Abstract
The mechanisms of modulating milk production traits remain largely unknown. Based on our previous RNA-seq, DDIT3 was presumed as a novel, promising candidate gene for regulating milk protein and fat traits in dairy cattle. To further detect the genetic effect of DDIT3 and its potential molecular mechanisms in regulating milk production traits in dairy cattle, here, we performed a genotype-phenotype association study. Two SNPs, g.-1194 C>T and g.-128 C>T, were significantly associated with MY (p = 0.0063), FY (p = 0.0001) and PY (p = 0.0216), respectively. A luciferase assay demonstrated that the allele T of g.-128 C>T increased the promoter activity by binding the HSF2, while allele C did not. To further reveal the molecular regulatory mechanisms, the DDIT3-knockdown MAC-T cells were established. It was observed that DDIT3 silencing could induce apoptosis and increase the number of PI-positive cells. Meanwhile, DDIT3 silencing led to increased expression of inflammatory markers, such as IL-6, IL6R, IL1B, IL7R, IL1RL2, IL1A, STAT1-5, MYC, IGFBP4, and IGFBP5, and especially for IL-6 (log2FC = 4.22; p = 3.49 × 10−112). Additionally, compared with the control group, increased lipid accumulation was found in the DDIT3-knockdown MAC-T cells. Thus, our results proved that lower expression of DDIT3 could result in increased lipid accumulation and apoptosis via up-regulating the expression of IL-6. These findings provided clues about the regulatory mechanisms of milk production traits in dairy cattle.
1. Introduction
The milk production traits of dairy cattle, including milk yield (MY), fat yield (FY), protein yield (PY), fat percentage (FP), and protein percentage (PP), are the most important economical traits, which are influenced by many genes []. Therefore, the most profitable breeding goal for milk-producing traits remains improvement []. As is known, RNA sequencing (RNA-seq) has enabled gene discovery and expedited the genetic improvement of dairy cattle [,]. Our previous RNA-seq study identified 31 differentially expressed genes associated with milk production traits in the mammary glands of lactating Holstein cows []. Of these, DNA damage-inducible transcript 3 (DDIT3) was significantly down-regulated (p = 4.01 × 10−5) in the Holstein cows []. Moreover, it was found that DDIT3 located only 0.46 Mb, from the SNP (ARS-BFGL-NGS-14781), which was significantly associated with fat percentage []. In addition, DDIT3 was also adjacent to a previously reported QTL for PP and FP []. These findings indicated that the DDIT3 gene may be a promising key gene for milk production traits in dairy cattle.
DDIT3, known as the CAAT/enhancer binding protein homologous transcription factor (CHOP) or growth retardation and DNA damage-inducible gene 153 (GADD153), is involved in various biological processes, including differentiation, proliferation, expression, and energy metabolism []. As a transcription factor, DDIT3 has specifically been shown to regulate DNA damage and ER stress-induced apoptosis by regulating the transcription of antiapoptotic and proapoptotic genes [,]. Recent publications have indicated that an overexpression of DDIT3 promotes apoptosis in several cell lines [,,]. Overexpression of DDIT3 in human cells led to cell death and growth arrest at the G1/S phase in murine 3T3 cells []. DDIT3-/- mouse experiments revealed that DDIT3-mediated apoptosis contributes to the pathogenesis of ER stress-related diseases in murine 3T3 cells []. Current evidence suggests that DDIT3 is an important mediator of cellular functions. However, the effects of DDIT3 on cellular functions have not been tested and clarified in dairy cattle.
To explore the potential causal genetic variants associated with milk production traits, we detected the genetic effects with an independent dairy cattle population. We also generated DDIT3 knock-out bovine MAC-T cells using RNA interference (RNAi) and applied RNA sequencing technology to investigate the biological function of DDIT3. Subsequently, the differentiation model was established to understand the molecular mechanisms of DDIT3. Our results may lead to a better understanding of the biochemical mechanisms by which DDIT3 regulates milk production ability and can be used via marker-assisted breeding.
2. Materials and Methods
2.1. Ethics Statement
The experimental programs and protocols were approved by the Institutional Animal Care and Use Committee of Hefei University of Technology (approval number HFUT20200518005).
2.2. Animals and Phenotypic Data
A daughter design was used in this study. A total of 717 Chinese Holstein cows from 12 corresponding sires were collected to construct the study population. The dairy cattle were from 16 dairy farms in Sanyuanlvhe Dairy Farming Center (Beijing, China). The estimated breeding values (EBVs) for milk yield (MY), protein yield (PY), fat yield (FY), protein percentage (PP), and fat percentage (FP) were predicted by the Dairy Data Center of the Dairy Association of China.
2.3. DNA Extraction and SNP Identification
Blood samples were collected from the dairy cows and stored at −20 °C. Genomic DNA was extracted using the Tiangen DP (318) Blood DNA Kit (Beijing, China). A DNA pool was constructed with equal DNA concentration for each. The results of the isolated DNA were detected by a NanoDrop™ ND-2000c Spectrophotometer (Thermo Scientific, Waltham, MA, USA). Based on the genomic sequence of the bovine DDIT3 gene (ENSBTAT00000044712.2), 8 pairs of primers were designed by Primer Premier 5.0 to amplify all four exons and 1500 bp of 5’ flanking sequences (Table S1).
2.4. Genotyping and Haplotype Analysis
The SNPs of all individuals were genotyped using the SEQUENOM MassArray MALDI-TOF mass spectrometer (Sequenom, San Diego, CA, USA). To further explore the LD extent, haplotypes of the DDIT3 gene were inferred using the software BEAGLE 3.2 program, and, where necessary, sporadic missing genotype data were also imputed. The measure of pairwise LD for all three SNPs was performed using the software Haploview 4.2 (Broad Institute of MIT and Harvard, Cambridge, MA, USA). Accordingly, the LD block was generated using the linkage disequilibrium coefficient D’. Haplotypes within these blocks were applied to test their associations with the milk production traits.
2.5. Association Analyses
The pedigrees of the dairy cows detected in the present study were traced back for three generations to create a genetic matrix. Based on the estimated matrices, associations between genotypes and/or haplotype combinations and the five milk production traits were evaluated by the mixed model in SAS 9.1.2 (University of Notre Dame, Notre Dame, IN, USA). A linear mixed regression model was shown as follows:
where y is the EBVs value of the five milk production traits, and μ is the overall mean for the values. B is the regression coefficient, x is a design vector of the SNP genotype or haplotype combination, a is the vector of residual polygenetic effects, Z is a diagonal incidence (0 or 1) matrix that links each animal additive genetic value to its “phenotype”, and e is the vector of residual errors.
The additive (a), dominance (d), and allele substitution (α) effects were further evaluated as follows: a = (AA − BB)/2, d = AB − (AA + BB)/2, and α = a + d (q − p), where AB represents heterozygous genotype, AA and BB indicate the two homozygous genotypes, and p and q are the A and B frequencies, respectively (Falconer and Mackay, 1996). For both analyses above, the Bonferroni method was used according to the number of SNP or haplotype blocks tested. A p value < 0.05/N was considered significant; here, N is the SNP loci numbers or haplotype blocks tested in the analyses.
2.6. Construction of Recombinant Plasmid and Luciferase Assay
To directly detect the allele-specific effects of the SNP g.-128 C>T on the promoter activity, three luciferase fragments, which correspond to the DDIT3 promoter (−281 to +52, +1 represents transcription start site) were synthesized. Two restriction enzyme recognition sites, KpnI and BglII, were used at the 5’ and 3’ termini, respectively. The fragments were then cloned into the Luciferase Assay Vector pGL4.14 (Promega, Madison, WI, USA).
Then, these recombinant plasmids were co-transfected to HEK-293 cells. Transfection reactions were performed using 490 ng of plasmid DNAs and 10 ng of pRL-TK as an internal control, containing Lipofectamine 2000 (Invitrogen, CA, USA), with pGL4.14 vector as a negative control. After 48 h transfection, the luciferase activities were measured using a Dual-Luciferase Reporter Assay System (Promega).
2.7. siRNA Design, Synthesis, and Assessment
Three siRNAs’ sequences targeting DDIT3 mRNA and a non-specific siRNA were designed and synthesized by GenePharma (GenePharma, Suzhou, China) (Table 1). MAC-T bovine mammary epithelial cells were cultured in DMEM/F12 basal medium (Gibco Life Technologies, Grand Island, NY, USA) supplied with 10% FBS (Gibco) and maintained with 5% CO2 at 37 °C. When the cells were 60–80% confluent, 100 pmol of siRNA was transfected into cells in one well using 5 μL of LipofectamineTM 2000 (Invitrogen), and the cells were then incubated (37 °C, 5% CO2) for 48 h. Next, the total RNA of the MAC-T cells was extracted with a TRIzol reagent (Invitrogen), and a qRT-PCR analysis was explored to detect the DDIT3 expression in the cells transfected by siRNAs. Each of the siRNAs was detected in triplicate.

Table 1.
Associations of the two SNPs in the DDIT3 gene with the five traits (LSM ± SE).
2.8. Construction of shRNA Vectors and Transfection in MAC-T cells
Based on the qRT-PCR results, the siRNA DDIT3-287 was found to reduce the DDIT3 expression by at least 60%. Hence, the DNA sense and antisense sequences for this shRNA were synthesized, and restriction sites were used for connecting synthetic DNA fragments into the LV3 shRNA lentiviral expression vectors (GenePharma)—namely LV3-287. With the appropriate lentiviral expression vectors, the HEK-293T cells (GenePharma) were transiently transfected, then following packaging vectors pGag/Pol, pRev, and pVSV-G to obtain lentiviral particles.
The MAC-T cells were propagated and incubated in DMEM/F12, with 10% FBS (Gibco), and 1×Penicillin-Streptomycin (Gibco) at 37 °C and 5% CO2. One day before infection, the cells were seeded in T25 culture flasks and infected when reaching approximately 80% confluence. The 500 μL mixed solution was as follows: 50 μL of lentiviral particles (1 × 109 TU/ml) carrying shRNA for DDIT3-287, 25 μg of Polybrene (Sigma), and 450 μL of Opti-MEM I Reduced-Serum Medium (Gibco). After one day, the cells were cultured in fresh growth media without lentiviral particles for another two days.
2.9. RNA Sequencing
Three parallel samples (cells from 3 culture flasks) of the experimental group (DDIT3-287 shRNA) and control group (NC shRNA) were prepared, respectively. The integrity of the obtained RNA was confirmed using Agilent BioAnalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). A total of 5 μg of RNA with RIN value >= 7.0 was used to standardize between the RNA samples. Using the Kit NEBNext® Ultra™ RNA Library Prep Kit (NEB), a sequencing library was prepared. Then, clustering of the index-coded samples was carried out by the TruSeq PE Cluster Kit v3-cBot-HS (Illumina). After cluster generation, these paired-end reads were generated, and RNA-seq libraries were sequenced using an Illumina HiSeq 2000 platform. Meanwhile, FASTQ sequence files were generated by CASAVA ver.1.8.2 (Illumina), accompanied by the removal of failed reads.
2.10. Differential Expression Analysis and Pathway Enrichment Analysis
The mapping of reads to the reference genome (Bos Taurus ARS-UCD1.2) was performed, and a differential expression analysis of the data was then performed using the package Bowtie v0.12.8 and paired-end and TopHat v2.0.0. To make estimates and determine differentially expressed genes, DESeq2 relies on the negative binomial distribution by using a generalized linear regression model. The Log2 fold-change (L2FC) values, p-values, and q-values (p-adjusted) of the differentially expressed genes were obtained in the output data. A q-value of 0.05 was considered significant.
Gene Ontology (GO) enrichment was analyzed by the GOseq R package. The enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were evaluated by KOBAS software. The GO term and KEGG pathway analysis results with corrected p values < 0.05 were considered to be significantly enriched.
2.11. Validation of RNA-Seq Results by qPCR
Eight DEGs were randomly selected to validate the RNA sequencing data (Table S1 shows the primers that were used in sequencing). A qRT-PCR was carried out using a SYBR Green I Master Kit using a LightCycler 96 real-time PCR system (Roche Applied Science, Penzberg, Germany), and the program was 95 °C for 5 min, 40 cycles of 95 °C for 12 s, 58 °C for 12 s, 72 °C for 12 s, and 72 °C for 6 min. The relative expression level was normalized to the GAPDH and ACTB by the 2−ΔΔCt method.
2.12. Oil Red O Staining and TG Content Measurement
After washing with PBS, the cells in the chamber slides were then fixed with 4% paraformaldehyde for 30 min, incubated for 5 min with Oil Red O at room temperature, and then visualized using ECLIPSE Ti-S light microscopy (Nikon Corporation, Tokyo, Japan). Additionally, the TG content was determined by a triglyceride detection kit (Sangon Biotech Co., Ltd., Shanghai, China).
2.13. Calcein-AM/Propidium Iodide (PI) Staining
After different stimulation, the MAC-T cells were then collected by trypsinization, centrifuged at 1500× g for 2 min at room temperature to collect the cell pellet, and washed once with 1× Assay Buffer. Then, the cells were mixed with 1× assay buffer and stained with the mixture by 2 μM of calcein-AM and 4.5 μM of PI per well for 15 min at room temperature. The images of the cells were collected immediately and analyzed using a Nikon eclipse Ti-S fluorescence microscope.
3. Results
3.1. SNP Identification and Its Associations with the Five Milk Traits
Through resequencing of the whole coding and 5’ regulatory regions, two SNPs were discovered. These two variants, g.-1194 C>T and g.-128 C>T, were in the 5’ regulatory regions (Figure 1a,b). Chi-squared testing showed that the SNPs of the DDIT3 gene were in Hardy–Weinberg equilibrium (p > 0.05). The allele frequencies and locations of the SNPs are shown in Table S2.
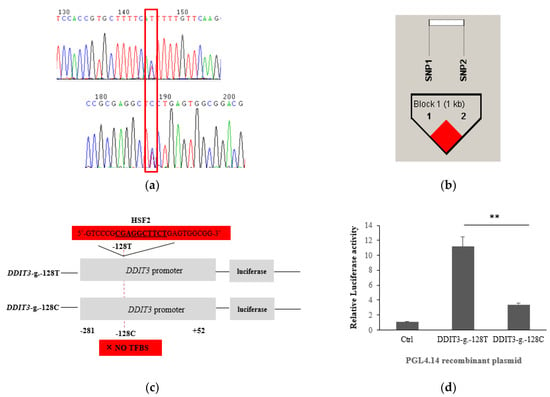
Figure 1.
The variants of DDIT3 and luciferase activity analysis in HEK293 cells. Note: (a) The variants g.-1194 C>T and g.-128 C>T in the 5’ regulatory regions. (b) The haplotype block and LD pattern formed by g.-1194 C>T and g.-128 C>T in the DDIT3; the darker shading indicates higher LD. The length of the block is provided in kilobases (kb), and pairwise linkage disequilibrium (D’) is given for each SNP combination. (c) TFBS modified from TFSEARCH software output; underlined nucleotides denote the TFBS sequences. (d) Luciferase activity analysis; the value was shown as the mean ± SEM. p values are from a t-test (two-tailed). ** p < 0.01.
The results of association between the EBVs of the five milk production traits and the two identified SNPs are shown in Table 1. After applying the Bonferroni correction for multiple t-testing, both the SNPs, g.-1194 C>T and g.-128 C>T, were significantly associated with MY (p = 0.0063), FY (p = 0.0001) and PY (p = 0.0216), and these two SNPs showed the same results. Correspondingly, significant dominant effects, additive effects, and allele substitution effects were observed (Table S3). On the other hand, one haplotype block from the HAPLOVIEW program, composed of g.-1194 C>T and g.-128 C>T, was observed in Figure 1a.
3.2. Prediction of TFBSs and Its Effect on Transcriptional Activity
The online software TFSEARCH was conducted to predict the putative differential TFBSs of the two regulatory SNPs, g.-1194 C>T and g.-128 C>T. It was shown that the allele of T in g.-128 C>T generated a putative TFBS for the transcription factors HSF2 (Heat shock factor protein 2), while that was abolished upon its substitution by the C allele (Figure 1c). However, there were no significant differences between the allele of T or C for the SNP g.-1194 C>T. Meanwhile, as shown in Figure 1d, the allele of T showed 68% higher luciferase activity compared with the allele of C (p < 0.01). Therefore, g.-128 C>T mutation can be considered as a functional SNP for gene expression via the alteration of transcription factor binding sites.
3.3. Interference Efficiency of Designed siRNAs in Bovine MAC-T Cells
A total of three siRNAs were designed against DDIT3 mRNA (Table 2; Figure S1). With the qRT-PCR assay, only siRNA D287 was found to decrease the mRNA expression of the DDIT3 gene in the MAC-T cells compared with the controls (p < 0.05), while D82 and D394 siRNAs did not have an obvious effect (p > 0.05) (Figure 2A). Of them, D287 was chosen for further RNAi experiments, which reduced the DDIT3 expression by about 50%. The shRNA lentiviral vector for D287, LV3-287, was constructed and transfected into the MAC-T cells. Green fluorescence signals of GFP were seen after 72 h of culturing by transient transfection of shRNA virus particles. As shown in Figure 2B, the surviving cells simultaneously expressed GFP and shRNA. Correspondingly, the mRNA levels of the DDIT3 gene in the knockdown lines were found to decrease by about 50% compared with the NC group using the qRT-PCR (p < 0.01).

Table 2.
Sequences and positions of 3 siRNAs against DDIT3 gene.
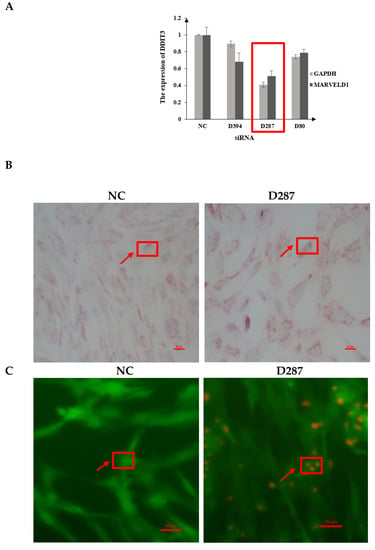
Figure 2.
shRNA interference of DDIT3 and its effect on MAC-T cells. Note: (A) The shRNA interference effects of LV3-DDIT3-D287 in MAC-T cells. (B) Lipid accumulation of NC and LV3-DDIT3-D287 groups in MAC-T cells by Oil Red O staining; NC-versus Ctrl group and D287-LV3-DDIT3-D287 group. (C) Calcein-AM/PI staining assays.
3.4. DEGs and Functional Annotation
In total, 3838 differentially expressed genes were identified (Table S4). Of these, 1940 genes were down-regulated and 1898 were genes up-regulated. The volcano plot of the DEGs is displayed in Figure S2. Moreover, the top 10 genes associated with the regulation of fat metabolism are shown in Table 3. Of them, IL6 was the most significantly up-regulated gene (Log2fold change = 4.22). The 3838 common DEGs were then processed by GO and KEGG enrichment analysis. We found that the expression pattern significantly changed the genes involved in cellular senescence, cell cycle and death, DNA replication and lysosome, etc. (Table S5).

Table 3.
Top 10 differentially expressed genes between knockdown and control groups.
Meanwhile, these results indicated that not only the regulation of autophagy, cellular senescence, and cell cycle, but also those involved in lipid metabolic signaling pathways could be regulated by DDIT3, for their role in cell growth, proliferation, and differentiation.
3.5. DDIT3 Knocking down Caused Lipid Accumulation and Apoptosis in Bovine MAC-T Cells
The differentiation of MAC-T cells was detected using Oil Red O staining and triglycerides content assay. It showed that the number and size of the lipid droplets in the DDIT3 knock-down group were significantly increased compared with the NC group (Figure 2B). Furthermore, the concentration of TG was significantly lower in the DDIT3 knock-down group than that of the control group. We then determined the role of DDIT3 in cell viability and apoptosis. Calcein-AM/propidium iodide (PI) staining showed that the transfection of DDIT3 siRNA could induce apoptosis and increase the number of PI-positive cells (Figure 2C). DDIT3 silencing by D287 led to increased expression of inflammatory markers.
3.6. The Expression Alteration of the Key Genes
Here, we first detected the expression of these genes (IL-6, IL6R, IL1B, IL7R, IL1RL2, IL1A, STAT1-5, MYC, IGFBP4, and IGFBP5), which were acquired by our RNA-seq and pathway analysis. Compared with the control group, the mRNA expression levels of these genes in the DDIT3 knock-down group altered significantly. As shown in Figure 3A,C,D, DDIT3 knocking down can markedly increase the mRNA expressions of IL-6, IL6R, IL1B, IL7R, MYC, and IGFBP4 (p < 0.01), especially for IL-6, which was known as pro-inflammatory cytokine. Moreover, the expression levels of STAT1-5, IL1RL2, IL1A, and IGFBP5 mRNA were lower in the DDIT3 knock-down group than those in the control group (p < 0.01) (Figure 3A,B,E). All the expressions of these detected genes were consistent with our RNA-seq data. Altogether, these results indicated that these key genes may play a critical role in regulating milk composition traits, which were induced by knocking down DDIT3. Then, based on the mRNA expression, we proposed that DDIT3 might modulate the lipid metabolism and apoptosis in dairy cattle through regulating the expression of IL6. Thus, these results demonstrated that DDIT3 knocking down caused lipid accumulation and apoptosis in bovine MAC-T cells.
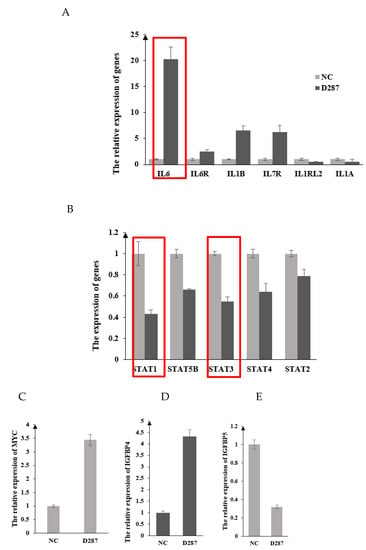
Figure 3.
Effects of DDIT3 knocking down on the mRNA expression of genes regulating apoptosis in MAC-T cells. Note: (A). The mRNA expression of genes regulating inflammatory response. (B). The expression of STAT1-5. (C–E). The mRNA expression of MYC, IGFBP4, and IGFBP5, respectively.
4. Discussion
In the present study, we confirmed that the DDIT3 gene was significantly associated with five milk production traits. DDIT3 was initially identified in our previous RNA-seq, where it was found to be significantly differentially expressed in the high and low PP and FP of lactating Holstein cows []. Genetic variations in key promoter regions may cause significant potential phenotype diversity []. In the present study, we found that the SNPs g.-1194 C>T and g.-128 C>T, which are located in the promoter region, were significantly associated with MY, FY, and PY. Cows with the TT genotype had lower MY, FY, and PY than those with the TT genotype. In the association analyses, the EBVs of daughters were used as phenotypic observations. By using phenotypic values for association analyses, Li et al. (2019) found that, in the first lactation, the SNP g.56284880C>T (which was g.-128 C>T in our study) was significantly associated with milk MY, FY, and PY; likewise, in the second lactation, it was significantly associated with milk MY and PY. These results were consistent with our current study. Hence, the findings for the association analyses based on the two different variables (phenotypic values and EBVs) basically overlapped. Thus, DDIT3 was considered to be a major gene influencing milk production traits.
Meanwhile, the promoter activity analysis clearly demonstrated that the allele T of g.-128 C>T of the DDIT3 gene was influenced by the increased promoter activity by binding the HSF2 factor, while allele C did not, indicating that the HSF2 (heat shock factor) transcription factor may up-regulate the expression of DDIT3. HSF2, as a transcription factor, participated in the expression of heat shock genes by interacting directly with HSF1 or HSF4 [,,]. Additionally, HSF2 plays important roles in regulating the HSF1-mediated stress response []. Numerous studies have shown that the SNPs in the promoter region are associated with the changes of gene expression by altering putative transcription factor-binding sites []. Considering the significant association effects of g.-128 C>T on the milk fat and protein production traits, it is possible that this SNP regulates the DDIT3 expression by changing the binding status of the transcription factor, HSF2, to affect the formation of milk and protein traits. Additionally, there was no TFBS for the loci g.-1194 C>T; the significant association of g.-1194 C>T with MY, FY, or PY may be due to the very strong LD between g.-1194 C>T and g.-128 C>T (D’ = 1.00). Therefore, the locus g.-128 C>T is directly responsible for the DDIT3 expression. Based on these findings, HSF2 can be used for marker-assisted selection to modulate the expression of the DDIT3 gene in dairy cattle to improve milk production.
It has been widely reported that DDIT3 affects ER stress and induces cell apoptosis or autophagy [,,], but its relationship with milk production traits remains unclear in dairy cattle. In the present study, for the loci g.-128 C>T, cows with the TT genotype had lower MY, FY, and PY than those with the TT genotype. Therefore, DDIT3 negatively regulated milk production traits, which were all consistent with our previous studies [,]. Furthermore, the concentration of TG in the DDIT3 knock-down group showed significantly higher content than that of the NC group. In addition, DDIT3-knockdown induced an increased number of PI-positive cells and could induce apoptosis by calcein-AM/ PI staining in the MAC-T cell line. This is the first report on DDIT3 inhibiting lipid accumulation and inducing apoptosis in the bovine MAC-T cell line.
To further exploit the effects of DDIT3 on the apoptosis of the bovine MAC-T cell line, we knocked down DDIT3 in bovine MAC-T cells and found that interleukin-6 (IL6) was regulated by DDIT3. It has been reported that IL6 regulates inflammatory responses, can be produced for a short period of time after the body is stimulated by inflammation and independently of stimulation by tumor necrosis factor-alpha (TNFα) and accompanied by the increase in other inflammatory substances [,]. In addition, IL6 mediates cellular hypertrophy through the activation of the JAK/STAT pathway in different types of cells [,]. In our present study, DDIT3 knockdown promoted the expression of IL6, and induced apoptosis of the bovine MAC-T cell line, indicating that the regulatory effects of DDIT3 on the vitality of MAC-T cells is mediated by IL6. As is known, the JAK/STAT pathway is stimulated in the early stage of diabetic nephropathy [,,]. Our present results only showed that IL6 could further stimulate STAT3-related pathways, resulting in an aggravating effect on inflammatory responses, whereas JAK2 was not induced significantly. The different results probably stemmed from the differences between the in vivo and in vitro studies, as well as the heterogeneity of the cells used in the study. Collectively, these findings and our present results suggest that the DDIT3/IL6/STAT3 signaling pathway may be involved in affecting milk production traits. Thus, further studies of animal models with IL6/STAT3 double deficiency will be required to provide additional evidence for this possibility.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/agriculture13010117/s1, Table S1. Primers used for pooled DNA sequencing for the DDIT3 gene; Table S2. Genotypic and allelic frequencies of the two SNPs in the DDIT3 gene; Table S3. Genetic effects of g.-1194 C>T and g.-128 C>T on milk production traits in Chinese Holsteins; Table S4. DEGs within two different comparison groups; Table S5. Significantly enriched KEGG terms of DEGs; Figure S1. Sequencing of recombinant plasmid; Figure S2. DEGs among the DDIT3-KDvsNC groups Note: FC presented in this figure is fold change.
Author Contributions
Conceptualization, X.C. and S.Y.; methodology, C.L.and F.Z.; software, X.C., C.L., Z.W. and H.M.; validation, X.C., C.L., Z.W. and H.M.; formal analysis, X.C., and C.L.; investigation, F.Z.; resources, C.W. and S.Y.; data curation, F.Z and Y.L..; writing—original draft preparation, X.C. and S.Y.; writing—review and editing, X.C., Y.L., C.W. and S.Y.; visualization, S.Y.; supervision, C.W. and S.Y.; project administration, C.W. and S.Y.; funding acquisition, C.W. and S.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Science Foundation Project of Anhui (1908085MC63), the Beijing Research and Technology Program (D121100003312001), and the Program of Introducing Talents of Discipline to Universities under grant (D21004).
Acknowledgments
The authors also appreciate the kind help from the official Dairy Data Center of China in providing the official EBVs data.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Caroli, A.M.; Chessa, S.; Erhardt, G.J. Invited review: Milk protein polymorphisms in cattle: Effect on animal breeding and human nutrition. J. Dairy Sci. 2009, 92, 5335–5352. [Google Scholar] [CrossRef] [PubMed]
- Bovenhuis, H.; Van Arendonk, J.A.M.; Korver, S. Associations between milk protein polymorphisms and milk production traits. J. Dairy Sci. 1992, 75, 2549–2559. [Google Scholar] [CrossRef] [PubMed]
- McCabe, M.; Waters, S.; Morris, D.; Kenny, D.; Lynn, D.; Creevey, C. RNA-seq analysis of differential gene expression in liver from lactating dairy cows divergent in negative energy balance. BMC Genom. 2012, 13, 193. [Google Scholar] [CrossRef] [PubMed]
- Wickramasinghe, S.; Rincon, G.; Islas-Trejo, A.; Medrano, J.F. Transcriptional profiling of bovine milk using RNA sequencing. BMC Genom. 2012, 13, 45. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Hou, Y.; Yang, S.; Xie, Y.; Zhang, S.; Zhang, Y.; Zhang, Q.; Lu, X.; Liu, G.E.; Sun, D. Transcriptional profiling of mammary gland in Holstein cows with extremely different milk protein and fat percentage using RNA sequencing. BMC Genom. 2014, 15, 226. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.B.; Wiggans, G.R.; Ma, L.; Sonstegard, T.S.; Lawlor, T.J., Jr.; Crooker, B.A.; Van Tassell, C.P.; Yang, J.; Wang, S.; Matukumalli, L.K.; et al. Genome-wide association analysis of thirty one production, health, reproduction and body conformation traits in contemporary U.S. Holstein cows. BMC Genom. 2011, 12, 408. [Google Scholar] [CrossRef]
- Bagnato, A.; Schiavini, F.; Rossoni, A.; Maltecca, C.; Dolezal, M.; Medugorac, I.; Sölkner, J.; Russo, V.; Fontanesi, L.; Friedmann, A.; et al. Quantitative trait loci affecting milk yield and protein percentage in a three-country Brown Swiss population. J. Dairy Sci. 2008, 91, 767–783. [Google Scholar] [CrossRef]
- Marola, O.J.; Syc-Mazurek, S.B.; Libby, R.T. DDIT3 (CHOP) contributes to retinal ganglion cell somal loss but not axonal degeneration in DBA/2J mice. Cell Death Discov. 2019, 5, 140. [Google Scholar] [CrossRef]
- Nashine, S.; Liu, Y.; Kim, B.J.; Clark, A.F.; Pang, I.H. Role of C/EBP homologous protein in retinal ganglion cell death after ischemia/reperfusion injury. Investig. Ophthalmol. Vis. Sci. 2014, 56, 221–231. [Google Scholar] [CrossRef]
- Silva, R.M.; Ries, V.; Oo, T.F.; Yarygina, O.; Jackson-Lewis, V.; Ryu, E.J.; Lu, P.D.; Marciniak, S.J.; Ron, D.; Przedborski, S.; et al. CHOP/GADD153 is a mediator of apoptotic death in substantia nigra dopamine neurons in an in vivo neurotoxin model of parkinsonism. J. Neurochem. 2005, 95, 974–986. [Google Scholar] [CrossRef]
- Liao, Y.; Fung, T.S.; Huang, M.; Fang, S.G.; Zhong, Y.; Liu, D.X. Upregulation of CHOP/GADD153 during coronavirus infectious bronchitis virus infection modulates apoptosis by restricting activation of the extracellular signal-regulated kinase pathway. J. Virol. 2013, 87, 8124–8134. [Google Scholar] [CrossRef] [PubMed]
- Abreu, M.M.; Sealy, L. The C/EBPβ isoform, liver-inhibitory protein (LIP), induces autophagy in breast cancer cell lines. Exp. Cell Res. 2010, 316, 3227–3238. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, J.; Wang, L.; Ho, C.T.; Zhang, K.; Liu, Q.; Zhao, H. Garcinol from Garcinia indica Downregulates Cancer Stem-like Cell Biomarker ALDH1A1 in Nonsmall Cell Lung Cancer A549 Cells through DDIT3 Activation. J. Agric. Food Chem. 2017, 65, 3675–3683. [Google Scholar] [CrossRef] [PubMed]
- Barone, M.V.; Crozat, A.; Tabaee, A.; Philipson, L.; Ron, D. CHOP (GADD153) and its oncogenic variant, TLS-CHOP, have opposing effects on the induction of G1/S arrest. Genes Dev. 1994, 8, 453–464. [Google Scholar] [CrossRef]
- Wang, X.; Tomso, D.J.; Liu, X.; Bell, D.A. Single nucleotide polymorphism in transcriptional regulatory regions and expression of environmentally responsive genes. Toxicol. Appl. Pharmacol. 2005, 207, 84–90. [Google Scholar] [CrossRef]
- He, H.; Soncin, F.; Grammatikakis, N.; Li, Y.; Siganou, A.; Gong, J.; Brown, S.A.; Kingston, R.E.; Calderwood, S.K. Elevated expression of heat shock factor (HSF) 2A stimulates HSF1-induced transcription during stress. J. Biol. Chem. 2003, 278, 35465–35475. [Google Scholar] [CrossRef]
- Ostling, P.; Björk, J.K.; Roos-Mattjus, P.; Mezger, V.; Sistonen, L. Heat shock factor 2 (HSF2) contributes to inducible expression of hsp genes through interplay with HSF1. J. Biol. Chem. 2007, 282, 7077–7086. [Google Scholar] [CrossRef]
- Sandqvist, A.; Björk, J.K.; Akerfelt, M.; Chitikova, Z.; Grichine, A.; Vourc’h, C.; Jolly, C.; Salminen, T.A.; Nymalm, Y.; Sistonen, L. Heterotrimerization of heat-shock factors 1 and 2 provides a transcriptional switch in response to distinct stimuli. Mol. Biol. Cell 2009, 20, 1340–1347. [Google Scholar] [CrossRef]
- Yang, L.N.; Ning, Z.Y.; Wang, L.; Yan, X.; Meng, Z.Q. HSF2 regulates aerobic glycolysis by suppression of FBP1 in hepatocellular carcinoma. Am. J. Cancer Res. 2019, 9, 1607–1621. [Google Scholar]
- Zi, C.; Wu, Z.; Wang, J.; Huo, Y.; Zhu, G.; Wu, S.; Bao, W. Transcriptional activity of the FUT1 gene promoter region in pigs. Int. J. Mol. Sci. 2013, 14, 24126–24134. [Google Scholar] [CrossRef]
- Posey, K.L.; Coustry, F.; Veerisetty, A.C.; Liu, P.; Alcorn, J.L.; Hecht, J.T. Chop (Ddit3) is essential for D469del-COMP retention and cell death in chondrocytes in an inducible transgenic mouse model of pseudoachondroplasia. Am. J. Pathol. 2012, 180, 727–737. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Sun, H.; Song, F.; Fu, D.; Wang, J. DDIT3 overexpression increases odontoblastic potential of human dental pulp cells. Cell Prolif. 2014, 47, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Hou, P.; Pan, W.; He, W.; He, D.C.; Wang, H.; He, H. DDIT3 Targets Innate Immunity via the DDIT3-OTUD1-MAVS Pathway To Promote Bovine Viral Diarrhea Virus Replication. J. Virol. 2021, 95, e02351-20. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Han, B.; Liu, L.; Zhao, F.; Liang, W.; Jiang, J.; Yang, Y.; Ma, Z.; Sun, D. Genetic association of DDIT3, RPL23A, SESN2 and NR4A1 genes with milk yield and composition in dairy cattle. Anim. Genet. 2019, 50, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Bobbo, V.C.; Engel, D.F.; Jara, C.P.; Mendes, N.F.; Haddad-Tovolli, R.; Prado, T.P.; Sidarta-Oliveira, D.; Morari, J.; Velloso, L.A.; Araujo, E.P. Interleukin-6 actions in the hypothalamus protects against obesity and is involved in the regulation of neurogenesis. J. Neuroinflam. 2021, 18, 192. [Google Scholar] [CrossRef]
- Jo, H.A.; Kim, J.Y.; Yang, S.H.; Han, S.S.; Joo, K.W.; Kim, Y.S.; Kim, D.K. The role of local IL6/JAK2/STAT3 signaling in high glucose-induced podocyte hypertrophy. Kidney Res. Clin. Pract. 2016, 35, 212–218. [Google Scholar] [CrossRef]
- Serrano, A.L.; Baeza-Raja, B.; Perdiguero, E.; Jardí, M.; Muñoz-Cánoves, P. Interleukin-6 is an essential regulator of satellite cell-mediated skeletal muscle hypertrophy. Cell Metab. 2008, 7, 33–44. [Google Scholar] [CrossRef]
- Begue, G.; Douillard, A.; Galbes, O.; Rossano, B.; Vernus, B.; Candau, R.; Py, G. Early activation of rat skeletal muscle IL-6/STAT1/STAT3 dependent gene expression in resistance exercise linked to hypertrophy. PloS ONE 2013, 8, e57141. [Google Scholar] [CrossRef]
- Mir, S.A.; Chatterjee, A.; Mitra, A.; Pathak, K.; Mahata, S.K.; Sarkar, S. Inhibition of signal transducer and activator of transcription 3 (STAT3) attenuates interleukin-6 (IL-6)-induced collagen synthesis and resultant hypertrophy in rat heart. J. Biol. Chem. 2012, 287, 2666–2677. [Google Scholar] [CrossRef]
- Ropelle, E.R.; Flores, M.B.; Cintra, D.E.; Rocha, G.Z.; Pauli, J.R.; Morari, J.; de Souza, C.T.; Moraes, J.C.; Prada, P.O.; Guadagnini, D.; et al. IL-6 and IL-10 anti-inflammatory activity links exercise to hypothalamic insulin and leptin sensitivity through IKKβ and ER stress inhibition. PLoS Biol. 2010, 8, e1000465. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).