A Novel PCR-Based Functional Marker of Rice Blast Resistance Gene Pi25
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Primer Design
2.3. DNA Extraction and Marker Assay
2.4. Rice Blast Resistance Evaluation
3. Results
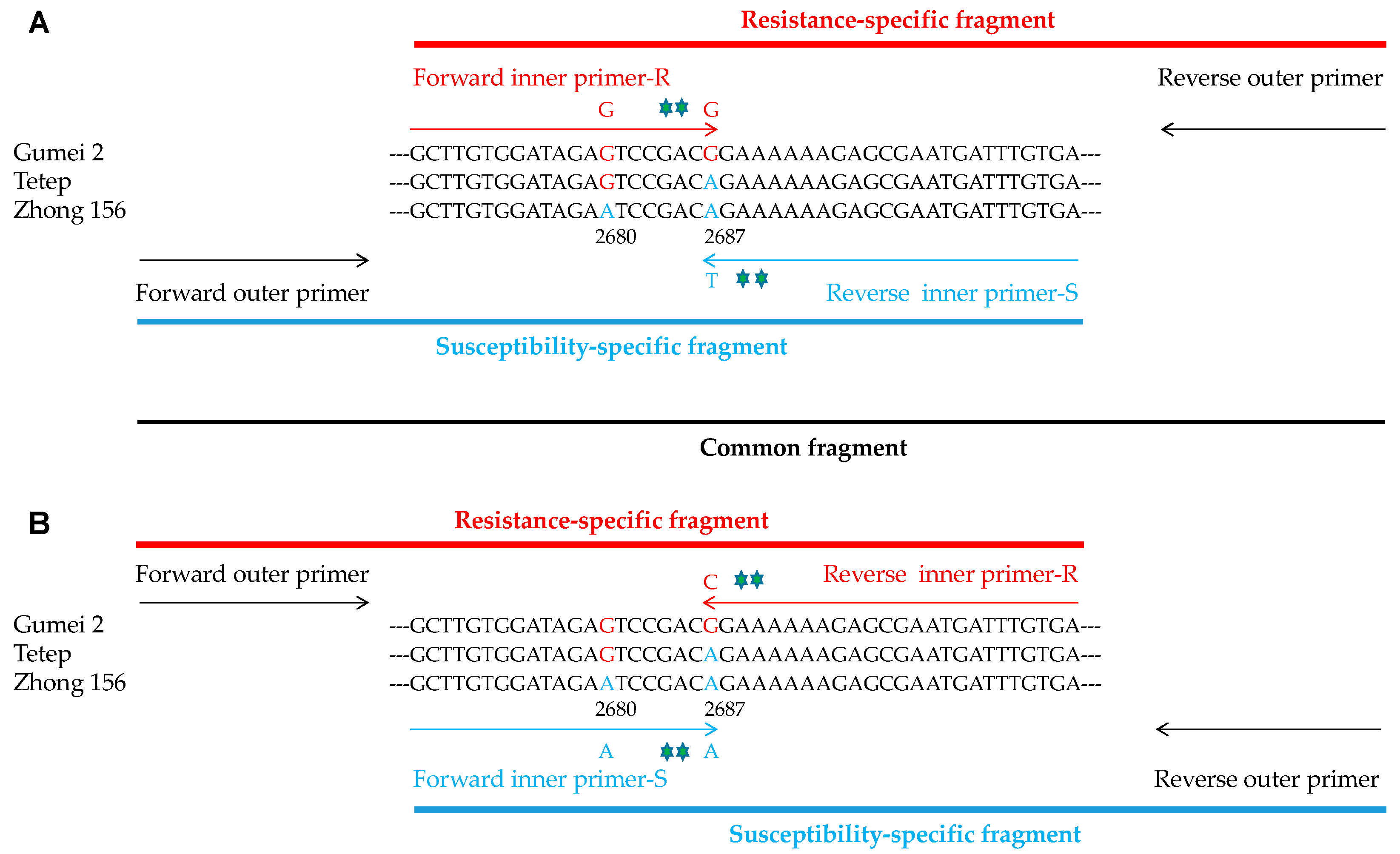
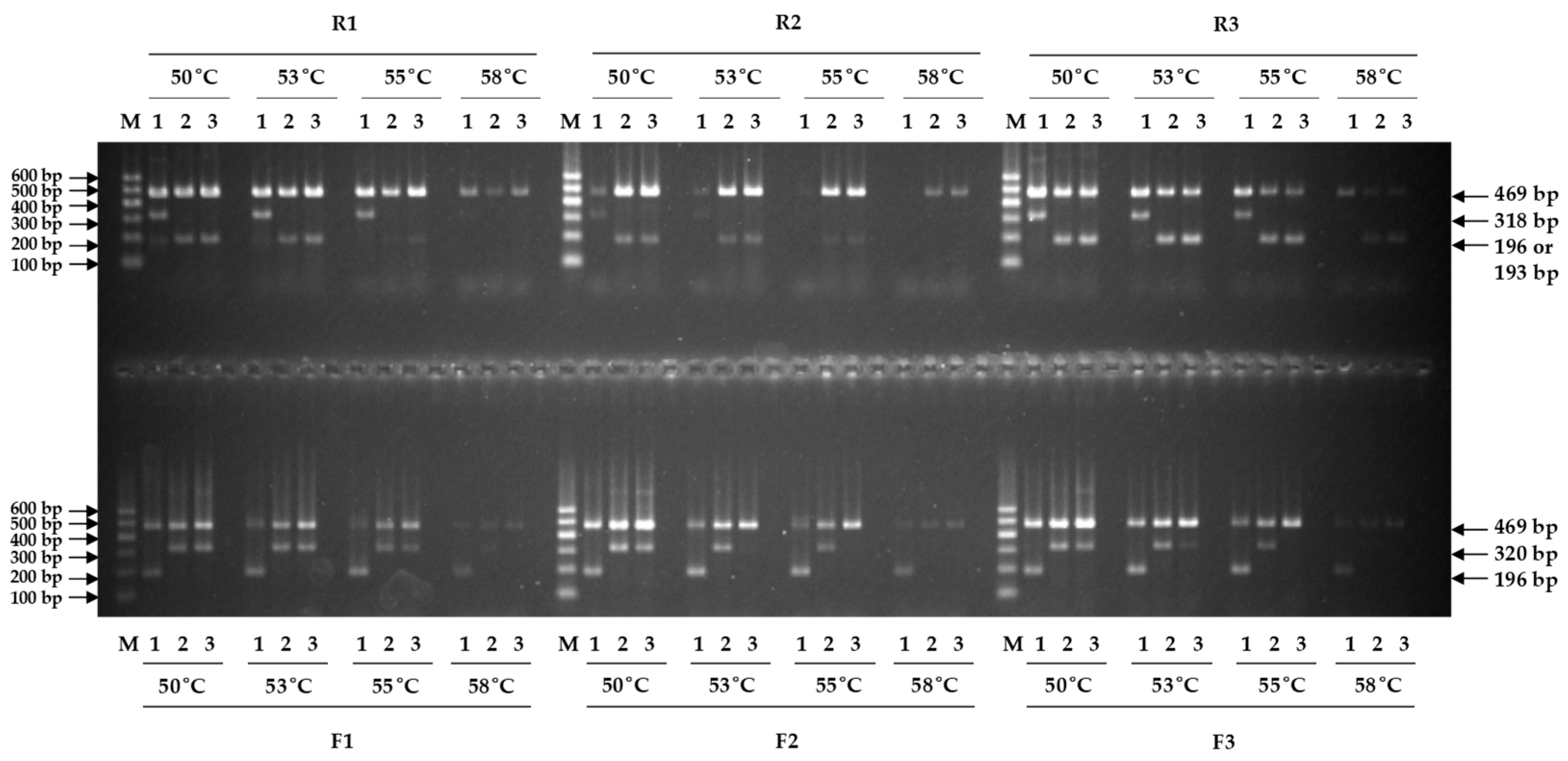
3.1. Selection of Primer Pairs
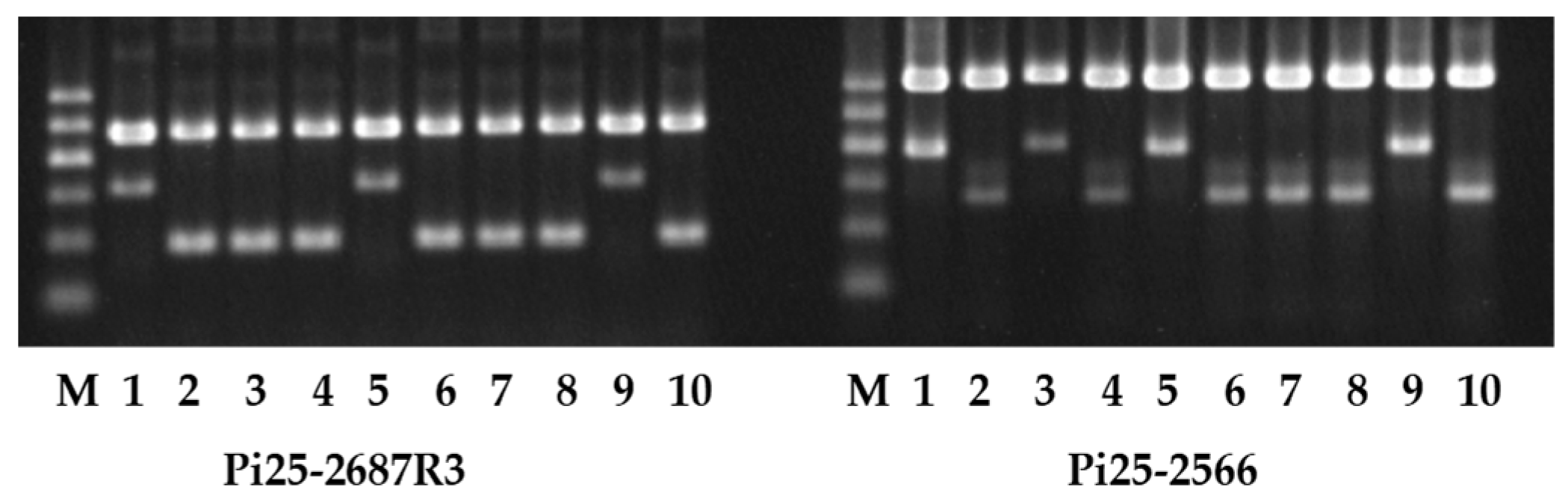
3.2. Validation of Newly Developed Marker
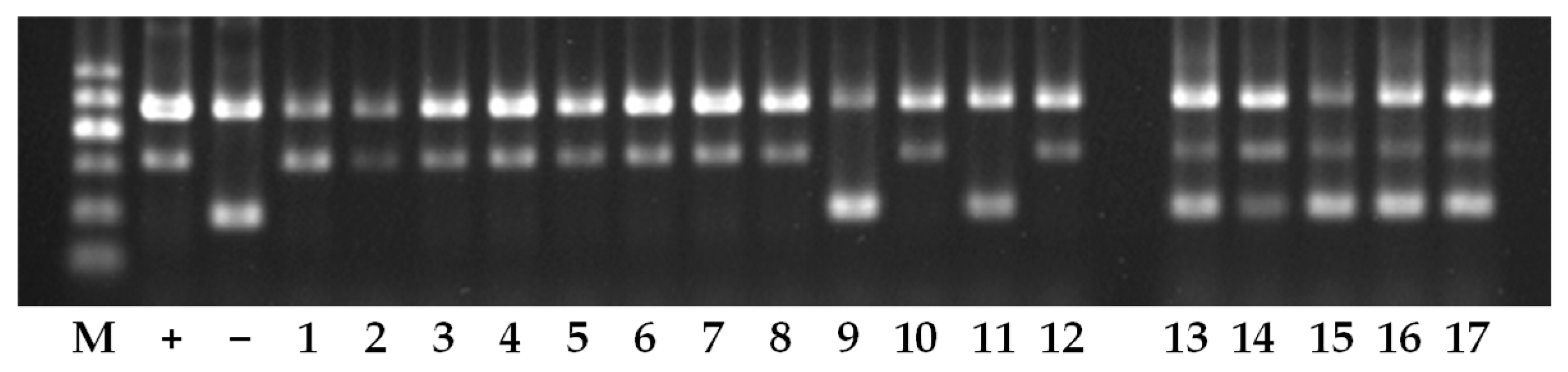
3.3. Genotyping Germplasm Using Allele-Specific Markers
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dean, R.; Van Kan, J.A.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef]
- Skamnioti, P.; Gurr, S.J. Against the grain: Safeguarding rice from rice blast disease. Trends Biotechnol. 2009, 27, 141–150. [Google Scholar] [CrossRef]
- Khush, G.S.; Jena, K.K. Current status and future prospects for research on blast resistance in rice (Oryza sativa L.). In Advances in Genetics, Genomics and Control of Rice Blast Disease; Wang, G.-L., Valent, B., Eds.; Springer: New York, NY, USA, 2009; pp. 1–10. [Google Scholar] [CrossRef]
- Li, W.; Chern, M.; Yin, J.; Wang, J.; Chen, X. Recent advances in broad-spectrum resistance to the rice blast disease. Curr. Opin. Plant Biol. 2019, 50, 114–120. [Google Scholar] [CrossRef]
- Cobb, J.N.; Biswas, P.S.; Platten, J.D. Back to the future: Revisiting MAS as a tool for modern plant breeding. Theor. Appl. Genet. 2019, 132, 647–667. [Google Scholar] [CrossRef]
- Zhuang, J.-Y.; Ma, W.-B.; Wu, J.-L.; Chai, R.-Y.; Lu, J.; Fan, Y.-Y.; Jin, M.-Z.; Leung, H.; Zheng, K.-L. Mapping of leaf and neck blast resistance genes with resistance gene analog, RAPD and RFLP in rice. Euphytica 2002, 128, 363–370. [Google Scholar] [CrossRef]
- Wu, J.-L.; Fan, Y.-Y.; Li, D.-B.; Zheng, K.-L.; Leung, H.; Zhuang, J.-Y. Genetic control of rice blast resistance in the durably resistant cultivar Gumei 2 against multiple isolates. Theor. Appl. Genet. 2005, 111, 50–56. [Google Scholar] [CrossRef]
- Chen, J.; Shi, Y.; Liu, W.; Chai, R.; Fu, Y.; Zhuang, J.; Wu, J. A Pid3 allele from rice cultivar Gumei2 confers resistance to Magnaporthe oryzae. J. Genet. Genom. 2011, 38, 209–216. [Google Scholar] [CrossRef]
- Wang, H.-M.; Chen, J.; Shi, Y.-F.; Pan, G.; Shen, H.-C.; Wu, J.-L. Development and validation of CAPS markers for marker-assisted selection of rice blast resistance gene Pi25. Acta Agron. Sini. 2012, 38, 1960–1968. [Google Scholar] [CrossRef]
- Zhou, H.; Zhan, X.; Chai, R.; Cheng, S.; Cao, L. Breeding of rice restorer lines carrying blast resistance gene Pi25 by marker-assisted selection. Chin. J. Rice Sci. 2008, 22, 590–596, (In Chinese with English Abstract). [Google Scholar] [CrossRef]
- Zhan, X.; Zhou, H.; Chai, R.; Zhuang, J.; Cheng, S.; Cao, L. Breeding of R8012, a rice restorer line resistant to blast and bacterial blight through marker-assisted selection. Rice Sci. 2012, 19, 29–35. [Google Scholar] [CrossRef]
- Yu, S.; Zheng, X.; Fan, T.; Huang, Y.; Chen, H.; Chen, S.; Hong, X.; Ruan, G. Breeding of PTGMS lines with blast resistance gene Pi25 by molecular marker-assisted selection. China Rice 2013, 19, 15–17, (In Chinese with English Abstract). [Google Scholar]
- Tu, S.; Zhou, P.; Zheng, Y.; Dong, R.; Zhang, S.; Huang, T.; Zheng, J. Breeding blast-resistant male sterile line of three line hybrid by Pi25 gene marker assisted selection. Mol. Plant Breed. 2015, 13, 1911–1917, (In Chinese with English Abstract). [Google Scholar] [CrossRef]
- Dong, R.; Wang, H.; Dong, L.; Zhou, P.; Tu, S.; You, Q.; Liao, F.; Huang, T. Improving the rice blast resistance for a CMS line of rice Zhenda A and its hybrids using molecular marker-assisted selection. J. Plant Genet. Resour. 2017, 18, 573–586, (In Chinese with English Abstract). [Google Scholar] [CrossRef]
- Liu, W.; Li, X.; Li, Y.; Pan, X.; Sheng, X.; Duan, Y. Improvement of rice blast resistance of Xiangwanxian No.13 with high quality by molecular marker-assisted selection. Mol. Plant Breed. 2017, 15, 3063–3069, (In Chinese with English Abstract). [Google Scholar] [CrossRef]
- Cao, N.; Ji, Z.; Zeng, Y.; Zheng, A.; Fang, J.; Wang, S.; Wu, K.; Chen, H.; Yang, C.; Liang, Y. Improving the blast resistance of super early-season rice variety Zhongzao39. J. Plant Genet. Resour. 2020, 21, 809–818, (In Chinese with English Abstract). [Google Scholar] [CrossRef]
- Mei, W.; Liu, P.; Hong, B.; Mu, H.; Lv, Q.; Sha, A.; Guo, S. Molecular identification of blast resistance genes Pi25, Pi56(t), Pit, and Pita from rice varieties. Hubei Agri. Sci. 2016, 55, 6604–6607, (In Chinese with English Abstract). [Google Scholar] [CrossRef]
- Xu, H.; Zhou, L.; Jiang, Y.; Li, S.; Zha, W.; Liu, K.; Chen, Z.; Yang, G.; Li, P.; Fang, G.; et al. Distribution of some disease and insect resistance genes and quality related genes in rice core germplasm resources. Mol. Plant Breed. 2018, 16, 1121–1126, (In Chinese with English Abstract). [Google Scholar] [CrossRef]
- Wang, Y.; Chen, X.; Yin, H.; Wang, Y.; Yang, R.; Zang, Z.; Wang, S. Molecular detection of rice blast resistance gene in the main rice germplasms in Henan Province. Mol. Plant Breed. 2018, 16, 3203–3212, (In Chinese with English Abstract). [Google Scholar] [CrossRef]
- Qi, C.; Xu, X.; Ma, J.; Wang, S.; Tian, P.; Meng, L.; Yan, W.; Zhao, Z.; Wang, J.; Wang, J.; et al. Distribution of blast resistance genes Pib, Pita, Pi5, Pi25 and Pi54 in mini-core collection of Chinese rice germplasm. J. Plant Genet. Resour. 2019, 20, 1240–1246+1254, (In Chinese with English Abstract). [Google Scholar] [CrossRef]
- Zhang, Y.; Gao, Q.; Zhao, Y.; Liu, R.; Fu, Z.; Li, X.; Sun, Y.; Jin, X. Evaluation of rice blast resistance and genetic structure analysis of rice germplasm in Heilongjiang Province. Sci. Agri. Sini. 2022, 55, 625–640, (In Chinese with English Abstract). [Google Scholar] [CrossRef]
- Zhou, L.; Cai, H.; Dai, F.; Xu, H.; Liu, K.; Zha, W.; Li, S.; Yang, G.; Chen, Z.; Li, P.; et al. Development and application of a functional SNP marker of the blast resistant gene Pi25 in rice. Mol. Plant Breed. 2016, 14, 2680–2685, (In Chinese with English Abstract). [Google Scholar] [CrossRef]
- Ye, S.; Dhillon, S.; Ke, X.; Collins, A.R.; Day, I.N. An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res. 2001, 29, E88. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Qiu, H.; Chai, R.; Mao, X.; Wang, Y.; Sun, G. Distribution and evaluation of 6 blast genes in major cultivated rice varieties in Zhejiang. Fujian J. Agri. Sci. 2019, 34, 214–222, (In Chinese with English Abstract). [Google Scholar] [CrossRef]
- Zheng, K.; Huang, N.; Bennett, J.; Khush, G.S. PCR-Based Marker-Assisted Selection in Rice Breeding; IRRI Discussion Paper Series No.12; International Rice Research Institute: Los Baños, Philippines, 1995. [Google Scholar]
- Zhou, B.; Qu, S.; Liu, G.; Dolan, M.; Sakai, H.; Lu, G.; Bellizzi, M.; Wang, G. The eight amino-acid differences within three leucine-rich repeats between Pi2 and Piz-t resistance proteins determine the resistance specificity to Magnaporthe grisea. Mol. Plant Microbe Interact. 2006, 19, 1216–1228. [Google Scholar] [CrossRef]
- Peng, M.; Lin, X.; Xiang, X.; Ren, H.; Fan, X.; Chen, K. Characterization and evaluation of transgenic rice pyramided with the Pi genes Pib, Pi25 and Pi54. Rice 2021, 14, 78. [Google Scholar] [CrossRef]
- Iyer-Pascuzzi, A.S.; McCouch, S.R. Functional marker for xa5-mediated resistance in rice (Oryza sativa L.). Mol. Breed. 2007, 19, 291–296. [Google Scholar] [CrossRef]
- Medrano, R.F.; de Oliveira, C.A. Guidelines for the tetra-primer ARMS-PCR technique development. Mol. Biotechnol. 2014, 56, 599–608. [Google Scholar] [CrossRef]
- Pereira, A.; Tcacenco, F.A.; Klabunde, G.H.F.; de Andrade, A. Detecting acetyl-coenzyme a carboxylase resistance gene in rice (Oryza sativa L.). Mol. Biol. Rep. 2019, 46, 6271–6276. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, A.; Wang, F.; Wang, J.; Bi, J.; Kong, D.; Zhang, F.; Luo, L.; Liu, G.; Yu, X. Development and validation of a PCR-based functional marker system for identifying the low amylose content-associated gene Wx hp in rice. Breed. Sci. 2019, 69, 702–706. [Google Scholar] [CrossRef]
- Mao, T.; Zhu, M.; Ahmad, S.; Ye, G.; Sheng, Z.; Hu, S.; Jiao, G.; Xie, L.; Tang, S.; Wei, X.; et al. Superior japonica rice variety YJ144 with improved rice blast resistance, yield, and quality achieved using molecular design and multiple breeding strategies. Mol. Breed. 2021, 41, 65. [Google Scholar] [CrossRef]
- Mesrian Tanha, H.; Mojtabavi Naeini, M.; Rahgozar, S.; Rasa, S.M.; Vallian, S. Modified tetra-primer ARMS PCR as a single-nucleotide polymorphism genotyping tool. Genet. Test. Mol. Biomark. 2015, 19, 156–161. [Google Scholar] [CrossRef] [PubMed]
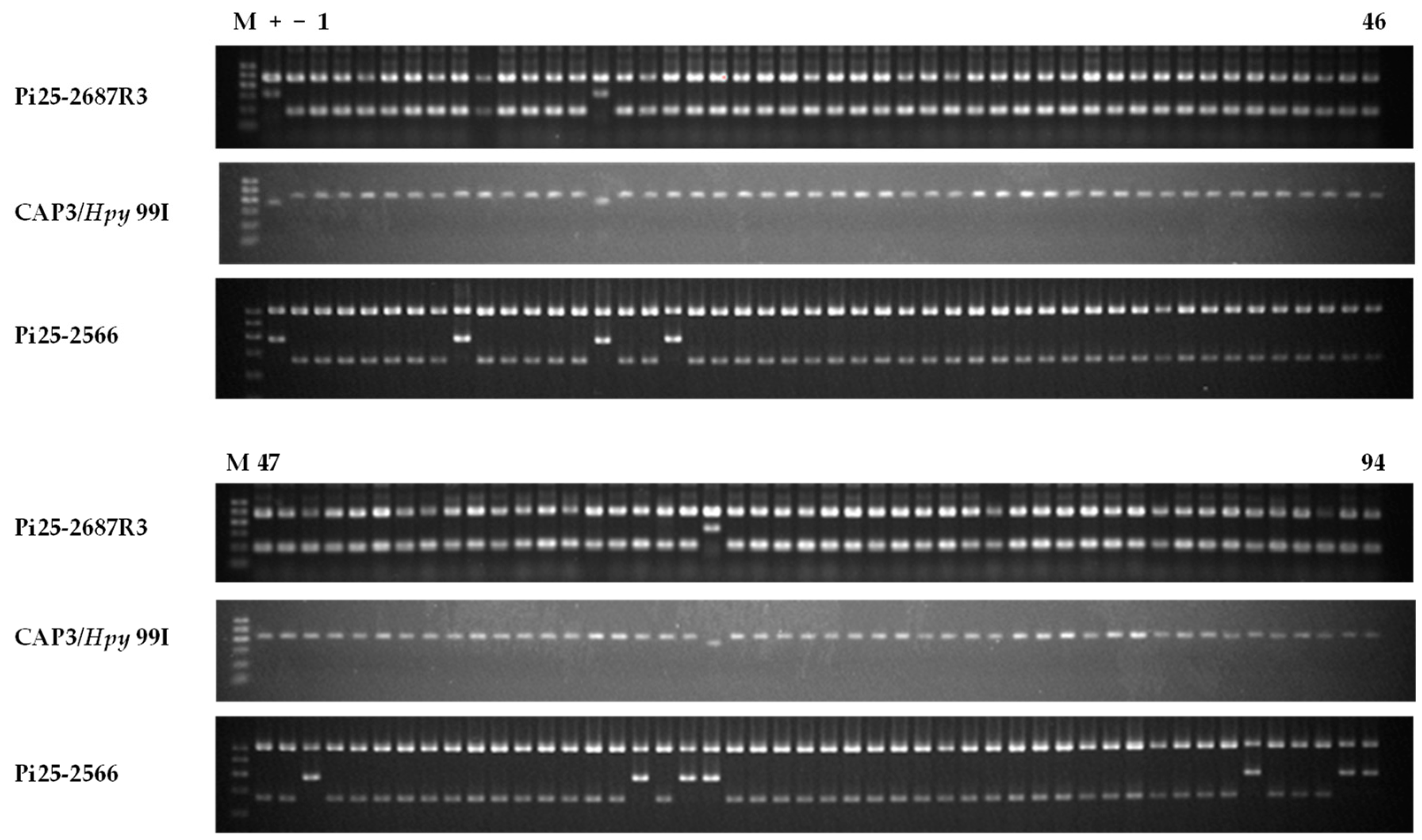
| No. | Accession Name | Country | Pi25-2687R3 | CAP3/Hpy 99I | Pi25-2566 |
|---|---|---|---|---|---|
| 1 | AKHNI SAIL | Bangladesh | − | − | − |
| 2 | AUS 43 | Bangladesh | − | − | − |
| 3 | BERI | Bangladesh | − | − | − |
| 4 | BR 601-3-3-2-4 | Bangladesh | − | − | − |
| 5 | CHAKULA | Bangladesh | − | − | − |
| 6 | RANGPUR | Bangladesh | − | − | − |
| 7 | MA HNAN THU KA | Burma | − | − | + |
| 8 | X72-7-1 | Burma | − | − | − |
| 9 | IB 28 | Burundi | − | − | − |
| 10 | IB 42 | Burundi | − | − | − |
| 11 | NEANG CHHOUK | Cambodia | − | − | − |
| 12 | NEANG SAN | Cambodia | − | − | − |
| 13 | 5173 | Colombia | + | + | + |
| 14 | RT 1031-69 | Congo | − | − | − |
| 15 | AMISTAD 82-8 | Cuba | − | − | − |
| 16 | C.CORTO 1 | Cuba | − | − | + |
| 17 | GU2725 | Cuba | − | − | − |
| 18 | SELECCION VG-5 | Cuba | − | − | − |
| 19 | GIZA 181 | Egypt | − | − | − |
| 20 | CSR 11 | India | − | − | − |
| 21 | CSR 12 | India | − | − | − |
| 22 | CSR 13 | India | − | − | − |
| 23 | CSR 5 | India | − | − | − |
| 24 | CSR 9 | India | − | − | − |
| 25 | DULAR | India | − | − | − |
| 26 | HARWA | India | − | − | − |
| 27 | HIMDHAN | India | − | − | − |
| 28 | HPA 74 | India | − | − | − |
| 29 | JC 195 | India | − | − | − |
| 30 | JW 42 | India | − | − | − |
| 31 | JW 60 | India | − | − | − |
| 32 | K 428-25 | India | − | − | − |
| 33 | KAJARU | India | − | − | − |
| 34 | KALING A2 | India | − | − | − |
| 35 | NDR 4012 | India | − | − | − |
| 36 | NDR 89 | India | − | − | − |
| 37 | OR 79-21 | India | − | − | − |
| 38 | PR32-PD47-PD4 | India | − | − | − |
| 39 | RP 1017-76-1-3-2 | India | − | − | − |
| 40 | T 23 | India | − | − | − |
| 41 | TM 10265 | India | − | − | − |
| 42 | VL DHAN 16 | India | − | − | − |
| 43 | B 6397F-MR-7-5M-1-1 | Indonesia | − | − | − |
| 44 | BAHBUTONG | Indonesia | − | − | − |
| 45 | BATANG PANE | Indonesia | − | − | − |
| 46 | BOYO | Indonesia | − | − | − |
| 47 | CIMANUK | Indonesia | − | − | − |
| 48 | CIPUNEGARA | Indonesia | − | − | − |
| 49 | PADI SEGUTUK | Indonesia | − | − | + |
| 50 | DOMSIAH 138 | Iran | − | − | − |
| 51 | HABATAKI | Japan | − | − | − |
| 52 | SANJIANG | Japan | − | − | − |
| 53 | SHINKWANG | Korea | − | − | − |
| 54 | JHALI | Nepal | − | − | − |
| 55 | ITA 304 | Nigeria | − | − | − |
| 56 | MILYANG 70 | North Korea | − | − | − |
| 57 | SHUIYUAN299 | North Korea | − | − | − |
| 58 | 368 | Pakistan | − | − | − |
| 59 | BASMATI 370 | Pakistan | − | − | − |
| 60 | BASMATI 685 | Pakistan | − | − | − |
| 61 | GANJAY | Pakistan | − | − | − |
| 62 | JIJAI NIKI | Pakistan | − | − | − |
| 63 | KHARA GANJA | Pakistan | − | − | + |
| 64 | RATRIA | Pakistan | − | − | − |
| 65 | TAK RATIA | Pakistan | − | − | + |
| 66 | PANAMA 1537 | Panama | + | + | + |
| 67 | 75-1-120 | Philippines | − | − | − |
| 68 | 97A-M54 | Philippines | − | − | − |
| 69 | A1452 | Philippines | − | − | − |
| 70 | BINICOL | Philippines | − | − | − |
| 71 | PIRURUTONG | Philippines | − | − | − |
| 72 | PR 23631-98 | Philippines | − | − | − |
| 73 | PRATAO PRECOCE | Philippines | − | − | − |
| 74 | SINABA | Philippines | − | − | − |
| 75 | SINANDOMENG | Philippines | − | − | − |
| 76 | BATHKIRIEL | Sri Lanka | − | − | − |
| 77 | BG 1165-1 | Sri Lanka | − | − | − |
| 78 | BG 170 | Sri Lanka | − | − | − |
| 79 | BG 915 | Sri Lanka | − | − | − |
| 80 | KATAHATA HAMBA | Sri Lanka | − | − | − |
| 81 | MALKORA | Sri Lanka | − | − | − |
| 82 | MURUNGAWEE | Sri Lanka | − | − | − |
| 83 | SINNA SIVAPPU | Sri Lanka | − | − | − |
| 84 | NIAW SANPAH TAWNG | Thailand | − | − | − |
| 85 | XIANLUOSICHI | Thailand | − | − | − |
| 86 | DAWN CI9534 | USA | − | − | − |
| 87 | NOVA 66 CI9481 | USA | − | − | − |
| 88 | BAU HUONG DOONG | Vietnam | − | − | − |
| 89 | CHAH NONG NGHE AN | Vietnam | − | − | + |
| 90 | CHIEM BAC | Vietnam | − | − | − |
| 91 | LAI TRANG | Vietnam | − | − | − |
| 92 | QUCM | Vietnam | − | − | − |
| 93 | RE BAU | Vietnam | − | − | + |
| 94 | RE CHANH | Vietnam | − | − | + |
| Type | Name | Sequence (5′-3′) |
|---|---|---|
| Forward outer primer | 2687-FO | CACACCTGAATGAAATTAAGATGACA |
| Reverse outer primer | 2687-RO | ATATACAATATTGAGGGTATGGAAC |
| Forward inner primer-R | 2687-FI-R | GCTTGTGGATAGAGTCCTTCG |
| Reverse inner primer-S | 2687-RI-S1 | CAAATCATTCGCTCTTTTCCCT |
| 2687-RI-S2 | CAAATCATTCGCTCTTTTGGCT | |
| 2687-RI-S3 | TCACAAATCATTCGCTCTTTTAACT | |
| Forward inner primer-S | 2687-FI-S1 | GAGCTTGTGGATAGAATCCTCCA |
| 2687-FI-S2 | GAGCTTGTGGATAGAATCCTGCA | |
| 2687-FI-S3 | GAGCTTGTGGATAGAATCCTTCA | |
| Reverse inner primer-R | 2687-RI-R | TCACAAATCATTCGCTCTTTTAACC |
| No. in Figure 4 | Line/Variety | Type | Genotype | Phenotype |
|---|---|---|---|---|
| 1 | R173 | Restorer line | + | R |
| 2 | R1909 | Restorer line | + | R |
| 3 | R1913 | Restorer line | + | R |
| 4 | R1925 | Restorer line | + | MR |
| 5 | R1927 | Restorer line | + | R |
| 6 | R1932 | Restorer line | + | MR |
| 7 | R1933 | Restorer line | + | MR |
| 8 | R1934 | Restorer line | + | R |
| 9 | R1937 | Restorer line | − | S |
| 10 | ZP174 | Restorer line | + | R |
| 11 | ZP175 | Restorer line | − | MS |
| 12 | ZP178 | Restorer line | + | R |
| 13 | Maoxiangyou 61 | Hybrid variety | +/− | R |
| 14 | Mixiangyou 153 | Hybrid variety | +/− | R |
| 15 | Huangyou 153 | Hybrid variety | +/− | R |
| 16 | Zhongxiangyou 173 | Hybrid variety | +/− | R |
| 17 | Zhiliangyou 173 | Hybrid variety | +/− | R |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, Y.; Zhang, Z.; Huang, D.; Huang, T.; Wang, H.; Zhuang, J.; Zhu, Y. A Novel PCR-Based Functional Marker of Rice Blast Resistance Gene Pi25. Agriculture 2023, 13, 1926. https://doi.org/10.3390/agriculture13101926
Fan Y, Zhang Z, Huang D, Huang T, Wang H, Zhuang J, Zhu Y. A Novel PCR-Based Functional Marker of Rice Blast Resistance Gene Pi25. Agriculture. 2023; 13(10):1926. https://doi.org/10.3390/agriculture13101926
Chicago/Turabian StyleFan, Yeyang, Zhenhua Zhang, Derun Huang, Tingxu Huang, Hongfei Wang, Jieyun Zhuang, and Yujun Zhu. 2023. "A Novel PCR-Based Functional Marker of Rice Blast Resistance Gene Pi25" Agriculture 13, no. 10: 1926. https://doi.org/10.3390/agriculture13101926
APA StyleFan, Y., Zhang, Z., Huang, D., Huang, T., Wang, H., Zhuang, J., & Zhu, Y. (2023). A Novel PCR-Based Functional Marker of Rice Blast Resistance Gene Pi25. Agriculture, 13(10), 1926. https://doi.org/10.3390/agriculture13101926







