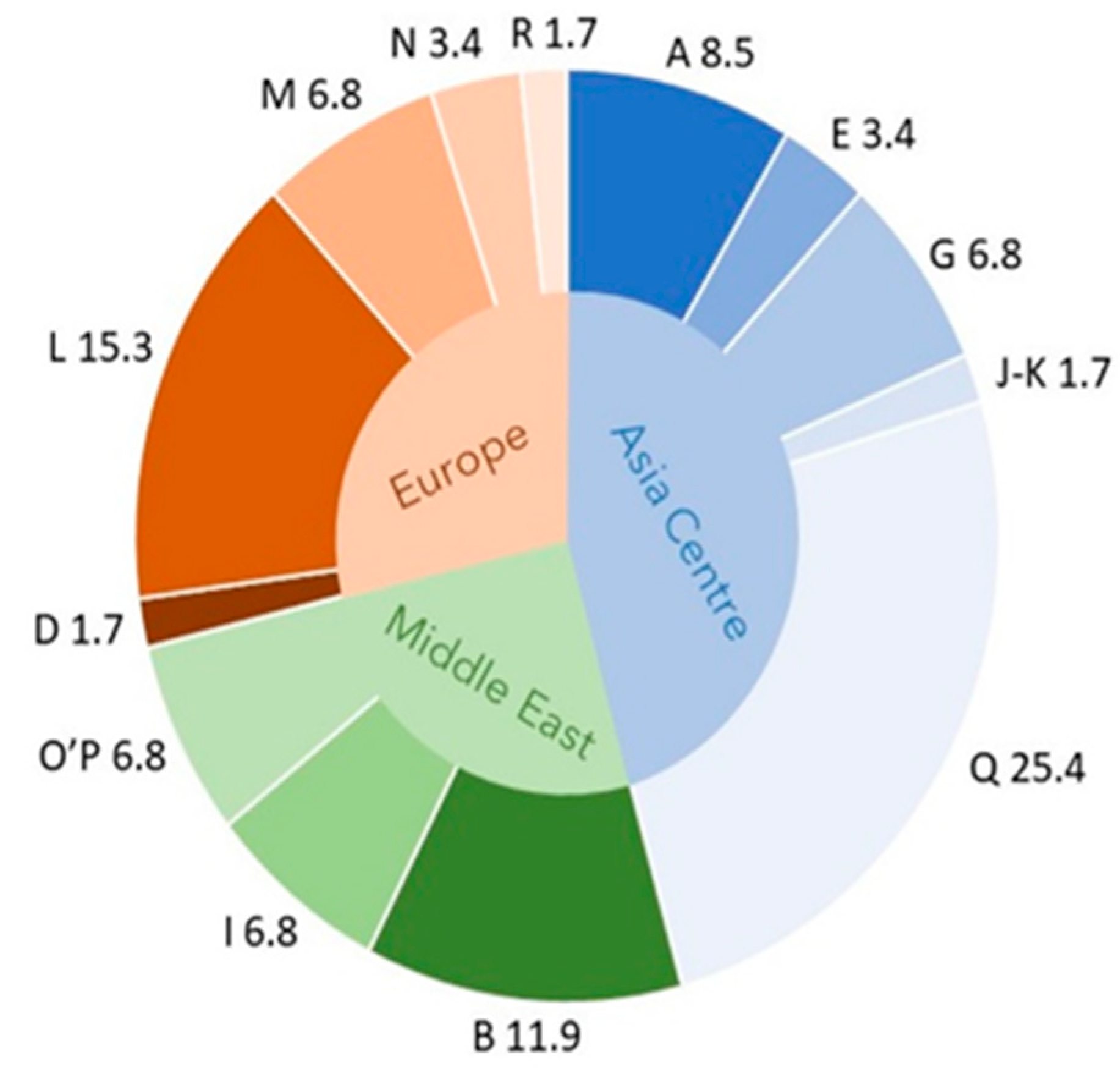
The additional circle graph (Figure 2) shows the composition of the matrilineal background of the Szekler horse according to the haplogroup most characteristic of the geographical areas. The diagram also illustrates that the Szekler horse shares the genetic pattern of the horse populations living in the three geographical areas on the one hand, with all of its haplogroups and, on the other hand, with its three most populous haplogroups as well [8]. According to the HG (e.g., Q) characteristic of Asian horses (Akhal-Teke), the examined stock is, to a remarkable extent, of Eastern origin (presumably, like other Hungarian horses brought from the East). The stock is complemented by the gene pool of horses from Europe (e.g., L) and the Middle East (e.g., B).
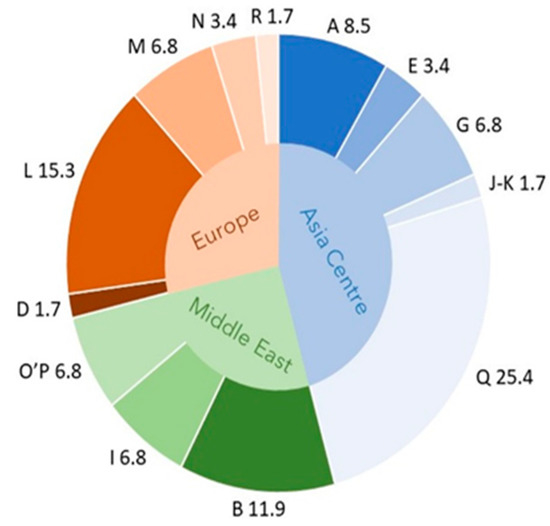
Figure 2.
Percentage of haplogroup (A–R) composition (outer ring) of the Szekler horse according to the haplogroup most characteristic of the geographical areas (inner ring). In Central Asian horses marked in blue, haplogroups A, E, G, J–K, and Q are more common than in other horses. In the Szekler horse, these haplogroups are present with 45.8% of the total. By contrast, the haplogroups characteristic of European and Middle Eastern horses appear with 28.8 and 25.4% of the total, respectively. From these values, we proportionally infer the origin of the reconstructed stock of Szekler horse.
The result of the Tajima D test was −0.26556, which was not statistically significant (p > 0.10). Fu’s Fs statistic was −6.566, with a low probability of the error <0.001. According to Fu and Li’s D* test statistic, which gave the value −0.44031, it was not significant, as non-significant p > 0.10. Regarding Fu and Li’s F* test, a result of −0.44723 was also not significant, as likewise p > 0.10.
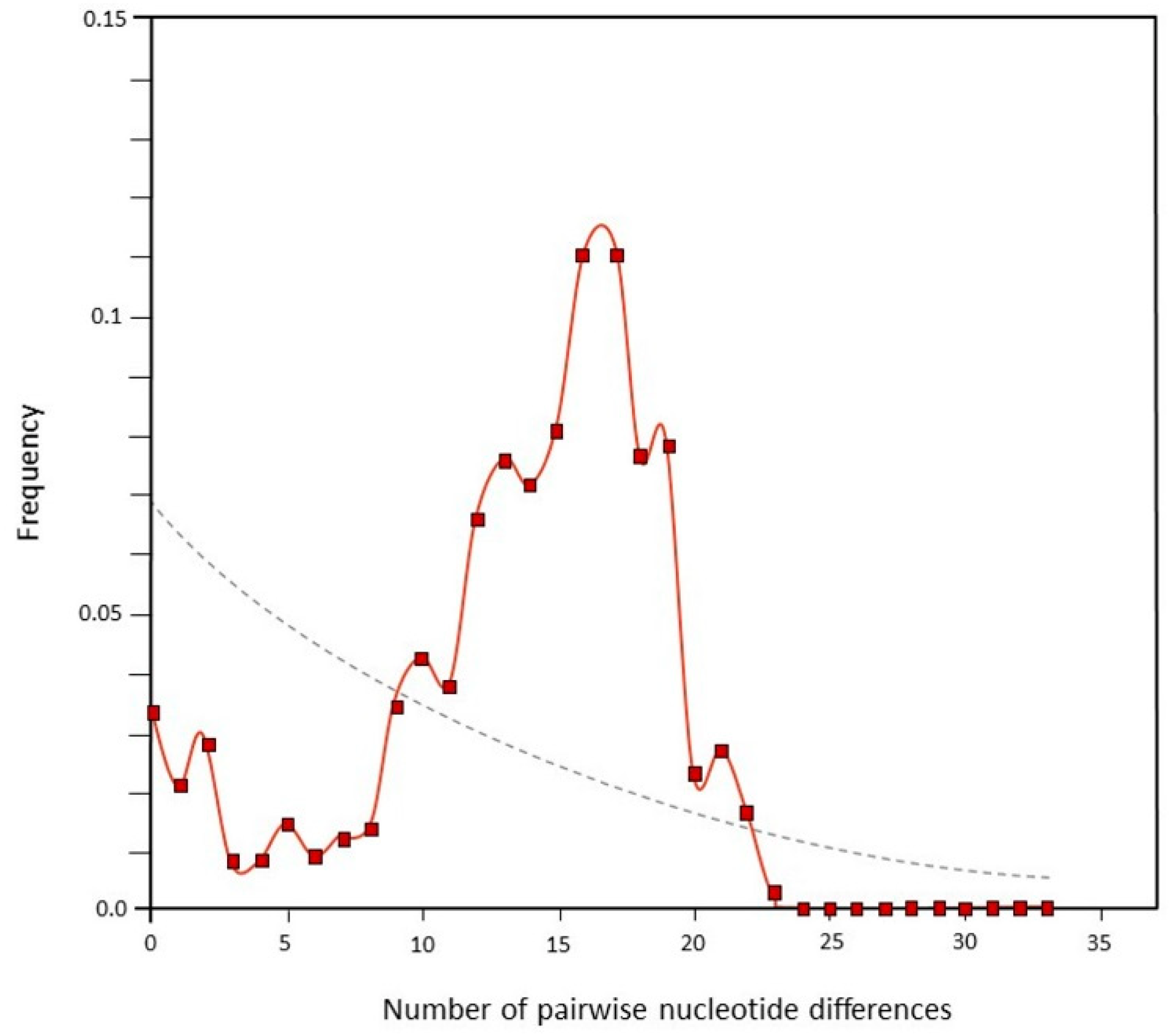
The points connected by a solid line reflect an intermediate (transition between mul-timodal and unimodal) observed distribution of sequence mismatches (Figure 3). The dotted line represents the expected distribution for a constant population size; its course is moderate.
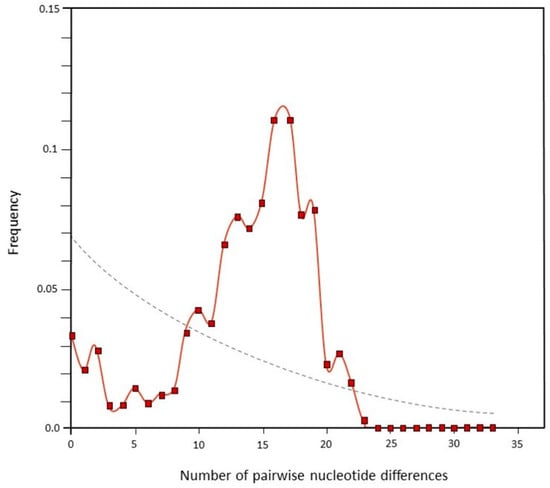
Figure 3.
Frequency distribution of the number of sequence mismatches between pairwise combinations of Szekler horse shown by solid red line (the dotted grey line represents the expected distribution for a constant population size).
4. Discussion
We determine that the examined stock of Szekler horse is of Eastern origin in almost half proportion, supplemented to a smaller extent by the gene pools of horses from Europe and the Middle East. It is confirmed to be separate from the horses used in comparison with its particularly common haplogroup Q (p < 0.006).
We see another example of how the distant, marked maternal background remains despite intensive grading-up. The Eastern Bulgarian horse breed, as a modern riding horse, was created based on native mares (Kamchia) by crossing Arabian and English thoroughbreds [18]. However, in the maternal background, a high proportion of individuals (35%) are carrying the Q haplotype group typical of Asian horses.
Sequence analysis of five Iranian native horse breeds (n = 95, 247-bp segment) established 27 haplotypes with 38 polymorphic sites and 12 haplogroups [19]. These indicators all took on higher values in our study (34, 68, and 13, respectively). However, the most common haplotypes in the Iranian breeds were found within haplogroup X2, which follows the nomenclature of Cieslak [20]. Nevertheless, the X2 was identified by Elsner et al. (2016) [21] in Late Iron Age horses in Switzerland. These ancient horse remains (Celtic and Roman) in Central Europe are represented in the major haplogroups (B, D, F, I, X2, X3) except for ones typical of today’s horses in Central Asia (e.g., A, G, Q). In the Carpathian Basin, according to Priskin et al. (2010) [22], none of the ancient (dating to the Avar and Hungarian conquest) and recent (Hucul in Hungary and Akhal Teke in Turkmenistan) horse haplotypes formed separate clusters (after Jansen et al., 2002, [23]). One-third of the Avar samples were grouped into haplogroup D and one-quarter into A. Half of the old Hungarian and modern Akhal Teke individuals were grouped in A. Half of the Huculs belonged to F. In the survey by Kusza et al. (2013) [24], the Hungarian Hucul horses (n = 71) typically carried haplogroups A, D, and F, which the authors explained by their close relationship with modern, Iberian peninsula and Far Eastern horses. They emphasize the 22 distant maternal lineages and the unique haplogroups of some of them, which provide a basis for preserving diversity. These observations suggest that most European horses are descended from ancestors that came directly through the Mediterranean region.
In the Cleveland Bay breed (n = 96; A, B, C, D), the authors [25] derive the presence of haplogroups A, B, and D from Spanish and North African horses. According to them, the breed maternally evolved from the native British pony only with regards to C. Its C1 cluster, determined by Jansen et al. (2002) [23], is associated with Exmoor, Fjord, Icelandic, and Scottish Highland ponies. In the Polish cold-blood horse (73 mares, 46 segregating sites, 15 distinct haplotypes), two main haplogroups (A, B) were found [26]. This synthetic breed is the result of crossing local mares with imported stallions of Western European cold-blood breeds. According to the investigation of Ivanković et al. (2009) [27], Posavina horse (n = 20), Croatian cold blood (n = 20), and Murinsulaner (n = 15) displayed the dominance of Jansen’s haplogroups [23] D, F, and B, H, and C, A, respectively. These breeds are endangered autochthonous breeds of Croatia and have been proven to display many ancient maternal lineages with high diversity in mtDNA. Sequences of all the observed horses stand closest to those of Belgian and Arabian horses. In particular, the Murinsulaner can be compared to the Szekler horse, since this breed is also undergoing breed reconstruction, which started a few decades ago.
From the study of Hristov et al. (2016) [28] on D-loop diversity in autochthonous mountain horse breeds from Bulgaria, it became clear that their haplogroup distribution showed the expected mixed genetic picture. In addition, the Rila-Pirin breed undoubtedly displayed Asian-specific haplogroups (A, E, Q, J, and C), which together accounted for half of the maternal background. In the Zabaikalskaya horse (n = 31) of the Trans-Baikal Region in Russia, 31 haplotypes and 8 haplogroups (B, C, G, H, L, M, Q, and R) were detected by Khrabrova et al. (2021) [29]. The most common haplogroups were G (16.3%) and Q (25.81%). A similar dominating prevalence of haplogroup Q was also found in the Szekler horse.
The genetic diversity of the reconstructed stock of Szekler horse is also revealed by the high number of polymorphic sites and haplotypes. Furthermore, this is confirmed by the high haplotype diversity (Hd = 0.966) and low nucleotide diversity (π = 0.02232). However, the estimated values of the applied tests were negative in all cases (Tajima D test, Fu and Li’s F* test, and Fu’s Fs statistics −0.26556, −0.44723, and −6.566, respectively), a result which was not statistically significant (p > 0.10). Only Fu’s Fs statistics showed a higher negative value and proved significant (<0.001). Its significant negative value indicates an abundance of rare haplotypes or recent selective mutations. Furthermore, comparatively, a significant positive result could be due to genetic diminution, such as the effects of a bottleneck. The pattern of the pairwise nucleotide differences in the nonrecombining CR of reconstructed stock may be used to ascertain the current demographic composition (status praesens) instead of to detect past population growth. As such, we can deduce that none of these animals are so closely maternally related.
In cooperation with the Szekler Horse Breeders’ Association, we laid the foundations of the breed’s DNA archive. The future goal is to enrich it with new samples (12 more samples, 2022). The breeding of mares and the selection of stallions can now be carried out within the breed based on the available mtDNA information of maternal lineages. An Arabian stallion (Reg. No. 001 Hamad B) is also involved in the program, because such melioration crossing is felt to promote the stabilization of the type with significant oriental blood, due to ancient breeding traditions in this remote area of Transylvania (the Szekler Land).
Furthermore, from a scientific point of view, it is very important to carry out genetic and phenotypic surveillance during the breed reconstruction process. In addition to the traditional recording of body measurements, we have started to achieve this by use of still images of video recording (Supplementary Material S2).
Regarding the environment of breed maintenance, attention must be drawn to the changes occurring in it. Our domestic animals are affected by global climate change–the Szekler horse is an animal with a close relationship with nature. Meteorological observations suggest that the temperature in the mountainous, tree-covered parts of Transylvania was higher than the average temperature of the 20th century (8.7 °C) almost every year during the last two decades. Climate change entails the reduction of forest vegetation and the erosion of mountain slopes in Transylvania [30,31], i.e., changes in the traditional usage environment. The Szekler horse is being used in both traditional and newer ways, keeping pace with technical development in equipment and the artificial environment. Such changes may also require accommodation from individuals and microevolutionary adaptation from the breed. A suitable and diverse environment should be in harmony with the breed’s abilities.
5. Conclusions
In our study, the maternal background of the reconstructed stock of the Szekler horse was successfully determined for the first time, using the hyper variable D-loop sequence according to the new haplogroup nomenclature. D-loop sequencing revealed a complex matrilineal descent of that breed with a marked composition typical of Central Asian horses. We are confident that a selection work can begin with the direct molecular genetic knowledge that we have just acquired, which will contribute to the sustainable breeding and utilization of the Szekler horse.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agriculture13020456/s1, Supplementary Material S1: Szekler horse_Agriculture_SuppMat1_Historical pictures.pdf, Supplementary Material S2: Szekler horse_Agriculture_SuppMat2_Conformation.pdf, Supplementary Material S3: Szekler horse_Agriculture_SuppMat3_Polymorphic_sites.pdf.
Author Contributions
A.G. and Á.M.-A. conceived and supervised the investigation and had substantial inputs into the completion of the manuscript. D.F. and M.H. participated in collecting the biological samples. Z.W. and I.B. provided an organizational and economic background for the implementation of the investigation. They collected background information to prepare the first draft of the manuscript. H.L., Á.M.-A. and A.G. had a substantial role in the sequencing and statistical analysis of sequences. All authors have read and agreed to the published version of the manuscript.
Funding
This project was funded by the European Union and co-financed by the European Regional Development Fund (ERDF) (Grant Contract No: VEKOP-2.3.2.-16-2016-00012).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The sequences were published in GenBank (https://www.ncbi.nlm.nih.gov/) under accession numbers: OL445148-OL445206.
Acknowledgments
Authors thank the Mereklye Association and the horse owners for enabling the research!
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wrangel, C.G. Siebenbürgen. In Ungarns Pferdezucht in Wort und Bild; Die Ungarische Landespferdezucht. Die Privatgestüte,Vierter Band; Verlag von Schickhardt & Ebner: Stuttgart, Germany, 1895; pp. 242–313. (In German) [Google Scholar]
- Hankó, B. Székely Lovak; Nagy Jenő és Fia Könyvnyomdája: Kolozsvár, Hungary, 1943; p. 26. (In Hungarian) [Google Scholar]
- Bodó, I.; Hecker, W. Lótenyésztők Kézikönyve; Mezőgazda Kiadó: Budapest, Hungary, 1992; p. 158. (In Hungarian) [Google Scholar]
- Hankó, B. Magyar Háziállataink; Ángyán Pál Tipográfiai Műintézet: Budapest, Hungary, 1943; p. 40. (In Hungarian) [Google Scholar]
- Bodó, I.; Hecker, W.; Surján, G.Y. A Székely Ló; Pharma Press Nyomdaipari Kft.: Budapest, Hungary, 2021; pp. 50–53. (In Hungarian) [Google Scholar]
- Ficsor, C.S. A Székely ló Fajta-Regenerálását Támogató Küllemtani Összehasonlító Vizsgálat. Master’s Thesis, Faculty of Agricultural- and Environmental Sciences, Szent István University, Gödöllő, Magyarország, 2016. (In Hungarian). [Google Scholar]
- Gáspárdy, A.; Ficsor, C.S.; Simon, L.; Bodó, I. A Possible Rescue of Székely Horse. The Breed “Székely ló” is Extinct, but the Type “Székely ló” Lives on (in Remote Areas of the Carpathian Mountains). Paper on “Unrecognised and Isolated Populations of rare Breeds and Varieties” 9th European Seminar on Agrobiodiversity and Annual Meeting of the SAVE Network, Lake Kerkini National Park, Greece, 11–13 September 2015. Available online: https://www.save-foundation.net/images/konferenzen/2015/Andras_Gaspardy_Szekely_Horse_2015.pdf (accessed on 5 January 2017).
- Achilli, A.; Olivieri, A.; Soares, P.; Lancioni, H.; Hooshiar Kashani, B.; Perego, U.A.; Nergadze, S.G.; Carossa, V.; Santagostino, M.; Capomaccio, S.; et al. Mitochondrial genomes from modern horses reveal the major haplogroups that underwent domestication. Proc. Natl. Acad. Sci. USA 2012, 109, 2449–2454. [Google Scholar] [CrossRef] [PubMed]
- Cardinali, I.; Giontella, A.; Tommasi, A.; Silvestrelli, M.; Lancioni, H. Unlocking Horse Y Chromosome Diversity. Genes 2022, 13, 2272. [Google Scholar] [CrossRef] [PubMed]
- Directive 2010/63/EU of the European Parliament and of the Council of 22 September 2010 on the Protection of Animals Used for Scientific Purposes. Available online: http://data.europa.eu/eli/dir/2010/63/oj (accessed on 8 July 2022).
- Giontella, A.; Sarti, F.M.; Cardinali, I.; Giovannini, S.; Cherchi, R.; Lancioni, H.; Silvestrelli, M.; Pieramati, C. Genetic Variability and Population Structure in the Sardinian Anglo-Arab Horse. Animals 2020, 10, 1018. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-Delbarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large datasets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Fu, Y.X. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 1997, 147, 915–925. [Google Scholar] [CrossRef]
- TIBCO Software Inc. Data Science Workbench, Version 14. 2020. Available online: http://tibco.com (accessed on 1 September 2021).
- Xu, X.; Arnason, U. The complete mitochondrial DNA sequence of the horse, Equus caballus: Extensive heteroplasmy of the control region. Gene 1994, 148, 357–362. [Google Scholar]
- Hristov, P.; Yordanov, G.; Vladov, V.; Neov, B.; Palova, N.; Radoslavov, G. Mitochondrial Profiles of the East Bulgarian and the Pleven Horse Breeds. J. Equine Vet. Sci. 2020, 88, 102933. [Google Scholar] [CrossRef]
- Moridi, M.; Masoudi, A.A.; Vaez Torshizi, R.; Hill, E.W. Mitochondrial DNA D-loop sequence variation in maternal lineages of Iranian native horses. Anim. Genet. 2013, 44, 209–213. [Google Scholar] [CrossRef]
- Cieslak, M.; Pruvost, M.; Benecke, N.; Hofreiter, M.; Morales, A.; Reissmann, M.; Ludwig, A. Origin and History of Mitochondrial DNA Lineages in Domestic Horses. PLoS ONE 2010, 12, e15311. [Google Scholar] [CrossRef]
- Elsner, J.; Deschler-Erb, S.; Stopp, B.; Hofreiter, M.; Schibler, J.; Schlumbaum, A. Mitochondrial d-loop variation, coat colour and sex identification of Late Iron Age horses in Switzerland. J. Archaeol. Sci. Rep. 2016, 6, 386–396. [Google Scholar] [CrossRef]
- Priskin, K.; Szabó, K.; Tömöry, G.; Bogácsi-Szabó, E.; Csányi, B.; Eördögh, R.; Downes, C.S.; Raskó, I. Mitochondrial sequence variation in ancient horses from the Carpathian Basin and possible modern relatives. Genetica 2010, 138, 211–218. [Google Scholar] [CrossRef]
- Jansen, T.; Forster, P.; Levine, M.A.; Oelke, H.; Hurles, M.; Renfrew, C.; Weber, J.; Olek, K. Mitochondrial DNA and the origins of the domestic horse. Proc. Natl. Acad. Sci. USA 2002, 99, 10905–10910. [Google Scholar] [CrossRef]
- Kusza, S.; Priskin, K.; Ivanković, A.; Jedrzejewska, B.; Podgorski, T.; Jávor, A.; Mihók, S. Genetic characterization and population bottleneck in the Hucul horse based on microsatellite and mitochondrial data. Biol. J. Linn. Soc. 2013, 109, 54–65. [Google Scholar] [CrossRef]
- Dell, A.C.; Curry, M.C.; Yarnell, K.M.; Starbuck, G.R.; Wilson, P.B. Mitochondrial D-loop sequence variation and maternal lineage in the endangered Cleveland Bay horse. PLoS ONE 2020, 15, e0243247. [Google Scholar] [CrossRef]
- Myćka, G.; Klecel, W.; Stefaniuk-Szmukier, M.; Jaworska, J.; Musiał, A.D.; Ropka-Molik, K. Mitochondrial Whole D-Loop Variability in Polish Draft Horses of Sztumski Subtype. Animals 2022, 12, 1870. [Google Scholar] [CrossRef]
- Ivanković, A.; Ramljak, J.; Konjačić, M.; Kelava, N.; Dovč, P.; Mijić, P. Mitochondrial D-loop sequence variation among autochthonous horse breeds in Croatia. Czech J. Anim. Sci. 2009, 54, 101–111. [Google Scholar] [CrossRef]
- Hristov, P.; Yordanov, G.; Ivanova, A.; Mitkov, I.; Sirakova, D.; Mehandzyiski, I.; Radoslavov, G. Mitochondrial diversity in mountain horse population from the South-Eastern Europe. Mitochondrial. DNA Part A 2016, 28, 787–792. [Google Scholar] [CrossRef]
- Khrabrova, L.A.; Blohina, N.V.; Bazaron, B.Z.; Khamiruev, T.N. Variability of mitochondrial DNA D-loop sequences in Zabaikalskaya horse breed. Vavilov J. Genet. Breed. 2021, 25, 486–491. [Google Scholar] [CrossRef]
- Rusu, T.; Moraru, P.; Coste, C.; Cacovean, H.; Chetan, F.; Chetan, C. Impact of climate change on climatic indicators in Transylvanian Plain, Romania. J. Food Agric. Environ. 2014, 12, 469–473. [Google Scholar]
- Cheval, S.; Bulai, A.; Croitoru, A.-E.; Dorondel, Ș.; Micu, D.; Mihăilă, D.; Sfîcă, L.; Tișcovschi, A. Climate change perception in Romania. Theor. Appl. Climatol. 2022, 149, 253–272. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).