Abstract
The livestock of Pakistan includes different animal breeds utilized for milk farming and exporting worldwide. Buffalo have a high milk production rate, and Pakistan is the third-largest milk-producing country, and its production is increasing over time. Hence, it is essential to recognize the best Buffalo breed for a high milk- and meat yield to meet the world’s demands and breed production. Pakistan has the second-largest number of buffalos among countries worldwide, where the Neli-Ravi breed is the most common. The extensive demand for Neli and Ravi breeds resulted in the new cross-breed “Neli-Ravi” in the 1960s. Identifying and segregating the Neli-Ravi breed from other buffalo breeds is the most crucial concern for Pakistan’s dairy-production centers. Therefore, the automatic detection and classification of buffalo breeds are required. In this research, a computer-vision-based recognition framework is proposed to identify and classify the Neli-Ravi breed from other buffalo breeds. The proposed framework employs self-activated-based improved convolutional neural networks (CNN) combined with self-transfer learning. Moreover, feature maps extracted from CNN are further transferred to obtain rich feature vectors. Different machine learning (Ml) classifiers are adopted to classify the feature vectors. The proposed framework is evaluated on two buffalo breeds, namely, Neli-Ravi and Khundi, and one additional target class contains different buffalo breeds collectively called Mix. The proposed research achieves a maximum of 93% accuracy using SVM and more than 85% accuracy employing recent variants.
1. Introduction
The livestock of Pakistan includes goats, sheep, buffalos, cattle, and their variants. They have been used for milk, meat, wool, and hidden purposes. Rearing animals for milk and meat has always been a desirable task for our farmers [1]. However, lots of aspects of red meat are still to be illuminated, and there is a lot more to do in that sector; on the other hand, the milk sector of Pakistan is prosperous so far, and the need of the hour is to sustain this opulence and success [2]. According to the IFCN (international farms comparison network) report, Pakistan is the third-largest milk-producing country globally, and the figures are expected to mount in upcoming years [3]. Interestingly, Pakistan’s buffalos carry much of this burden as they constitute about 65% of total milk production in the entire country. Buffalos are the most widely used animal in South Asia, especially in Pakistan; Buffalo have been bred here for a long time, and Pakistan is fortunately rich in many world-class breeds of buffalo, such as Nili Ravi and Kundi [3].
Before the late nineties, a revolution occurred in Pakistan’s milk sector before almost 60% of animals were kept by rural people in small units. The grazing fulfilled the feed requirements and other green fodder [3]. In the last two decades, the dairy sector of Pakistan has witnessed rapid growth; lots of dairy farms have been established, and especially Kundi is a vital player [4]. Worldwide, there is a dearth of buffalos; previously, the developed world has primarily considered cattle as a principal source for getting milk requirements. The USA’s milk industry relies chiefly on dairy cattle, but now it is moving forward toward the utilization of Buffalo milk [5]. There are many reasons behind this shift, such as the low percentage of fat in cattle milk (buffalos have twice as much as cattle), its low viscosity (buffalo milk is much more viscous, thick, and creamy than that of cattle), and the inability of it to be rendered into some refining kind of cheese such as Mozzarella cheese and some varieties of Cheddar cheese, etc. To manufacture these from cattle milk, millions of dollars have been invested, but the results have been negative [6].
These buffalo breeds tolerate wide variations in fodder supply, feed quality, and adaptation to low-input monitoring systems. Their home track is in central Punjab’s canal-irrigated areas, where they are fed plenty of green fodder [7]. They produce much less milk in their home tract despite their capacity, which may be due to (1) a long calving interval, (2) silent heat, (3) late maturity, and (4) a lack of attention paid to the past to improvement by selection and progeny testing [7].
The classification of native buffalo breeds has traditionally been based on morphological differences, but regional variations are insufficient to distinguish breeds closely related [8]. For their effective and meaningful improvement and conservation, the detailed characterization and evaluation of differences among these breeds using modern technology tools is required.
The technological developments in the field of artificial intelligence (AI) [9], blockchain [10], IoT [11], cloud technologies [12], and data science [13] are going to transform the practice of traditional dairy farming into smart dairy farming. The latest challenge in dairy farming, which is in focus and an essential aspect of dairy farming, is to achieve animal health and welfare at the best possible standards, keeping in mind the high-performance potential of animals [14]. It is necessary for animal sciences to meet these challenges faced by the farmer’s introduction of technological advancement.
Artificial neural networks (ANNs) are one of the advanced computing methods used mainly for classification and prediction [15]. ANN is a multi-layer connected neural network that looks like a web. The basic ANN unit consists of an input, a hidden layer, and an output layer. Each layer has neurons or nodes, which are interconnected to each other in the next layer. Each node in the hidden layer has some weight and bias values. ANN is a nonlinear technique that can take any input. Some of the advantages of ANN include its ability to learn and model nonlinear and complex associations between inputs and outputs, as well as its ability to generalize as it can derive unperceived connections after learning from the primary inputs and their correlation with the outputs, making the model capable of generalizing and predicting undiscovered data [16].
Pakistan is the world’s second-most buffalo-populated country, accounting for 16.83% of the global buffalo population, with Nili-Ravi constituting the majority [17]. Due to extensive crossbreeding, the Nili and Ravi breeds have been combined into a single Nili-Ravi since the 1960s [18]. Compared to native breeds, this single Nili-Ravi is now considered the best-performing breed for meat and milk production. However, the differentiation between the Nili-Ravi breed and other native breeds is complex, and only experts can identify it. Visual feature-based classification is an exciting research topic with emerging AI-based techniques. The visual features include the shape of the head, horns, texture, and color, and the formation of the udder, foot, and tail [18].
Correct identification of the Nili-Ravi breed will help improve the number of quality animals, ultimately helping to elevate poverty in rural areas of Punjab, where most of the house expenses are met by selling buffalo milk. Using morphological markers and visual features to identify animals remains an expensive and time-consuming manual task by researchers. As a result, we demonstrate that a deep convolutional neural network can detect different breeds.
1.1. Objectives
The objective of this study is to propose a computer-vision-based recognition system for the identification and classification of Neli-Ravi buffalo breeds from other buffalo breeds. The proposed framework used self-activated-based enhanced CNN coupled with self-transfer learning. Information-oriented feature vectors were obtained by transferring feature maps and then classified using machine learning-based classifiers. The proposed method was tested on Neli-Ravi, Khundi, and Mix (a group of several breeds).
1.2. Related Work
As per our knowledge, no work has been done so far to segregate the Neli-Ravi breed from other buffalo breeds using machine learning and deep learning. The morphological features of Nili-Ravi buffalo are given with details in Table 1.

Table 1.
Morphological features of Nili-Ravi buffalo.
The authors previously contributed in [19], which included establishing an experimental image processing framework on a sheep farm, creating a database containing 1642 sheep images of four breeds recorded on a farm and labeled by an expert with their breed, and training a sheep breed classifier using computer vision and machine learning to obtain an overall precision of 95.8% with 1.7 mean difference. Their method may help sheep farmers distinguish between breeds efficiently, allowing for more accurate meat yield estimation and cost control.
The authors in [20] presented a deep-learning-based method for identifying individual cattle based on their predominant muzzle point image sequence features, which tackles the issue of missing or switched livestock and false insurance claims. They developed a set of muzzle-point images that were not publicly accessible. They also used a deep-learning-based CNN to extract meaningful shape information and visualize a cattle’s muzzle point picture. The derived function of muzzle point images is encoded using the stacked denoising auto-encoder technique.
The classification of horse breeds in natural scene images is addressed in [21]. The author collected a data set by exploring the internet. The dataset includes 1693 photos of six different horse breeds. For the classification and identification of horse breeds, deep learning techniques, specifically, CNNs, were used. The proposed dataset is used to train and fine-tune well-known deep CNN architectures pre-trained on the ImageNet dataset. As pre-trained CNN classifiers, VGG architectures with 16 and 19 layers, InceptionV3, ResNet50, and Xception, are used. The average classification accuracy of ResNet50 was 95.90%.
In the other study [22], the authors tried to classify six distinct goat breeds using mixed-breed goat photos. The images of goat breeds were taken at various recorded goat herds in India. Without causing discomfort to the livestock, nearly 2000 digital images of individual goats were collected in limited and unregulated conditions. For the efficient recognition and localization of goat breeds, a pre-trained deep-learning-based object-detection model called Faster R-CNN was fine-tuned using transfer-learning on the acquired images. The fine-tuned model can locate the goat and identify its breed in the image. The model was assessed using the Pascal VOC object-detection assessment metrics.
The authors compare various deep learning models and propose an Android application [23] that uses a smartphone camera to determine a given cat’s position and breed. The finalized model’s accuracy rate was 81.74%. Another research [24] aims to propose an AI-driven system for fish species identification based on the CNN framework. The proposed CNN architecture has 32 deep layers, each very deep, to extract useful and distinguishing features from the image. The VGGNet architecture is subjected to deep supervision to improve classification efficiency by incorporating various convolutional layers into each level’s training. They built the Fish-Pak dataset to test the performance of the proposed 32-layer CNN architecture.
The authors used deep learning models [25] to assess the absolute maximum accuracy in detecting the Canchim breed species, which is virtually identical to the Nelore breed. They also mapped the optimal ground sample distance (GSD) for detecting the Canchim breed. There were 1853 UAV images with 8629 animal samples in the experiments, and 15 different CNN architectures were tested. A total of 900 models were trained (2 datasets, 10 spatial resolutions, K-fold cross-validation, and 15 CNN architectures), allowing for a thorough examination of the factors that influence cattle detection using aerial images captured by UAVs. Several CNN architectures were found to be robust enough to accurately detect animals in aerial images even under less-than-ideal conditions, suggesting the feasibility of using UAVs for cattle monitoring.
Another study [26] suggested using CNN to develop a recognition system for the Pantaneira cattle breed. The researchers examined 51 animals from the Aquidauana Pantaneira cattle center (NUBOPAN). The area is situated in Brazil’s Midwest area. Four surveillance cameras were mounted on the walls and took 27,849 photographs of the Pantaneira cattle breed from multiple distances and locations.
2. Materials and Methods
Deep-learning-based identification and recognition approaches are widely used in medical, natural, animal, and many other fields. The deep convolutional network inspires researchers to identify and classify different instances based on the visual features [24,27]. The proposed research offers a self-activated CNN for the identification of buffalos breeds. The proposed CNN contains a 23-layer architecture with 5 blocks of convolution, batch normalization, reLU, and a max-pooling layer. Feature vectors extracted from CNN layers without transfer learning and self-activation reach up to 77.61% validation accuracy. However, applying transfer learning from the fully connected layer of the proposed CNN tends to increase the overall performance. All of the extracted feature vectors are employed for classification using classical ML classifiers with various test and train ratios such as 50–50, 60–40, and 70–30. The K-fold cross-validation techniques with 5 and 10 folds are also used to validate different ML classifiers’ performance. The primary steps adopted in the proposed research are shown in Figure 1.
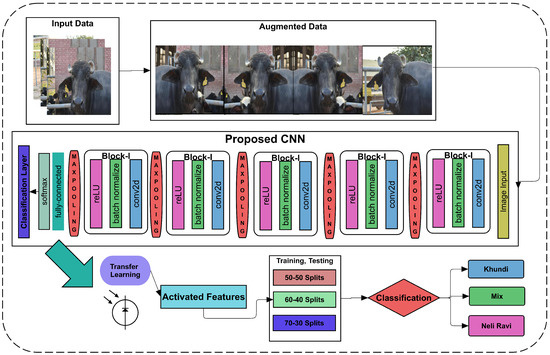
Figure 1.
Proposed framework used to identify and segregate Neli-Ravi breed from all other breeds.
Figure 1 determines that the input data are augmented with rotation, clipping, and the pixel range. The augmented data are further transmitted to the proposed CNN to classify buffalos breeds. To extract the rich feature vectors, we applied transfer learning to the 21st layer of CNN, which splits into various ratios to classify the breeds.
2.1. Data-Set Description
We collected data sets from Buffalo research center Pakistan. The acquired data-set is publically available at the data-set repository called Mendeley [28]. We normalized and preprocessed the data, including all class labels for self-activated CNN training. The size of the data set and details are shown in Table 2.

Table 2.
Data-set description.
Table 2 has shown three categories of used data-set in the proposed research. The three classes include Khundi, Mix, and Neli-Ravi. All images’ dimensions and formats are kept the same as given, 256 × 256 × 3, and JPG, respectively. However, data are primarily augmented before training and testing to avoid over-fitting and other deep learning issues.
2.2. Data Augmentation
The data augmentation is performed using various augmentation techniques. The bounding points of images are being transformed using reflection in the X and Y direction at random locations. The X and Y transformation specifies that the image has left, right, up, and down reflections. The random X, Y translation is set to [−4, 4]. It defines the random translation in X and Y dimensions with a specific range of pixel intensities on the image. The used pixel range is −4 to 4. Moreover, random cropping is applied, which performs a specific bitwise hide and seek operation, including cut-out and random erasing on the given image. The data samples are shown in Figure 2.

Figure 2.
Number of data samples increased for training through augmentation.
2.3. Proposed CNN
The self-activated CNN architecture principally depends upon 23 layers containing five convolutional blocks with a different number of filters and sizes. The first layer is an image () input layer having 256 × 256 × 3 input size of each given image instance. The 2nd layer is a convolutional layer with half the image size as several filters with a 3 × 3 kernel size. The padding and stride taken as one mean the sliding over the pixel to pixel for a given image are one so that each 3 × 3 kernel evolves on the whole image. The 2D convolutional computations are calculated as shown in Equations (2) and (3) by getting input from Equation (1) [29].
The Equation (2) represents the general kernel mask of 3 × 3, which later multiplies with the input image, where i and j represent the rows and columns; at the end of convolve operation, the output matrix is calculating in .
Each batch normalization layer is used in all convolutional blocks, where batch normalization is used for input normalization by calculating the variance and mean for each colour channel.
in Equation (4) is calculating the activated normalization, which is anticipated by using the division of difference between input and mean with the square root of the sum of squared variance and constant value. The constant value invariance is added to stable the minimal variance value. The re-consideration of this activation is taken as ; the zero mean and variance with the unit value are essential to scaling. The scaling is shown in Equation (5).
In Equation (5), the learnable factors and add to scale the activated output. The multiplication will shift the previous activated output into a new value with learnable parameters.
The max-pooling layer is down-sampling the given upper inputs into higher intensity values. The max-pooling for each convolve block is computed in Figure 3.
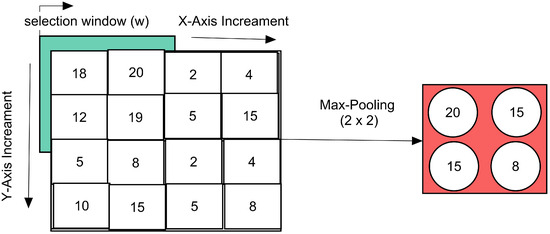
Figure 3.
Max-pooling operation employed for proposed CNN.
For iteration over the 2D matrix of an image, the input image selected the window, as explained in Equation (6).
The max-values from each incremented window ‘w’ are taken firstly from the left to right of the image, and then the next row is started; in this way, the y-direction increment starts, and lastly, a max-pooled downloaded max-window is gathered for each activated input of upper layer. The max-pooling is shown in Equation (7).
After getting the convolving data, the data are normalized using the batch normalization layer. In this way, the input batch data variations are reduced. After normalizing the data, the rectified linear unit (ReLU)-based activations are performed. The ReLU function works like a threshold value that specifies the negative value to zero and upper as it is for the same intensity values as calculated in Equation (8).
The normalized feature vectors are then passed to the max-pooling layer with a kernel size of 2, and the stride is also kept as 2 to slide over 2 points to take as the next input pool. All these layers’ parameters are shown in Table 3.

Table 3.
Convolution layers’ description of proposed CNN.
Table 3 has shown that only the convolutional layer changes the number of filter variations throughout the network where the batch normalization and ReLU activations are performed in each convolutional block. However, the architecture with each layer’s weights have been shown in CNN architect in Figure 4. Each layer’s weights are activated using the trained network to see the pooling effect in each architecture. Therefore, in the proposed architecture diagram, the weights of convolutions with batch normalization and reLU activations are shown as a stack. The convolve showing the colorful inputs is taken where the ongoing following batch normalization shows how the colors are being activated and changes for following layer input. Finally, in the fifth block, the convolve weights and batch normalization highlight more decision-making intensities. The final activated weights are passed on to the fully connected layer, taking it as the three classes and applying the soft-max operation-based activations.
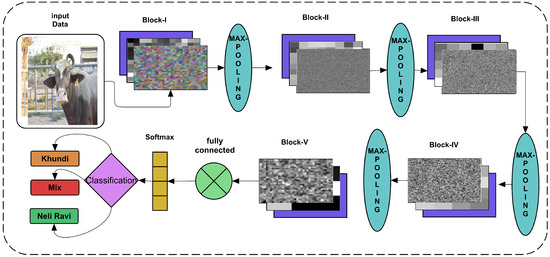
Figure 4.
Block diagram of proposed CNN architecture.
Figure 4 has shown the input data layer as augmented data input with all the layers’ flow towards final classification.
2.4. Self-Transfer Learning Features
Transfer learning is used in many recent studies [30], and it motivated us to use it for better classification results in terms of accuracy. However, many studies have employed state-of-the-art pre-trained ImageNet variants for transfer learning to get the numeric features. The proposed study does not get any promising results using the proposed CNN, but the transfer learning of data-specific CNN makes it more efficient. The proposed CNN’s use of self-transfer learning is also suitable for getting more accurate precision values.
2.5. Classification
Firstly, the proposed study self-activated CNN for classification purposes and then used seven classical ML classification methods. CNN is not performed well even on a higher number of iterations. It observes that a greater number of iterations tends to decrease the validation accuracy. The optimal number of parameters and their values are discussed in the results section. However, the seven used classifiers include 3 KNN variants fine-KNN, medium-KNN, and Coarse-KNN. The ensemble and boosting algorithms named total boost, LP-boost, and bag-ensemble are likewise applied. In order to make more certainty in the results, different data splits were applied using random instances selection.
3. Results and Discussion
Each image’s background has a static context since a static camera captures the various buffalo breeds’ images. Each pixel has its intensity, and the background of each buffalo image is generated by merging all pixels. Statistical concepts underpin the majority of context models. The context model can be divided into two groups based on various background descriptions: one is a reference model resulting in each pixel’s feature details. The other model is one that was generated by extracting the region function. Since image division requires high-level visual prior information, context models of this kind are typically complicated. Parameters setting to train proposed CNN on the acquired data set are described in Table 4. These parameters were found as the most optimal features via performance measures.

Table 4.
Parameters setting considered during the training and testing of proposed CNN.
Table 4 indicates a proposed CNN’s optimal parameters with the most accurate found CNN as 77.62% validation accuracy. The training time with different values is validated to improve the accuracy. Therefore, the performance-based selection of CNN is used with the given parameters of CNN. The training and validation graph is shown in Figure 5.
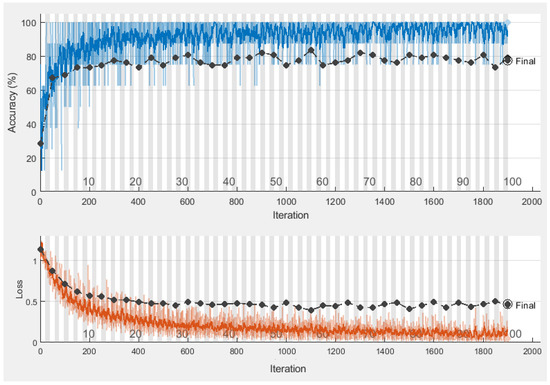
Figure 5.
Proposed CNN training and validation plots.
Figure 5 designates the absolute accuracy by finding the 77.62% as validation accuracy where the loss is up to 0.5 with a bit of variation. The data-specific learning of a given data set is more favorable as it covers all aspects of features, such as location, geometry, or other local features. At this stage, the feature vector is taken as transfer learning to get the numeric features from the CNN fully connected layer encodings. The activated features are then later used as a classification feature vector.
3.1. Data Splitting
The activated features are of size n × 3, where all image features are split up with randomization to remove the data’s biases. The used data split ratios are 50–50% (training, testing), 60–40% (training-testing), and 70–30% (training, testing). All data divisions employ seven different classifiers.
3.2. Experiment 1 (50–50)
In the first experimental phase, we split the features vectors as 50% for training and 50% for testing. The features matrix displays 225 × 3 and 225 × 3 for training and testing vectors. The trained classifiers-based predictions are then evaluated in terms of accuracy, precision, sensitivity, and F1 score. Moreover, a statistical evaluation metric such as Kappa has also been computed to evaluate the results. The prediction results are given in Table 5.

Table 5.
Results obtained using different ML classifiers on 50–50 Split.
Table 5 has bestowed that the maximum prediction accuracy is achieved by two classifiers, including 85.33% by coarse-knn and total-boost. The additional algorithms also performed well, with the accuracy range nearer to the two best-achieved algorithms. Their accuracy remains in the range of 80%+, where the accuracy obtained using the bag-ensemble method is not promising. Prediction plots obtained using different ML classifiers on 50–50 split are given in Figure 6.
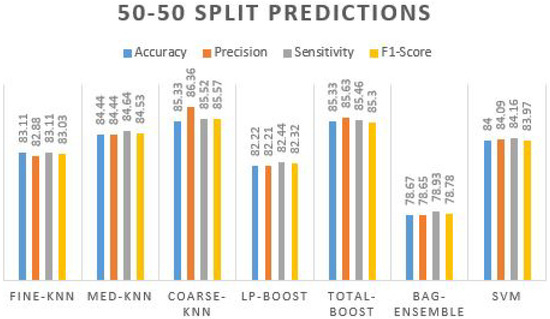
Figure 6.
Prediction plot obtained using different ML classifiers on 50–50 split.
We can observe that other performance measures of classification predictions are also higher as the precision value is 86.36% by coarse-knn, which is more reliable than the precision obtained using the total-boost method. Hence, in terms of more true optimistic predictions over the summation of true positives and false positives, = the sensitivity and F1s-core are also improved by the coarse-knn compared to other classifiers. The confusion matrix obtained using Coarse-KNN is presented in Table 6.

Table 6.
Confusion matrix obtained using Coarse-KNN.
The confusion matrix of coarse-knn indicated that the accurately predicted instances do not satisfy the Mix class as 20 instances are wrongly predicted in the khundi target class that belongs to the Mix class. However, the correctly predicted number of instances is 192 in total.
3.3. Experiment 2 (60–40)
Experiment 2 uses features vectors obtained from the last layer of self-activated CNN, splitting into 60–40% with 60% for training and 40% for testing. The overall results were almost identical to the 50–50 split strategy. Results obtained using different ML classifiers on 60–40 Split are presented in Table 7.

Table 7.
Results obtained using different ML classifiers on 60–40 split.
In Table 7, we can observe that the coarse-knn got the highest accuracy (84.44%) with slightly less accuracy precision, sensitivity, F1-score, and kappa values. Moreover, the other classifiers also perform in the range of 80%+. We can see in the 2nd row; the medium-knn achieved 84.07% accuracy. Similarly, SVM achieved the same accuracy value but with a lower precision rate of 84.54%. Fine-knn and bag-ensemble obtained the same accuracy value of 80.74% with all the same other evaluation measures. A prediction plot obtained using different ML classifiers on 60–40 split is displayed in Figure 7.
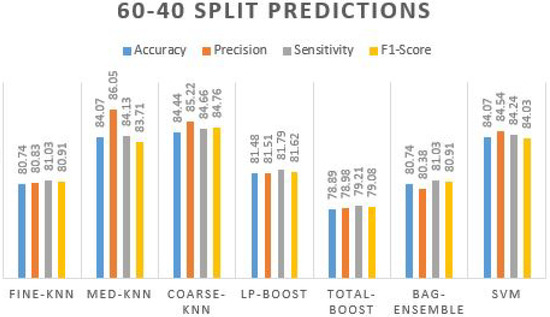
Figure 7.
Prediction plot obtained using different ML classifiers on 60-40 split.
The first and second phases of experimental chunks obtained almost the same results, which were more promising as different classifiers of different categories providing almost identical accuracy values—the most accurate classifier’s prediction-based confusion matrix is shown in Table 8. The primarily used conventional 70–30% split ratio is also utilized for more confident results.

Table 8.
Confusion matrix obtained using SVM.
The confusion matrix identified that the khundi class predictions are more accurate, whereas the ‘Mix’ class needs more improvement. The 76, 70, and 82 are the accurate predictions, whereas the wrongly predicted numbers are 13,1 and 23,3 and 2. Those wrongly predicted instances are on the upper and lower diagonal of the table.
3.4. Experiment 3 (70–30)
In the third experiment phase, we performed experiments using standard 70% random features splitting with 30% data testing. The evaluation measures are the same as in experimental phases one and two. The obtained results show gradual improvements as compared to the previous ones. Results obtained using different ML classifiers on 70–30 split are exhibited in Table 9.

Table 9.
Results obtained using different ML classifiers on 70–30 Split.
In Table 9, the maximum accuracy recorded using SVM is 93.33%, with the most accurate precision value. The given accuracy of this phase is justified and reportable. The other classifiers also display more than 87% accuracy. A prediction plot obtained using different ML classifiers on a 70–30 split is manifested in Figure 8.
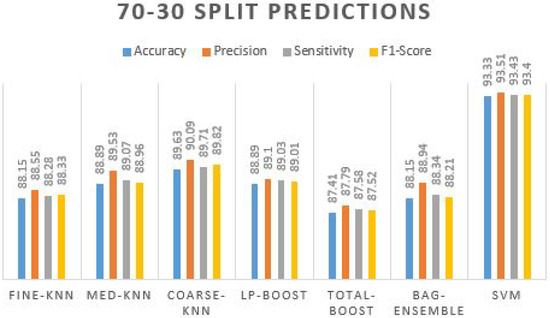
Figure 8.
Prediction plot obtained using different ML classifiers on 70-30 split.
It is observed that in all experiments, the performance measure does not vary. The confusion matrix for the best-achieved accuracy algorithm is also shown in Table 10.

Table 10.
Confusion matrix obtained using SVM on 70–30 split.
The confusion matrix of the most accurate SVM method shows that the Mix class still has more wrong predictions. The overall precision rate is improved to 93.51% over the summation of true positives and false positives. There are fewer wrong predictions in the other two classes; the F1-score provides both precision and a sensitivity response, which is also improved to the extent of 93.40%.
3.5. Experiment 4 (Five-Fold Cross Validation)
After the holdout validation techniques, the results are further evaluated on K-fold cross-validation. The primarily used values for K are 5 and 10 folds on the extracted feature vectors. The results, in this case, were also promising; the results were obtained using different ML classifiers on five-fold cross-validation in Table 11.

Table 11.
Results obtained using different ML classifiers on five-fold cross validation.
Table 11 confirms that coarse-knn and SVM produce the best accuracy and other effects. The accuracy value reaches up to 86.22% with 86.52% precision, 86.39% sensitivity, and 86.41% of the F1-score. By examining the precision value, the true positive value seems to be truer where the F1-score cross-validated it by taking sensitivity and precision over it and again received 86.41%. The prediction plot obtained using different ML classifiers on five-fold cross validation is manifested in Figure 9.
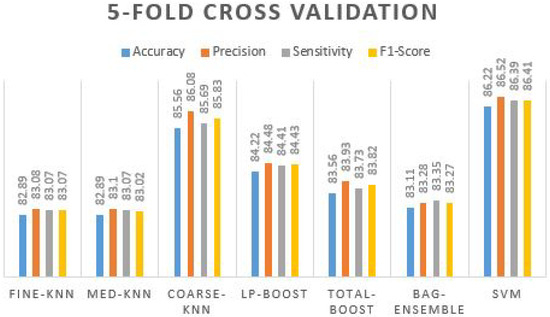
Figure 9.
Prediction plot obtained using different ML classifiers on 5-Fold Cross Validation.
We can see from the graph that all five other classifiers are above 80% to 84% of the accuracy values, where coarse-knn and SVM have accurate results. Furthermore, we evaluated the proposed framework on 10-fold validation to ensure the extracted feature vectors’ integrity.
3.6. Experiment 5 (10-Fold Cross Validation)
All extracted features are given to 10-fold validation, making each split during the training and validation tenfold. All the results have appeared approximately similar to the five-fold cross-validation; no significant degradation seems in any instance of performance. Results obtained using different ML classifiers on 10-fold cross-validation are presented in Table 12.

Table 12.
Results obtained using different ML classifiers on 10-fold cross validation.
In Table 12, again coarse-knn and SVM received the highest accuracy results, with more than 85% accuracy and more than 85% of precision, sensitivity, and F1-score. The prediction plot obtained using different ML classifiers on 10-fold cross validation is displayed in Figure 10.
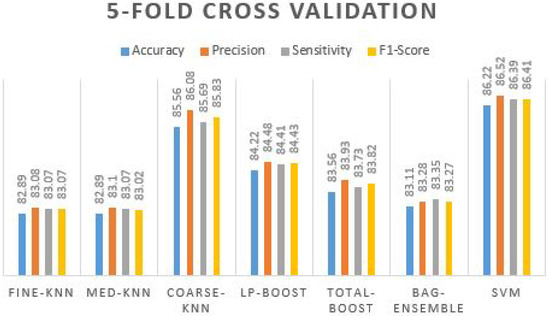
Figure 10.
Prediction plot obtained using different ML classifiers on 10-fold cross validation.
It is observed that the performance of overall classifiers is improved. This indicates that higher training folds can increase the performance of all the methods used. The SVM confusion matrix is shown in Table 13 and Table 14.

Table 13.
Confusion matrix obtained using SVM on five-fold cross validation split.

Table 14.
Confusion matrix obtained using SVM on 10-fold cross validation split.
It seems that the ‘Mix’ class is devising inaccuracy, which suggests it may contain both types of features of Neli-Ravi and Khundi. However, the Neli-Ravi and Khundi were classified more accurately.
The evaluation metrics used for the experiments are given in the Equations (9)–(12).
In evaluation measures, one statistical coefficient is used to validate the results. The value of kappa shows the kappa agreement on predicted values compared to the actual values. The kappa value remains from 0 to 1, showing confidence in the given rules. It provides no agreement on the 0–0.20 value and shows minimal agreement on 0.21 to 0.39. Additionally, it shows a weak agreement from 0.40 to 0.59. The substantial ranges start from 0.60 to 0.90 or above. This shows confident agreement upon a 0.90 to 1 value. However, the calculation is performed using a confusion matrix by obtaining predicted and actual class numbers [31].
The margin values of the calculated confusion matrix are used for the calculation of Pr (e), which is actually the major nominator value for Equation (16).
Equation (15) shows the actual calculation of Pr(e). The shows the confusion matrix margin values of all classes, whereas shows the row margin values of the confusion matrix. However, the cohen-kappa shows confidence in its 0.8 to 0.9 strong agreement. We can see from the table that the SVM has a 0.85 value of kappa coefficient, which is a far more substantial value to rely on data content or features.
4. Conclusions
Pakistan’s dairy production centers face issues in identifying and separating the Neli-Ravi breed from other buffalo breeds. As a result, automatic buffalo breed identification and classification are needed. The proposed study utilized the Pakistani buffalos data set for computer-vision-based recognition and Neli-Ravi classification from other buffalo breeds. The proposed deep CNN architecture deduced the spatial information from the buffalo images and assembled the rich feature vectors. Furthermore, a learning-based approach transfers rich features into activated layers for better classification. Several ML-based classifiers are adopted to classify the instances into relevant target classes. The confidence in the data set and features are validated using different data splits, which do not abruptly vary. All results are normalized and reliable, making the proposed research an accurate recognition method for different buffalo breeds. The proposed framework achieves a maximum accuracy of 93%, and using recent versions, it achieves more than 85% accuracy.
In the future, we intend to employ optimized deep-learning-based architectures using optimization algorithms such as genetic algorithm (G.A), bee-swarm optimization (BSO), and the differential evolution algorithm (D.E) to enhance the accuracy.
Author Contributions
Conceptualization, Y.P., H.J., J.G. and H.T.R.; methodology, Y.P. and H.J.; software, Y.P., H.J., J.G. and H.T.R.; visualization, Y.P., H.J., J.G. and H.T.R.; writing—original draft, Y.P. and H.J.; and supervision, H.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China [61902156], Zhenjiang Science and Technology Bureau; Zhenjiang Finance Bureau high-tech enterprise storage cultivation funds; Jiangsu Science and Technology Department, Jiangsu Provincial Finance Department high-tech enterprise storage cultivation funds; and Zhenjiang Hongxiang Automation Financial support from Technology Co., Ltd.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Dataset is available here https://data.mendeley.com/datasets/vdgnxsm692/2. (accessed on 1 August 2022).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sikandar, B.A.; Shi, X.; Gou, X.; Zhaobing, G.; Qing, L.; Jamal, M.A.; Khederzadeh, S.; Talpur, M.Z.; Ming, M.H. Genetic relationship and diversity analysis in four buffalo breeds of Pakistan. Pak. J. Agric. Sci. 2020, 57. [Google Scholar]
- Bilal, M.; Suleman, M.; Raziq, A. Buffalo: Black gold of Pakistan. Livest. Res. Rural Dev. 2006, 18, 140–151. [Google Scholar]
- Shah, S.K. Buffaloes of Pakistan; Pakistan Agricultural Research Council (PARC): Istanbul, Turkey, 1991.
- Luo, X.; Zhou, Y.; Zhang, B.; Zhang, Y.; Wang, X.; Feng, T.; Li, Z.; Cui, K.; Wang, Z.; Luo, C.; et al. Understanding divergent domestication traits from the whole-genome sequencing of swamp-and river-buffalo populations. Natl. Sci. Rev. 2020, 7, 686–701. [Google Scholar] [CrossRef]
- Pasha, T. Comparison between bovine and buffalo milk yield in Pakistan. Ital. J. Anim. Sci. 2007, 6, 58–66. [Google Scholar] [CrossRef]
- Murtaza, M.A.; Pandya, A.J.; Khan, M.M.H. Buffalo milk. In Handbook of Milk of Non-Bovine Mammals, 2nd ed.; Park, Y.W., Haenlein, G.F.W., Wendorff, W.I., Eds.; John and Wiley and Sons: Hoboken, NJ, USA, 2017; pp. 261–368. [Google Scholar]
- Aujla, K.M.; Hussain, A. Economics of milk production of major dairy buffalo breeds by agro-ecological zones in Pakistan. Pak. J. Agric. Res. 2015, 28. [Google Scholar]
- Sajid, I.; Babar, M.; Javed, K. Genetic diversity of Nili-Ravi from Nili and Ravi buffalo breeds of Pakistan. Ital. J. Anim. Sci. 2007, 6, 314–317. [Google Scholar] [CrossRef]
- Murase, H. Artificial intelligence in agriculture. Comput. Electron. Agric. 2000, 29, 4377–4383. [Google Scholar] [CrossRef]
- Queiroz, M.M.; Telles, R.; Bonilla, S.H. Blockchain and supply chain management integration: A systematic review of the literature. Supply Chain. Manag. Int. J. 2020, 25, 241–254. [Google Scholar] [CrossRef]
- Hossain, M.S.; Rahman, M.H.; Rahman, M.S.; Hosen, A.S.; Seo, C.; Cho, G.H. Intellectual Property Theft Protection in IoT Based Precision Agriculture Using SDN. Electronics 2021, 10, 1987. [Google Scholar] [CrossRef]
- Gao, J.; Wang, H.; Shen, H. Machine Learning Based Workload Prediction in Cloud Computing. In Proceedings of the 2020 29th International Conference on Computer Communications and Networks (ICCCN), Honolulu, HI, USA, 3–6 August 2020; IEEE: New York, NY, USA, 2020; pp. 1–9. [Google Scholar]
- Lokhorst, C.; De Mol, R.; Kamphuis, C. Invited review: Big Data in precision dairy farming. Animal 2019, 13, 1519–1528. [Google Scholar] [CrossRef]
- Faye, B.; Konuspayeva, G. The sustainability challenge to the dairy sector–The growing importance of non-cattle milk production worldwide. Int. Dairy J. 2012, 24, 50–56. [Google Scholar] [CrossRef]
- Yegnanarayana, B. Artificial Neural Networks; PHI Learning Pvt. Ltd.: New Delhi, India, 2009. [Google Scholar]
- Mijwel, M.M. Artificial Neural Networks Advantages and Disadvantages. 2018. Available online: https://www.linkedin.com/pulse/artificial-neural-networks-advantages-disadvantages-maad-m-mijwel (accessed on 1 August 2022).
- Islam, S.; Reddy, U.K.; Natarajan, P.; Abburi, V.L.; Bajwa, A.A.; Imran, M.; Zahoor, M.Y.; Abdullah, M.; Bukhari, A.M.; Iqbal, S.; et al. Population demographic history and population structure for Pakistani Nili-Ravi breeding bulls based on SNP genotyping to identify genomic regions associated with male effects for milk yield and body weight. PLoS ONE 2020, 15, e0242500. [Google Scholar] [CrossRef]
- Mirza, R.; Waheed, A.; Akhtar, M.; Khan, M.; Dilshad, S.; Faraz, A. Correlation of linear type traits with milk yield in Nili Ravi buffaloes of Pakistan. JAPS J. Anim. Plant Sci. 2020, 30, 780–785. [Google Scholar]
- Jwade, S.A.; Guzzomi, A.; Mian, A. On farm automatic sheep breed classification using deep learning. Comput. Electron. Agric. 2019, 167, 105055. [Google Scholar] [CrossRef]
- Kumar, S.; Pandey, A.; Satwik, K.S.R.; Kumar, S.; Singh, S.K.; Singh, A.K.; Mohan, A. Deep learning framework for recognition of cattle using muzzle point image pattern. Measurement 2018, 116, 1–17. [Google Scholar] [CrossRef]
- Atabay, H.A. Deep Learning for Horse Breed Recognition. CSI J. Comput. Sci. Eng. 2017, 15, 45–51. [Google Scholar]
- Ghosh, P.; Mustafi, S.; Mandal, S.N. Image-Based Goat Breed Identification and Localization Using Deep Learning. Int. J. Comput. Vis. Image Process. 2020, 10, 74–96. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, L.; Sinnott, R. A Mobile Application for Cat Detection and Breed Recognition Based on Deep Learning. In Proceedings of the 2019 IEEE 1st International Workshop on Artificial Intelligence for Mobile (AI4Mobile), Hangzhou, China, 24 February 2019; IEEE: New York, NY, USA, 2019. [Google Scholar] [CrossRef]
- Rauf, H.T.; Lali, M.I.U.; Zahoor, S.; Shah, S.Z.H.; Rehman, A.U.; Bukhari, S.A.C. Visual features based automated identification of fish species using deep convolutional neural networks. Comput. Electron. Agric. 2019, 167, 105075. [Google Scholar] [CrossRef]
- Barbedo, J.G.A.; Koenigkan, L.V.; Santos, T.T.; Santos, P.M. A Study on the Detection of Cattle in UAV Images Using Deep Learning. Sensors 2019, 19, 5436. [Google Scholar] [CrossRef]
- De Lima Weber, F.; de Moraes Weber, V.A.; Menezes, G.V.; da Silva Oliveira Junior, A.; Alves, D.A.; de Oliveira, M.V.M.; Matsubara, E.T.; Pistori, H.; de Abreu, U.G.P. Recognition of Pantaneira cattle breed using computer vision and convolutional neural networks. Comput. Electron. Agric. 2020, 175, 105548. [Google Scholar] [CrossRef]
- Meraj, T.; Rauf, H.T.; Zahoor, S.; Hassan, A.; Lali, M.I.; Ali, L.; Bukhari, S.A.C.; Shoaib, U. Lung nodules detection using semantic segmentation and classification with optimal features. Neural Comput. Appl. 2019, 33, 10737–10750. [Google Scholar] [CrossRef]
- Rauf, H.T. Buffalo-Pak: Buffalo Breed Dataset from Pakistan for Visual Features Based Classification. Mendeley Data. 2021. Available online: https://data.mendeley.com/datasets/vdgnxsm692/2 (accessed on 1 August 2022).
- Gilg, M. 2 - Representation of Networks of Wireless Sensors with a Grayscale Image: Application to Routing. In Building Wireless Sensor Networks; Femmam, S., Ed.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 31–66. [Google Scholar] [CrossRef]
- Tan, C.; Sun, F.; Kong, T.; Zhang, W.; Yang, C.; Liu, C. A survey on deep transfer learning. In Proceedings of the International Conference on Artificial Neural Networks, Rhodes, Greece, 4–7 October 2018; Springer: Berlin/Heidelberg, Germany, 2018; pp. 270–279. [Google Scholar]
- McHugh, M.L. Interrater reliability: The kappa statistic. Biochem. Medica 2012, 22, 276–282. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).