Abstract
Seed germination is a key contributing factor to the yield of direct seeding cultivation in rice. Unraveling the genetic architecture underlying rice seed germination is pivotal for breeding elite direct-seeded rice varieties. However, only a limited number of genes regulating seed germination have been characterized in rice. In this study, we implemented a genome-wide association study (GWAS) to dissect the genetic structure of seed germination by using 131 Japonica rice accessions. We identified six stable loci (qGR1.1, qGR2.1, qGR3.1, qGR7.1, qGR8.1 and qGR9) associated with seed germination in two consecutive years, all of which were co-localized with previously reported quantitative trait loci (QTLs). OsGA2ox5, encoding a gibberellin 2-oxidase, was identified as the most plausible candidate gene of the major locus qGR7.1. Knockout of OsGA2ox5 led to delayed seed germination and retarded seedling growth. A non-synonymous variant (Chr7-218,245) within the coding region of OsGA2ox5 might be closely associated with variation in seed germination among Japonica accessions. Low nucleotide diversity at the OsGA2ox5 locus in Japonica could be a result of selection during rice improvement. Taken together, our results provide an important foundation for elucidating the molecular mechanism underlying seed germination and genetic improvement of rice seed vigor in the future.
1. Introduction
Seed germination represents a crucial event in the seed plant life cycle during which the emerging seedling relies upon reserves stored in the seed for continuous growth [1,2,3]. It is generally considered that seeds with low vigor germinate slowly and produce weak seedlings, whereas high-vigor seeds produce robust seedlings that are more tolerant of diverse environmental stresses in the field [4]. In recent years, the direct-seeding model has become increasingly popular in Asian countries due to its numerous advantages, such as low cultivation cost and simplicity of operation [5]. However, erratic seed germination and poor post-germination seedling growth are severe constraints limiting the production yield in the direct-seeding cultivation model [6]. Hence, identification of novel regulatory genes that regulate seed germination will facilitate the genetic improvement of rice by molecular breeding.
Seed germination is a complex trait that is controlled by numerous QTLs. Biparental linkage mapping strategy has been commonly employed to identify QTLs/genomic regions associated with genetic variation of seed germination/vigor in rice. For example, the QTL qLTG3-1 was shown to have a function on low-temperature germinability regulation by weakening of seed tissues during germination [7]. The QTL Seed dormancy 4 (Sdr4), encoding a novel protein, contributed substantially to differences in pre-harvest sprouting resistance between Japonica and Indica varieties [8]. The QTL qSD1-2 associated with primary dormancy and plant height was detected in different mapping populations and was identified as a gibberellin synthesis gene OsGA20ox2 [9]. Although biparental linkage mapping has proven useful in identifying candidate genes, this approach offered limited genomic resolution and allelic diversity [10]. More recently, GWAS approach has become feasible to interrogate causal SNPs (single nucleotide polymorphisms) linked to rice seed germination using populations of natural accessions. For example, three candidate genes OsOMT (encoding a 2-oxoglutarate/malate translocator), OsCDP3.10 (encoding a cupin domain protein) and OsPK5 (encoding a pyruvate kinase) associated with seed germination and vigor were identified by the GWAS approach and were validated by a knockout experiment using the clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 system [11,12,13]. An updated GWAS model with a genotype-by-environment interaction was employed to explore candidate genes responsible for seed germination under two different immersion temperatures, and one causal gene encoding a 14-3-3 protein, GF14h, was shown to mediate temperature-dependent germination [14]. Although some seed germination QTLs and a limited number of genes regulating seed germination have been characterized in rice, more diverse natural alleles for seed germination from natural rice populations need to be discovered.
Gibberellins (GAs) are a group of complex tetracyclic diterpenoid compounds, members of which function as essential phytohormones regulating numerous aspects of plant growth and development, including seed germination [15]. GA is synthesized from trans-geranylgeranyl diphosphate (GGDP), which is converted to GA12 (the common precursor of all GAs) by the successive action of four enzymes, including ent-copalyl diphosphate synthase (CPS), ent-kaurene synthase (KS), ent-kaurene oxidase (KO) and ent-kaurenoic acid oxidase (KAO) [16]. Bioactive GAs are synthesized from GA12 by GA 20-oxidase (GA20ox) and GA 3-oxidase (GA3ox), and the deactivation reaction for GAs is catalyzed by GA 2-oxidase (GA2ox) [17]. GA pathway has been a subject of intense investigation for many years, and the genes encoding key enzymes related to GA biosynthesis and catabolism have been characterized in many plants, such as rice [18]. However, allelic variations of GA pathway-related genes associated with seed germination have rarely been reported in natural rice populations.
In this study, we performed a GWAS on seed germination trait to identify QTLs using 131 Japonica rice accessions. A total of 22 loci associated with early seed germination were detected, and six loci were stably expressed in two consecutive years. Further in-depth analysis showed that a candidate gene belonging to GA2ox family, OsGA2ox5, was significantly associated with divergence in rice seed germination and validated through the CRISPR/Cas9 approach. These results will increase the current understanding of the genetic architecture of seed germination in Japonica rice and provide a vital theoretical basis for effectively improving rice seed vigor through molecular breeding in the future.
2. Materials and Methods
2.1. Plant Materials
Seeds of the 131 Japonica inbred rice accessions used in the present study was obtained from Rice Diversity Panel (RDP1) [19]. These accessions consisted of both landraces and elite cultivars, which were further divided into 62 TRJ (Tropical Japonica), 62 TEJ (Temperate Japonica), and 7 ARO (Aromatic) accessions (Table S1). The seeds harvested in 2016 and 2017 were used for germination tests, respectively, as descirbed by the previous study [13]. The osga2ox5 mutant was generated in the Japonica variety Zhonghua-11 background using the CRISPR/Cas9 system [20] and planted in the field at the Zhongkai University of Agriculture and Engineering (Guangdong Province, China). The mature seeds were harvested at 35 days after flowering, and then air-dried for three days (approximately 12 to 13% moisture content of the seeds) and stored for one month at room temperature before the germination assay.
2.2. Evaluation of Seed Germination
Fifty healthy seeds of each replicate were surface-sterilized with 0.5% (v/v) sodium hypochlorite (NaClO) solution for 10 min and then thoroughly washed with sterile deionized water for three times. Seeds were placed on the 9-cm-diameter Petri dishes containing two layers of filter paper soaked with 10 mL distilled water, and the Petri dishes were incubated in a 25°C constant temperature incubator for 7 days in a 12-h light, 12-h dark cycle. Germination was defined as radicle emergence through the hull by ≥2 mm, and a seedling was recorded as established when the root length reached or exceeded the seed length and the shoot length reached or exceeded half of the seed length [5]. The germinability of the seeds was assessed every day for 7 days and the germination rate and seedling percentage were calculated. The 4-day-old seedlings (15 seedlings for each replicate) were used for shoot length and root length measurement. Three biological replicates were included in this study. Mean phenotype values were used for subsequent association analysis.
2.3. GWAS
The high-SNP-density dataset (700,000 SNPs) used in this study has been published previously [21]. SNPs with a minor allele frequency (MAF) lower than 5% and missing ratio greater than 25% were eliminated [22] using TASSEL 5.2.40 software [23], resulting in 225,800 SNPs for association analysis (Figure S1). GWAS was implemented with SUPER (Settlement of MLM Under Progressively Exclusive Relationship) method using the R package GAPIT [24]. A significance threshold of p ≤ 10−4 (i.e., −log10 p ≥ 4) was used to identify significant associations according to the method used in the previous study [21]. The Manhattan plots for the GWAS results were produced by using the qqman implemented in the R package. A region containing significant SNP clusters (at least three consecutive SNPs) was defined as a single associated locus between any two significant SNPs within distances less than 170 kb, given linkage disequilibrium (LD) decay value of ~167 kb in Japonica population [25]. The most significant SNP (i.e., the highest -log10 p) in a single associated locus was chosen to represent lead SNP.
2.4. Identification of the Causal Gene for qGR7.1
The LD heatmap of qGR7.1 was created based on SNP data (between 0 to 500 kb on chromosome 7) using the LD heatmap implemented in the R package, and the LD block delineation was further determined by applying Haploview software, version 4.2 [26]. According to the previous study [22], the significant SNPs within the LD block of qGR7.1 were split into four functional groups based on the RiceVarMap v2.0 database (http://ricevarmap.ncpgr.cn/; accessed on 25 September 2021) [27]. Group I consisted of significant SNPs causing potentially functional amino acid changes (non-synonymous SNPs); Group II contained significant SNPs identified within the promoter or 5’ untranslated region (5’-UTR); Group III included significant SNPs located in coding sequence without changing amino acids (synonymous SNPs), or located in introns or 3’ untranslated regions (3’-UTR). Group IV included significant SNPs that were determined to be located in intergenic regions.
2.5. Population Genetics Analysis
The distribution frequencies of the key SNP allele at the OsGA2ox5 locus of each wild and Japonica rice subpopulation were collected from the RiceVarMap v2.0 database (http://ricevarmap.ncpgr.cn/; accessed on 20 November 2022) [27] and the ECOGEMS database (http://ecogems.ncpgr.cn/; accessed on 20 November 2022) [28]. To evaluate the evolution of OsGA2ox5, the genomic sequences of 118 accessions of O. rufipogon (wild rice), 150 varieties of Indica and 128 varieties of Japonica (including 41 TRJ and 87 TEJ) were obtained from previously published sequencing datasets [29,30], with a missing rate of ≤25%. Estimates of nucleotide diversity (π) and population fixation statistics (Fst) were determined using DnaSP software, version 6.12.03 [31]. Populations were considered to have strong differentiation if the Fst value is greater than 0.25 [32].
2.6. Data Analysis
Phenotype data and variance (ANOVA) were statistically analysed with Microsoft Excel 2017 software. The significant differences were assessed with Student’s t-test at the 5% and 1% probability levels. Broad-sense heritability was determined according to the previous method [33].
3. Results
3.1. Phenotypic Variation of Seed Germination in Japonica Rice
To assess the genetic diversity of rice seed germination, 131 Japonica rice accessions derived from RDP1 were selected for the phenotypic evaluation of their germination rate at three days after imbibition in 2016 and 2017 (Table S1). Statistical analysis of the phenotypic data measured across two years showed that germination rate ranged from 0 to 100%, and the coefficient of variations (CVs) were more than 0.5 (Table 1). The phenotypic values observed in two years showed a similar distribution pattern (Figure 1). The largest number of rice accessions fell within a range of germination rate from 90 to 100% (Figure 1). Two-way analysis of variance revealed that germination rate had a high heritability of more than 90%, and genotype-environment interaction (G × E) was significant (p < 0.001) (Table 1), indicating that genotype and environmental factors could significantly influence seed germination in Japonica. Moreover, 31 accessions had a high germination rate of more than 90% across two years (Table S1). Accordingly, these accessions may serve as good donor parents for improving seed vigor in breeding programs.

Table 1.
Descriptive statistics of germination rate at three days after imbibition in two years.
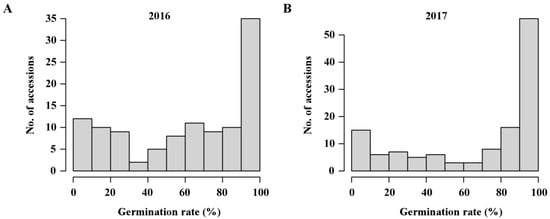
Figure 1.
Natural variation for seed germination in Japonica rice. The distribution of germination rate at three days after imbibition in 2016 (A) and 2017 (B).
3.2. Mining Loci for Seed Germination in Japonica Rice through GWAS
GWAS analysis was used to determine the genetic basis of seed germination in Japonica using SUPER method integrated in GAPIT for 225,800 SNPs (Figure S1). According to the previously described method [34], we defined a SNP peak as a locus that contained at least three consecutive SNPs associated with seed germination with −log10 p ≥ 4. By applying this approach, a total of 22 loci were identified, corresponding to 225 significant SNPs that associated with germination rate (Figure 2; Table 2, Table 3 and Table S2). Of them, six loci were commonly found in two consecutive years, including qGR1.1, qGR2.1, qGR3.1, qGR7.1, qGR8.1 and qGR9 (Figure 2; Table 2). The associated loci identified here were compared with the previously published QTLs, revealing that only one locus was newly identified in the present study and the other 21 loci were co-localized with previously described QTLs (Table S3). Interestingly, a great many significant SNPs were found in the candidate region of qGR7.1 in two consecutive years (Figure 2), suggesting that it is a stable major locus for seed germination in Japonica rice. Therefore, qGR7.1 was selected for further investigation.
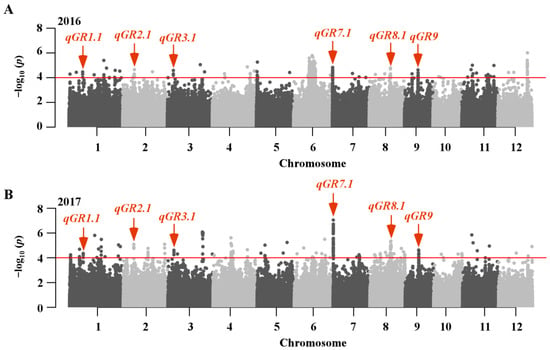
Figure 2.
GWAS of seed germination in Japonica rice. Manhattan plots for germination rate at three days after imbibition in 2016 (A) and 2017 (B). The horizontal red lines represent a statistical significance threshold of −log10 p = 4. The red arrows indicate that six stable loci were identified across two years.

Table 2.
Loci associated with germination rate in both two years.

Table 3.
Loci uniquely identified in 2016 or 2017.
3.3. OsGA2ox5 Is the Causal Gene of qGR7.1 for Seed Germination
To further narrow down the candidate region of qGR7.1, we conducted an LD block analysis surrounding the lead SNP (Chr7-192,333) using Haploview software. The candidate region was narrowed from the 456-kb region down to a 107.8-kb genomic region (Block 4: 132,222 to 240,062 bp) by using pairwise LD correlations (R2 ≥ 0.8) (Figure 3A,B). This block contained 15 annotated genes in the Nipponbare reference genome (http://rice.uga.edu/; accessed on 13 September 2021; Figure 3C). To further assess these candidate genes, 15 common significant SNPs in both years in this block were separated into four groups (I to IV) according to their locations [22]. We assigned two SNPs to group I, whereas we assigned five SNPs to group II, four to group III and four to group IV (Figure 3D). In group I, one non-synonymous SNP (Chr7-231,849) was annotated as retrotransposon protein (LOC_Os07g01360), while another one (Chr7-218,245) was annotated as gibberellin 2-beta-dioxygenase 7 (LOC_Os07g01340) and was identical to OsGA2ox5 (Figure 3D). Therefore, OsGA2ox5 was the most likely candidate gene for qGR7.1 and selected for further functional validation.
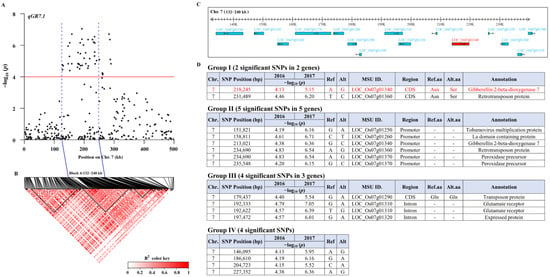
Figure 3.
Identification of candidate gene for qGR7.1. (A) Regional Manhattan plot surrounding the peak on chromosome 7 (0–500 kb); (B) LD heatmap for qGR7.1; (C) Fifteen annotated genes within the LD block (132–240 kb) of qGR7.1; (D) Fifteen common significant SNPs across two years within the LD block of qGR7.1 were divided into four functional groups. As a measure of LD, the color intensity of the box on the LD plot corresponds with the extent of R2 according to the legend. From an LD analysis, 11 blocks (inverted black triangles) were identified on qGR7.1. The lead SNP was located within an approximately 108-kb block (Block 4: 132,222 to 240,062 bp). The rectangles show the position of 15 annotated genes within the LD block (132–240 kb) of qGR7.1. The black arrow direction represents the orientation of transcription. The red rectangle indicates the causal gene (LOC_Os07g01340; OsGA2ox5) of qGR7.1. The non-synonymous SNP (Chr7-218,245) shown in red was located in the coding region of OsGA2ox5.
3.4. Functional Validation of OsGA2ox5 through CRISPR-Cas9 System
To better characterize the role of OsGA2ox5 in seed germination, the CRISPR/Cas9 system of genome editing was used to produce mutant in the Japonica variety Zhonghua-11. osga2ox5 contained a “G” deletion in exon of OsGA2ox5 (Figure 4A), which resulted in a frame-shift and thus a premature translation termination of OsGA2ox5 (Figure 4B). Homozygous osga2ox5 mutant was self-pollinated, and the progeny were selected to evaluate seed germination-related traits, including germination rate, seedling percentage, shoot length and root length. The results showed that the germination rates at day 2 to day 3 and seedling percentages at day 3 to day 4 of osga2ox5 mutant were significantly reduced compared with those in the wild type (WT) during early germination (Figure 4C,D,G). We also noted a significant reduction in shoot length and root length in osga2ox5 mutant relative to WT in the fourth day of germination (Figure 4E,F). These results indicated that the disruption of OsGA2ox5 led to low germination speed and seedling growth during seed germination.
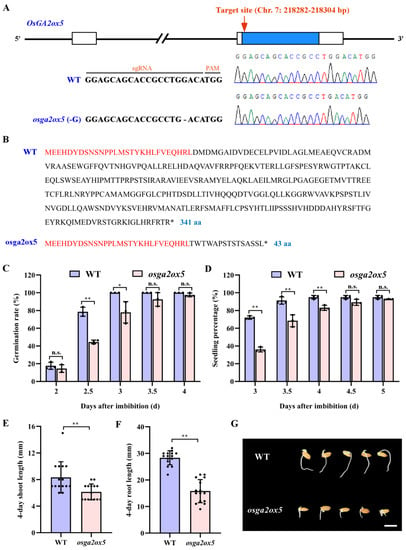
Figure 4.
Functional validation of OsGA2ox5. (A) Generation of osga2ox5 mutant using CRISPR/Cas9 system; (B) Comparison of inferred amino acid sequences between the WT and osga2ox5 mutant; (C) Comparison of germination rate between the WT and osga2ox5 mutant from 2 to 4 days after imbibition; (D) Comparison of seedling percentages between the WT and osga2ox5 mutant from 3 to 5 days after imbibition; Comparison of shoot length (E) and root length (F) between the WT and osga2ox5 mutant at four days after imbibition; (G) Germination phenotype of the WT and osga2ox5 mutant at three days after imbibition. Scale bars = 1 cm. Blue rectangle indicates exon; white rectangle indicates UTR. Identical amino acid sequence is highlighted in red. Each black dot represents a technical replicate. Statistical differences between the WT and osga2ox5 mutant were analysed using Student’s t-test. * and ** indicate a significant difference between the WT and osga2ox5 mutant at p < 0.05 and p < 0.01, respectively. n.s., non-statistically significant difference.
3.5. Population Genetic Analysis of OsGA2ox5 in Wild Rice and Cultivated Rice
As mentioned above, one non-synonymous SNP (Chr7-218,245) in OsGA2ox5 was significantly associated with germination rate in both years (Figure 3D), and we therefore further analyzed the effect of this key SNP on seed germination. The two allelic groups separated based on the base type of this SNP displayed statistically significant differences in seed germination across two years, with the G-type showing higher germination rate than the A-type (Figure 5A), suggesting that the G-type was potentially a favorable allele of OsGA2ox5 for improving rice seed vigor. To characterize the distribution of A-type and G-type in rice subpopulations, the SNP details were retrieved from the RiceVarMap database and the ECOGEMS database. We found a higher proportion of the allelic frequency of the G-type in Japonica (81.41%) accessions than in O. rufipogon (70.32%) (Figure 5B). These data suggested that the favorable allele of OsGA2ox5 gene was likely selected during rice improvement. To confirm this, average nucleotide diversity was estimated in OsGA2ox5 gene and its flanking regions (10 kb) based on sequencing data reported previously [29,30]. Compared with O. rufipogon, average π value in the genomic region harboring OsGA2ox5 locus was significantly reduced in Japonica, TEJ and TRJ (Figure 5C), suggesting that OsGA2ox5 has undergone selection during rice improvement. We also calculated population differentiation statistics for OsGA2ox5 and its flanking regions (10 kb) between different subpopulations. The results showed that pairwise measurements of Fst ranged from 0.13 to 0.54 in the region surrounding OsGA2ox5 (Figure 5D). Among them, the Fst values between Indica and Japonica were more than 0.25. Therefore, we propose that very strong genetic divergence in the OsGA2ox5 region might exist between Indica and Japonica.
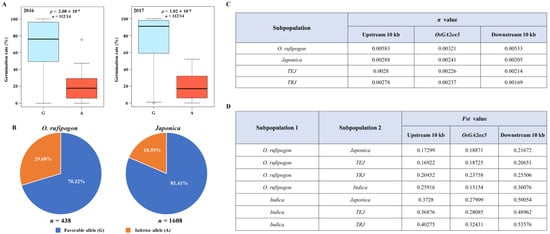
Figure 5.
Population genetic analysis of OsGA2ox5. (A) Box plots showing the genotype effects of SNP Chr7-218,245 on germination rate in Japonica rice; (B) The allelic frequencies of SNP Chr7-218,245 in diverse wild rice and Japonica rice; (C) Nucleotide diversity analysis of OsGA2ox5 and its flanking regions (10 kb); (D) Population differentiation analysis of OsGA2ox5 and its flanking regions (10 kb). Statistical differences between the G-type (light blue) and A-type (bright red) were analysed using Student’s t-test.
4. Discussion
Seed germination is an important indicator for the assessment of seed vigor, and rapid uniform germination is a key contributing factor to the success of direct-seeding rice [35]. Understanding the genetic architecture of seed germination is valuable for breeding high-vigor rice varieties suitable for direct seeding. In this study, we investigated the phenotypic variation for seed germination across two years using 131 varieties of Japonica rice. In this association panel, the phenotypic values of the germination rate had high variability with the CVs above 0.5 in both years, which was generally consistent with results from previous studies in rice [36], and other crops [37,38]. This finding suggested that the association population contains abundant genetic and phenotypic diversity and is suitable for dissecting the genetic architecture of seed germination.
Benefiting from cost-efficient sequencing technologies and improved statistical power, GWAS has been widely applied to dissect the genetic loci of complex quantitative traits in many crops [39]. In this study, a total of 22 candidate loci containing multiple signals of association for early seed germination were detected by GWAS in two years. Sixteen year-unstable loci identified in this study agreed with the previous finding that some of the QTLs controlling seed germination traits were significantly influenced by the environmental conditions [40]. Six common loci identified in both years, including qGR1.1, qGR2.1, qGR3.1, qGR7.1, qGR8.1 and qGR9, might have stable effects on rice seed germination in different environments. In addition, these stable loci were close to the QTLs reported in previous studies using association and biparental mapping populations. For example, qGR1.1 was found near microsatellite marker RM8095 associated with shoot length [41]; qGR2.1 coincided with the region of qSST2.3 for seed storability [42]; qGR3.1 and qGR7.1 were found to overlap with the intervals of qSD3BR and qDOM7.1 for seed dormancy, respectively [43,44]; qGR8.1 co-localized with the regions of qSURE8.1 for seed reserve utilization efficiency and qSG8.2 for seed germination [13,45]; qGR9 overlapped with the intervals of qDOR-9-1 for seed dormancy and qSSn-9 for seed storability [46,47]. The above observations overlap with previous findings not only validate the power of a GWAS for identifying significant loci associated with seed germination, but also suggest that the six stable loci detected in this study can be developed as functional molecular markers for rice breeding.
In this study, the region of qGR7.1 harboured multiple significant SNPs in both years, and therefore it was of interest to explore potentially functional genes at this locus. Recently, the approach of dividing the significant SNPs within candidate regions into different categories based on their locations has been widely successfully used for nominating candidate genes at GWAS-discovered loci [13,22,33,48,49]. By applying this approach, the significant SNP (Chr7-218,245) in OsGA2ox5 (LOC_Os07g01340) was found to be annotated as a non-synonymous variant, giving rise to the substitution of an amino acid, and we, therefore, speculated that OsGA2ox5 encoding a gibberellin 2-oxidase was the most plausible candidate gene of qGR7.1. Based on the allelic genotypes of SNP Chr7-218,245, we found that Japonica accessions with G-type displayed high germination rate while accessions with A-type exhibited low germination rate, indicating that this causal SNP in OsGA2ox5 gene was responsible for variation in germination rate among Japonica accessions. According to previously published data [50], a similar comparison study for 473 accessions from the rice 3K panel produced similar result (Figure S2). Therefore, this SNP could be converted into KASP (Kompetitive Allele Specific PCR) marker for marker-assisted selection in the future. In addition, OsGA2ox5 was found to be located in a previously reported candidate improvement region (200,001 to 290,000 bp) [51], this finding was in line with the results of nucleotide diversity analysis in the genomic region harboring OsGA2ox5. We, therefore, speculated that low nucleotide diversity of OsGA2ox5 in Japonica is likely due to a result of selection during rice improvement.
As one of the most important phytohormones, GA functions as an essential natural regulator during plant developmental processes [52]. The members of the GA20ox, GA3ox and GA2ox gene families are considered key enzymes involved in regulating GA homeostasis in plants [53]. Among them, GA2ox functions in reducing the amount of bioactive GA [54]. In previous studies, several ga2ox mutants have been reported to negatively regulate seed germination or early seedling growth in rice and Arabidopsis. For example, a loss-of-function mutation of Arabidopsis ga2ox2 was shown to increase GA level and partly promote seed germination during dark imbibition after irradiation with a far-red light pulse [55]. The Arabidopsis ga2ox7 mutant had longer primary roots than WT under high-salinity stress [56]. In rice, the knockout mutant osga2ox8 had an increase in shoot and root length at the seedling stage [18]. In contrast, in the present study we found that the mutation of OsGA2ox5 caused delayed seed germination and early seedling growth, and a possible explanation is that knockout of OsGA2ox5 might not lead to increase GA level due to the presence of multiple genes of OsGA2ox in rice. From the STRING database (https://string-db.org/; accessed on 6 October 2022), OsGA2ox5 was predicted to interact with GA biosynthesis enzymes (OsKAO, OsGA20ox1, OsGA20ox3, OsGA3ox1 and OsGA3ox2), GA deactivation enzyme (OsGA2ox1) and auxin response factor (OsARF22) (Figure 6). Therefore, we speculated that OsGA2ox5 might play an important and complex role during seed germination in rice. In addition, the osga2ox5 mutant also showed reduced grain length, grain width and 1000-grain weight compared to WT (Figure S3). However, the exact mechanisms underlying how this gene regulates seed germination and grain size need to be investigated in further studies.
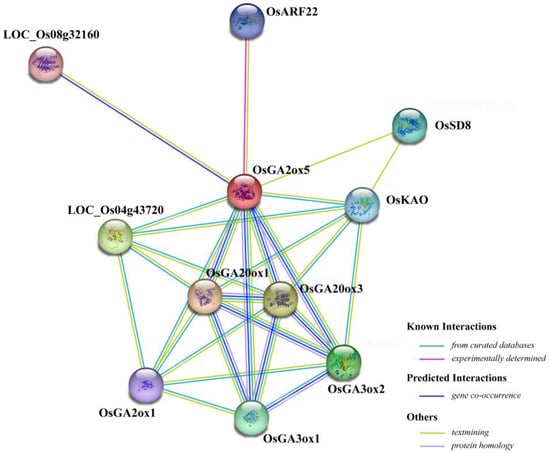
Figure 6.
Schematic prediction of the proteins that might interact with OsGA2ox5. Colored lines drawn between OsGA2ox5 and other proteins indicate protein interactions, which were identified through bioinformatics and experimental methods, including known interactions, predicted interactions, textmining and protein homology. The color key in the connecting lines is shown to the bottom-right corner of the figure.
5. Conclusions
In this study, six stable loci associated with early seed germination were detected across two years by GWAS. The causal gene of stable locus qGR7.1 was identified as OsGA2ox5, which was annotated as a gibberellin 2-oxidase. A causal SNP (Chr7-218,245) in the coding region of OsGA2ox5 might be linked to the difference in seed germination among Japonica accessions, and mutant analysis suggested that OsGA2ox5 may play a vital role in regulating rice seed germination. The association loci identified in this study provide a strong foundation for further decoding the molecular mechanism of seed germination.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agriculture13010118/s1, Figure S1: The remaining 225,800 SNPs were used for GWAS after filtering, Figure S2: Box plots showing the genotype effects of SNP Chr7-218,245 on germination rate in 473 accessions from the rice 3K panel. Statistical differences between the G-type and A-type were analysed using Student’s t-test, Figure S3: Comparison of grain size and grain weight between the WT and osga2ox5 mutant. Statistical differences between the WT and mutant were analysed using Student’s t-test; Table S1: List of Japonica rice accessions used for GWAS; Table S2: SNPs significantly associated with seed germination; Table S3: Comparison of GWAS loci with reported QTLs. Refs. [57,58,59,60,61,62,63,64,65,66,67,68,69,70] are cited in the Supplementary Materials.
Author Contributions
Conceptualization, X.W. and L.W.; Investigation, B.Y., J.Z., S.C. and S.L.; Writing—original draft preparation, B.Y.; Writing—review and editing, B.Y. and X.W.; Visualization, B.Y. and J.Z.; Supervision, B.Y., L.W. and X.W.; Project administration, B.Y. and X.W.; Funding acquisition, B.Y. and X.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (Grant No. 32101802, 32071737 and 32111530289), the Department of Education of Guangdong Province (Grant No. 2020ZDZX1013), the Department of Science and Technology of Guangdong Province (Grant No. 2021A050530073), and the Agricultural and Rural Department of Guangdong Province (Grant No. KB1708008).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data from the experiments presented in this study are available on request to the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gruis, D.F.; Selinger, D.A.; Curran, J.M.; Jung, R. Redundant proteolytic mechanisms process seed storage proteins in the absence of seed-type members of the vacuolar processing enzyme family of cysteine proteases. Plant Cell 2002, 14, 2863–2882. [Google Scholar] [CrossRef]
- Rajjou, L.; Duval, M.; Gallardo, K.; Catusse, J.; Job, D. Seed germination and vigor. Annu. Rev. Plant Biol. 2012, 63, 507–533. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zeng, J.; Zheng, Z.; Zhou, Q.; Chen, S.; Zheng, Y.; Wan, X.; Yang, B. Comparative transcriptome analysis reveals the mechanisms underlying differential seed vigor in two contrasting peanut genotypes. Agriculture 2022, 12, 1355. [Google Scholar] [CrossRef]
- Cheng, X.; Wu, Y.; Guo, J.; Du, B.; Chen, R.; Zhu, L.; He, G. A rice lectin receptor-like kinase that is involved in innate immune responses also contributes to seed germination. Plant J. 2013, 76, 687–698. [Google Scholar] [CrossRef] [PubMed]
- Xu, E.; Chen, M.; He, H.; Zhan, C.; Cheng, Y.; Zhang, H.; Wang, Z. Proteomic analysis reveals proteins involved in seed imbibition under salt stress in rice. Front. Plant Sci. 2017, 7, 2006. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Yang, B.; He, Y.; Zhan, C.; Cheng, Y.; Zhang, J.; Zhang, H.; Cheng, J.; Wang, Z. A quantitative trait locus, qSE3, promotes seed germination and seedling establishment under salinity stress in rice. Plant J. 2019, 97, 1089–1104. [Google Scholar] [CrossRef]
- Fujino, K.; Sekiguchi, H.; Matsuda, Y.; Sugimoto, K.; Ono, K.; Yano, M. Molecular identification of a major quantitative trait locus, qLTG3-1, controlling low-temperature germinability in rice. Proc. Natl. Acad. Sci. USA 2008, 105, 12623–12628. [Google Scholar] [CrossRef]
- Sugimoto, K.; Takeuchi, Y.; Ebana, K.; Miyao, A.; Hirochika, H.; Hara, N.; Ishiyama, K.; Kobayashi, M.; Ban, Y.; Hattori, T.; et al. Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice. Proc. Natl. Acad. Sci. USA 2010, 107, 5792–5797. [Google Scholar] [CrossRef]
- Ye, H.; Feng, J.; Zhang, L.; Zhang, J.; Mispan, M.S.; Cao, Z.; Beighley, D.H.; Yang, J.; Gu, X.Y. Map-based cloning of seed dormancy1-2 identified a gibberellin synthesis gene regulating the development of endosperm-imposed dormancy in rice. Plant Physiol. 2015, 169, 2152–2165. [Google Scholar]
- Prince, S.J.; Valliyodan, B.; Ye, H.; Yang, M.; Tai, S.; Hu, W.; Murphy, M.; Durnell, L.A.; Song, L.; Joshi, T.; et al. Understanding genetic control of root system architecture in soybean: Insights into the genetic basis of lateral root number. Plant Cell Environ. 2019, 42, 212–229. [Google Scholar] [CrossRef]
- Li, W.; Yang, B.; Xu, J.; Peng, L.; Sun, S.; Huang, Z.; Jiang, X.; He, Y.; Wang, Z. A genome-wide association study reveals that the 2-oxoglutarate/malate translocator mediates seed vigor in rice. Plant J. 2021, 108, 478–491. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Sun, S.; Yang, B.; Zhao, J.; Li, W.; Huang, Z.; Li, Z.; He, Y.; Wang, Z. Genome-wide association study reveals that the cupin domain protein OsCDP3.10 regulates seed vigour in rice. Plant Biotechnol. J. 2022, 20, 485–498. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Chen, M.; Zhan, C.; Liu, K.; Cheng, Y.; Xie, T.; Zhu, P.; He, Y.; Zeng, P.; Tang, H.; et al. Identification of OsPK5 involved in rice glycolytic metabolism and GA/ABA balance for improving seed germination via genome-wide association study. J. Exp. Bot. 2022, 73, 3446–3461. [Google Scholar] [CrossRef]
- Yoshida, H.; Hirano, K.; Yano, K.; Wang, F.; Mori, M.; Kawamura, M.; Koketsu, E.; Hattori, M.; Ordonio, R.L.; Huang, P.; et al. Genome-wide association study identifies a gene responsible for temperature-dependent rice germination. Nat. Commun. 2022, 13, 5665. [Google Scholar] [CrossRef] [PubMed]
- McGinnis, K.M.; Thomas, S.G.; Soule, J.D.; Strader, L.C.; Zale, J.M.; Sun, T.P.; Steber, C.M. The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 2003, 15, 1120–1130. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.P. The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr. Biol. 2011, 21, R338–R345. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, S.; Böhlenius, H.; Moritz, T.; Nilsson, O. GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell 2006, 18, 2172–2181. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Du, F.; Wang, J.; Li, Y.; Zhang, Y.; Zhao, X.; Zheng, T.; Li, Z.; Xu, J.; Wang, W.; et al. Molecular dissection of the gene OsGA2ox8 conferring osmotic stress tolerance in rice. Int. J. Mol. Sci. 2021, 22, 9107. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Tung, C.W.; Eizenga, G.C.; Wright, M.H.; Ali, M.L.; Price, A.H.; Norton, G.J.; Islam, M.R.; Reynolds, A.; Mezey, J.; et al. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat. Commun. 2011, 2, 467. [Google Scholar] [CrossRef]
- Lu, Y.; Ye, X.; Guo, R.; Huang, J.; Wang, W.; Tang, J.; Tan, L.; Zhu, J.K.; Chu, C.; Qian, Y. Genome-wide targeted mutagenesis in rice using the CRISPR/Cas9 system. Mol. Plant 2017, 10, 1242–1245. [Google Scholar] [CrossRef]
- McCouch, S.R.; Wright, M.H.; Tung, C.W.; Maron, L.G.; McNally, K.L.; Fitzgerald, M.; Singh, N.; DeClerck, G.; Agosto-Perez, F.; Korniliev, P.; et al. Open access resources for genome-wide association mapping in rice. Nat. Commun. 2016, 7, 10532. [Google Scholar] [CrossRef]
- Yano, K.; Yamamoto, E.; Aya, K.; Takeuchi, H.; Lo, P.C.; Hu, L.; Yamasaki, M.; Yoshida, S.; Kitano, H.; Hirano, K.; et al. Genome-wide association study using whole-genome sequencing rapidly identifies new genes influencing agronomic traits in rice. Nat. Genet. 2016, 48, 927–934. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Lipka, A.E.; Tian, F.; Wang, Q.; Peiffer, J.; Li, M.; Bradbury, P.J.; Gore, M.A.; Buckler, E.S.; Zhang, Z. GAPIT: Genome association and prediction integrated tool. Bioinformatics 2012, 28, 2397–2399. [Google Scholar] [CrossRef]
- Huang, X.; Wei, X.; Sang, T.; Zhao, Q.; Feng, Q.; Zhao, Y.; Li, C.; Zhu, C.; Lu, T.; Zhang, Z.; et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 2010, 42, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef]
- Zhao, H.; Yao, W.; Ouyang, Y.; Yang, W.; Wang, G.; Lian, X.; Xing, Y.; Chen, L.; Xie, W. RiceVarMap: A comprehensive database of rice genomic variations. Nucleic Acids Res. 2015, 43, D1018–D1022. [Google Scholar] [CrossRef]
- Yao, W.; Huang, F.; Zhang, X.; Tang, J. ECOGEMS: Efficient compression and retrieve of SNP data of 2058 rice accessions with integer sparse matrices. Bioinformatics 2019, 35, 4181–4183. [Google Scholar] [CrossRef]
- Huang, X.; Kurata, N.; Wei, X.; Wang, Z.X.; Wang, A.; Zhao, Q.; Zhao, Y.; Liu, K.; Lu, H.; Li, W.; et al. A map of rice genome variation reveals the origin of cultivated rice. Nature 2012, 490, 497–501. [Google Scholar] [CrossRef]
- Chen, W.; Gao, Y.; Xie, W.; Gong, L.; Lu, K.; Wang, W.; Li, Y.; Liu, X.; Zhang, H.; Dong, H.; et al. Genome-wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism. Nat. Genet. 2014, 46, 714–721. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Pandey, B.B.; Ratnakumar, P.; Kiran, B.U.; Dudhe, M.Y.; Lakshmi, G.S.; Ramesh, K.; Guhey, A. Identifying traits associated with terminal drought tolerance in sesame (Sesamum indicum L.) genotypes. Front. Plant Sci. 2021, 12, 739896. [Google Scholar] [CrossRef] [PubMed]
- Zhan, C.; Hu, J.; Pang, Q.; Yang, B.; Cheng, Y.; Xu, E.; Zhu, P.; Li, Y.; Zhang, H.; Cheng, J. Genome-wide association analysis of panicle exsertion and uppermost internode in rice (Oryza sativa L.). Rice 2019, 12, 72. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, H.; Xu, J.; Jiang, C.; Yin, Z.; Xiong, H.; Xie, J.; Wang, X.; Zhu, X.; Li, Y.; et al. Loci and natural alleles underlying robust roots and adaptive domestication of upland ecotype rice in aerobic conditions. PLoS Genet. 2018, 14, e1007521. [Google Scholar] [CrossRef]
- Mahender, A.; Anandan, A.; Pradhan, S.K. Early seedling vigour, an imperative trait for direct-seeded rice: An overview on physio-morphological parameters and molecular markers. Planta 2015, 241, 1027–1050. [Google Scholar] [CrossRef]
- Cheng, J.; He, Y.; Yang, B.; Lai, Y.; Wang, Z.; Zhang, H. Association mapping of seed germination and seedling growth at three conditions in indica rice (Oryza sativa L.). Euphytica 2015, 206, 103–115. [Google Scholar] [CrossRef]
- Li, X.; Guo, D.; Xue, M.; Li, G.; Yan, Q.; Jiang, H.; Liu, H.; Chen, J.; Gao, Y.; Duan, L.; et al. Genome-wide association study of salt tolerance at the seed germination stage in flax (Linum usitatissimum L.). Genes 2022, 13, 486. [Google Scholar] [CrossRef]
- Ma, L.; Wang, C.; Hu, Y.; Dai, W.; Liang, Z.; Zou, C.; Pan, G.; Lübberstedt, T.; Shen, Y. GWAS and transcriptome analysis reveal MADS26 involved in seed germination ability in maize. Theor. Appl. Genet. 2022, 135, 1717–1730. [Google Scholar] [CrossRef]
- Tian, D.; Wang, P.; Tang, B.; Teng, X.; Li, C.; Liu, X.; Zou, D.; Song, S.; Zhang, Z. GWAS Atlas: A curated resource of genome-wide variant-trait associations in plants and animals. Nucleic Acids Res. 2020, 48, D927–D932. [Google Scholar] [CrossRef]
- Geshnizjani, N.; Snoek, B.L.; Willems, L.A.J.; Rienstra, J.A.; Nijveen, H.; Hilhorst, H.W.M.; Ligterink, W. Detection of QTLs for genotype × environment interactions in tomato seeds and seedlings. Plant Cell Environ. 2020, 43, 1973–1988. [Google Scholar] [CrossRef]
- Dang, X.; Thi, T.G.; Dong, G.; Wang, H.; Edzesi, W.M.; Hong, D. Genetic diversity and association mapping of seed vigor in rice (Oryza sativa L.). Planta 2014, 239, 1309–1319. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Li, N.; Yu, Y.; Chen, W.; Yu, S.; He, H. Insights into the regulation of rice seed storability by seed tissue-specific transcriptomic and metabolic profiling. Plants 2022, 11, 1570. [Google Scholar] [CrossRef] [PubMed]
- Subudhi, P.K.; Parco, A.; Singh, P.K.; Deleon, T.; Karan, R.; Biradar, H.; Cohn, M.A.; Brar, D.S.; Sasaki, T. Genetic architecture of seed dormancy in U.S. weedy rice in different genetic backgrounds. Crop Sci. 2012, 52, 2564–2575. [Google Scholar] [CrossRef]
- Yuan, S.; Wang, Y.; Zhang, C.; He, H.; Yu, S. Genetic dissection of seed dormancy using chromosome segment substitution lines in rice (Oryza sativa L.). Int. J. Mol. Sci. 2020, 21, 1344. [Google Scholar] [CrossRef]
- Cheng, X.; Cheng, J.; Huang, X.; Lai, Y.; Wang, L.; Du, W.; Wang, Z.; Zhang, H. Dynamic quantitative trait loci analysis of seed reserve utilization during three germination stages in rice. PLoS ONE 2013, 8, e80002. [Google Scholar] [CrossRef]
- Cai, H.W.; Morishima, H. Genomic regions affecting seed shattering and seed dormancy in rice. Theor. Appl. Genet. 2000, 100, 840–846. [Google Scholar] [CrossRef]
- Lin, Q.; Wang, W.; Ren, Y.; Jiang, Y.; Sun, A.; Qian, Y.; Zhang, Y.; He, N.; Hang, N.T.; Liu, Z.; et al. Genetic dissection of seed storability using two different populations with a same parent rice cultivar N22. Breed. Sci. 2015, 65, 411–419. [Google Scholar] [CrossRef]
- Patishtan, J.; Hartley, T.N.; de Carvalho, R.F.; Maathuis, F.J.M. Genome-wide association studies to identify rice salt-tolerance markers. Plant Cell Environ. 2018, 41, 970–982. [Google Scholar] [CrossRef]
- Guo, H.; Zeng, Y.; Li, J.; Ma, X.; Zhang, Z.; Lou, Q.; Li, J.; Gu, Y.; Zhang, H.; Li, J.; et al. Differentiation, evolution and utilization of natural alleles for cold adaptability at the reproductive stage in rice. Plant Biotechnol. J. 2020, 18, 2491–2503. [Google Scholar] [CrossRef]
- Shi, Y.; Gao, L.; Wu, Z.; Zhang, X.; Wang, M.; Zhang, C.; Zhang, F.; Zhou, Y.; Li, Z. Genome-wide association study of salt tolerance at the seed germination stage in rice. BMC Plant Biol. 2017, 17, 92. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Wang, G.; Yuan, M.; Yao, W.; Lyu, K.; Zhao, H.; Yang, M.; Li, P.; Zhang, X.; Yuan, J.; et al. Breeding signatures of rice improvement revealed by a genomic variation map from a large germplasm collection. Proc. Natl. Acad. Sci. USA 2015, 112, E5411–E5419. [Google Scholar] [CrossRef]
- Kaneko, M.; Inukai, Y.; Ueguchi-Tanaka, M.; Itoh, H.; Izawa, T.; Kobayashi, Y.; Hattori, T.; Miyao, A.; Hirochika, H.; Ashikari, M.; et al. Loss-of-function mutations of the rice GAMYB gene impair α-amylase expression in aleurone and flower development. Plant Cell 2004, 16, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, S. Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 2008, 59, 225–251. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.G.; Phillips, A.L.; Hedden, P. Molecular cloning and functional expression of gibberellin 2- oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc. Natl. Acad. Sci. USA 1999, 96, 4698–4703. [Google Scholar] [CrossRef]
- Yamauchi, Y.; Takeda-Kamiya, N.; Hanada, A.; Ogawa, M.; Kuwahara, A.; Seo, M.; Kamiya, Y.; Yamaguchi, S. Contribution of gibberellin deactivation by AtGA2ox2 to the suppression of germination of dark-imbibed Arabidopsis thaliana seeds. Plant Cell Physiol. 2007, 48, 555–561. [Google Scholar] [CrossRef] [PubMed]
- Magome, H.; Yamaguchi, S.; Hanada, A.; Kamiya, Y.; Oda, K. The DDF1 transcriptional activator upregulates expression of a gibberellin-deactivating gene, GA2ox7, under high-salinity stress in Arabidopsis. Plant J. 2008, 56, 613–626. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.F.; Wang, J.F.; Bao, Y.M.; Wang, F.H.; Zhang, H.S. Quantitative trait loci analysis for rice seed vigor during the germination stage. J. Zhejiang Univ. Sci. B 2010, 11, 958–964. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Oh, C.S.; Suh, J.P.; McCouch, S.R.; Ahn, S.N. Identification of QTLs for domestication-related and agronomic traits in an Oryza sativa × O. rufipogon BC1F7 population. Plant Breed. 2005, 124, 209–219. [Google Scholar] [CrossRef]
- Zhou, Y.; Xie, Y.; Cai, J.; Liu, C.; Zhu, H.; Jiang, R.; Zhong, Y.; Zhang, G.; Tan, B.; Liu, G.; et al. Substitution mapping of QTLs controlling seed dormancy using single segment substitution lines derived from multiple cultivated rice donors in seven cropping seasons. Theor. Appl. Genet. 2017, 130, 1191–1205. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xinshun, Q.U.; Wan, S.; Chen, L.; Zhu, Y. Comparison of QTL controlling seedling vigour under different temperature conditions using recombinant inbred lines in rice (Oryza sativa). Ann. Bot. 2005, 95, 423–429. [Google Scholar] [CrossRef]
- Miura, K.; Lin, Y.; Yano, M.; Nagamine, T. Mapping quantitative trait loci controlling seed longevity in rice (Oryza sativa L.). Theor. Appl. Genet. 2002, 104, 981–986. [Google Scholar] [CrossRef]
- Dong, Y.; Tsuzuki, E.; Kamiunten, H.; Terao, H.; Lin, D.; Matsuo, M.; Zheng, Y. Identification of quantitative trait loci associated with pre-harvest sprouting resistance in rice (Oryza sativa L.). Field Crops Res. 2003, 81, 133–139. [Google Scholar] [CrossRef]
- Wang, L.; Cheng, J.; Lai, Y.; Du, W.; Huang, X.; Wang, Z.; Zhang, H. Identification of QTLs with additive, epistatic and QTL × development interaction effects for seed dormancy in rice. Planta 2014, 239, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Foley, M.E.; Gu, X.Y. New seed dormancy loci detected from weedy rice-derived advanced populations with major QTL alleles removed from the background. Plant Sci. 2010, 179, 612–619. [Google Scholar] [CrossRef]
- Marzougui, S.; Sugimoto, K.; Yamanouchi, U.; Shimono, M.; Hoshino, T.; Hori, K.; Kobayashi, M.; Ishiyama, K.; Yano, M. Mapping and characterization of seed dormancy QTLs using chromosome segment substitution lines in rice. Theor. Appl. Genet. 2012, 124, 893–902. [Google Scholar] [CrossRef] [PubMed]
- Dimaano, N.G.B.; Ali, J.; Mahender, A.; Cruz, P.C.S.; Baltazar, A.M.; Diaz, M.G.Q.; Pang, Y.L.; Acero, B.L.; Li, Z.K. Identification of quantitative trait loci governing early germination and seedling vigor traits related to weed competitive ability in rice. Euphytica 2020, 216, 159. [Google Scholar] [CrossRef]
- Gu, X.Y.; Kianian, S.F.; Foley, M.E. Phenotypic selection for dormancy introduced a set of adaptive haplotypes from weedy into cultivated rice. Genetics 2005, 171, 695–704. [Google Scholar] [CrossRef]
- Jiang, S.; Yang, C.; Xu, Q.; Wang, L.; Yang, X.; Song, X.; Wang, J.; Zhang, X.; Li, B.; Li, H.; et al. Genetic dissection of germinability under low temperature by building a resequencing linkage map in japonica rice. Int. J. Mol. Sci. 2020, 21, 1284. [Google Scholar] [CrossRef]
- Gu, X.Y.; Kianian, S.F.; Foley, M.E. Isolation of three dormancy QTLs as Mendelian factors in rice. Heredity 2006, 96, 93–99. [Google Scholar] [CrossRef][Green Version]
- Mao, F.; Wu, D.; Lu, F.; Yi, X.; Gu, Y.; Liu, B.; Liu, F.; Tang, T.; Shi, J.; Zhao, X.; et al. QTL mapping and candidate gene analysis of low temperature germination in rice (Oryza sativa L.) using a genome wide association study. PeerJ 2022, 10, e13407. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).