Genome-Wide Survey and Expression Analysis of B-Box Family Genes in Cucumber Reveal Their Potential Roles in Response to Diverse Abiotic and Biotic Stresses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification and Characterization of BBX Family Members in Cucumber
2.2. Multiple Sequence Alignment and Phylogenetic Analysis
2.3. Gene Structure and Conserved Motif Analysis
2.4. Chromosomal Distribution, Gene Duplication, and Cis-Acting Elements Analysis of CsBBX Genes
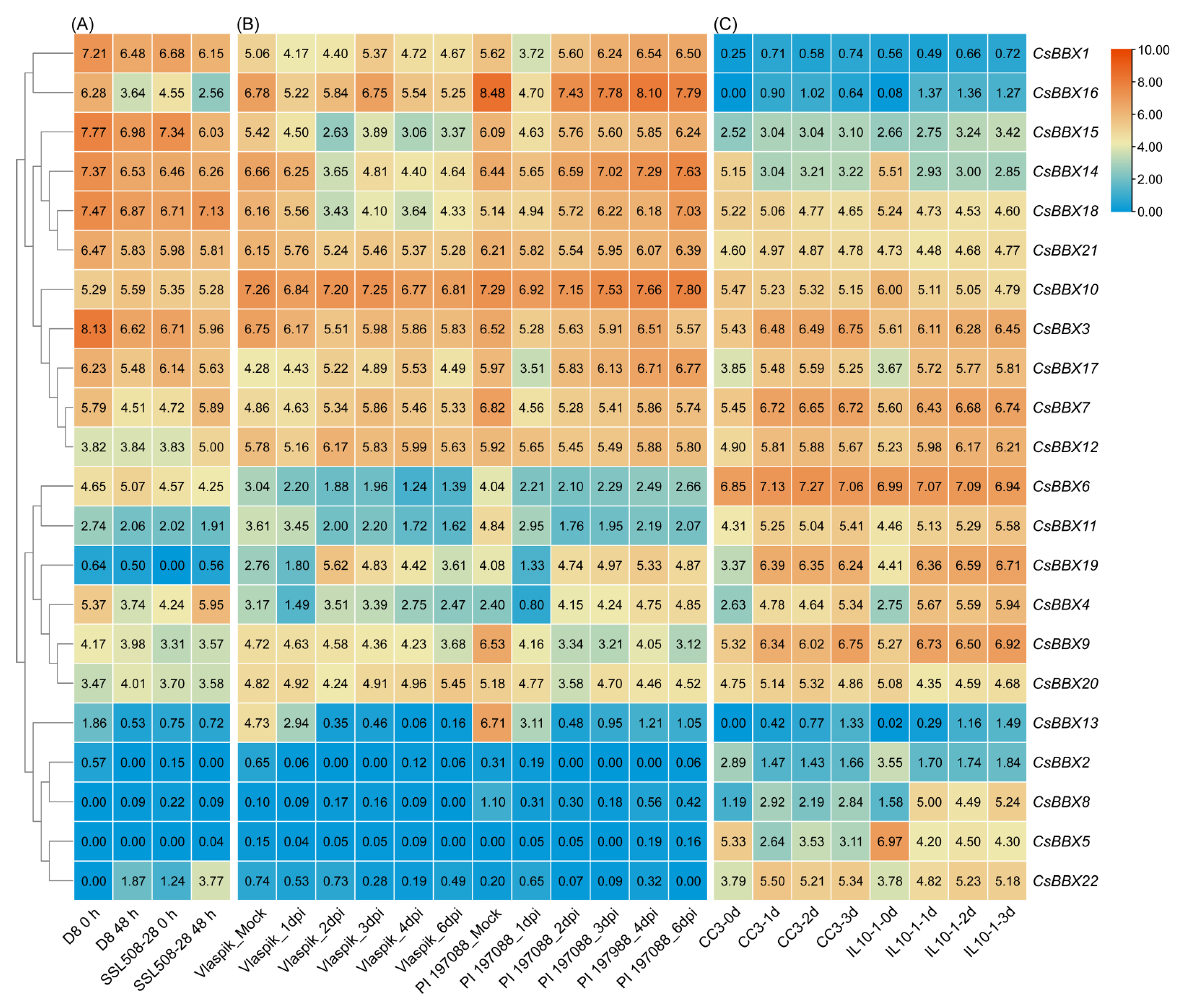
2.5. Expression Profiling of the CsBBX Genes via RNA-Seq Data
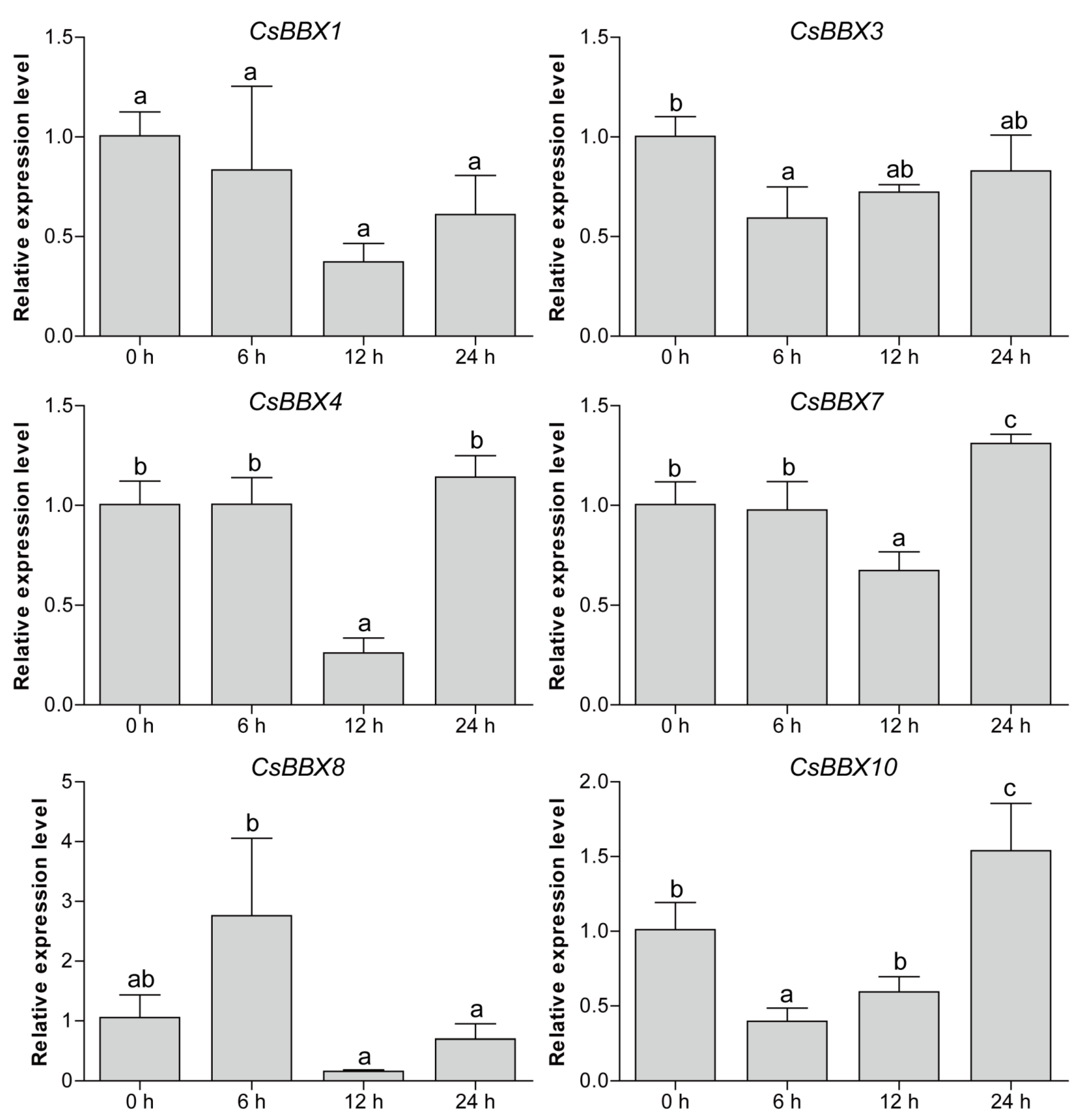
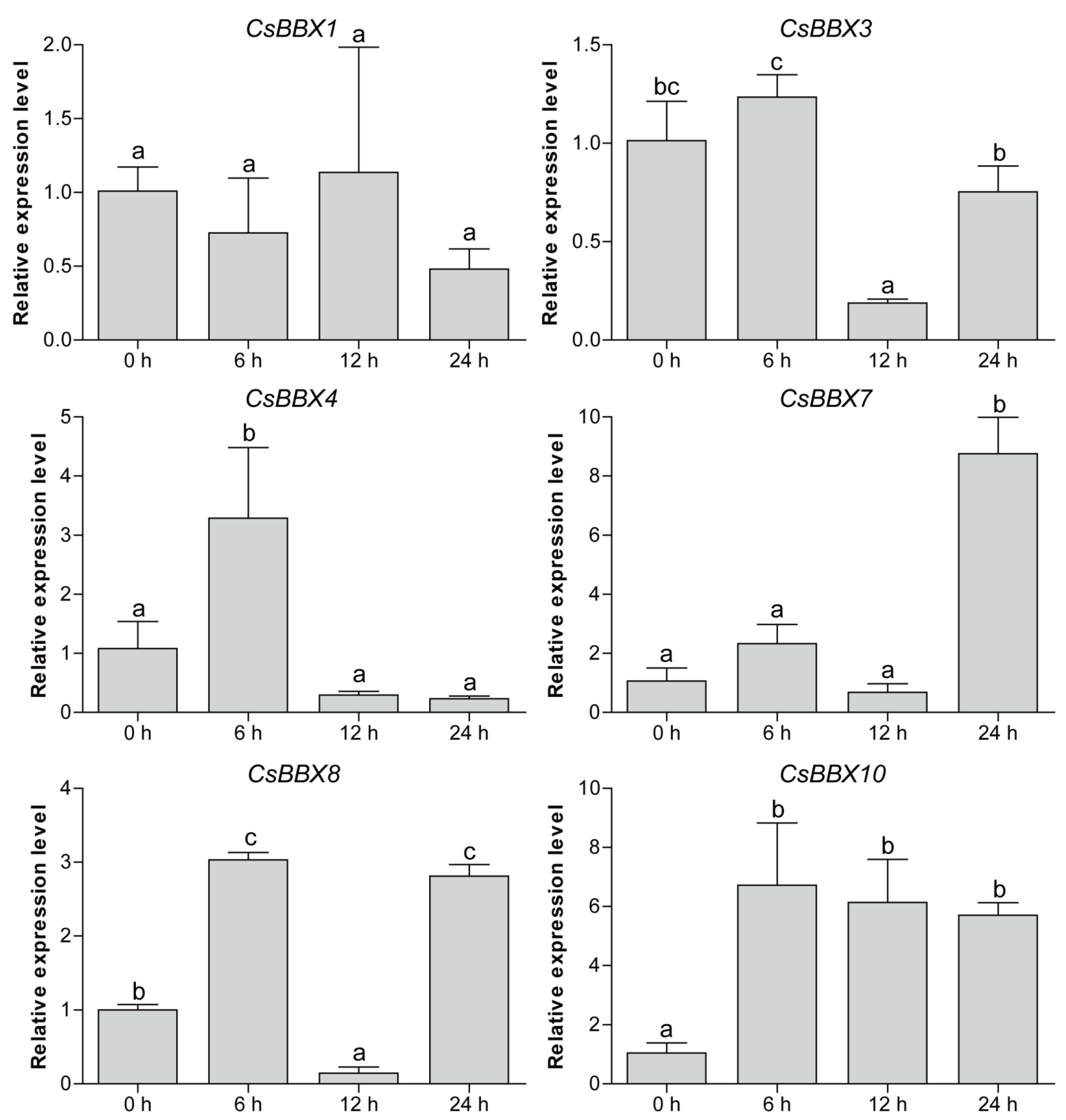
2.6. Plant Materials, Abiotic Stress Treatment, and qRT-PCR Analysis
3. Results
3.1. Identification and Characterization of CsBBX Genes in Cucumber
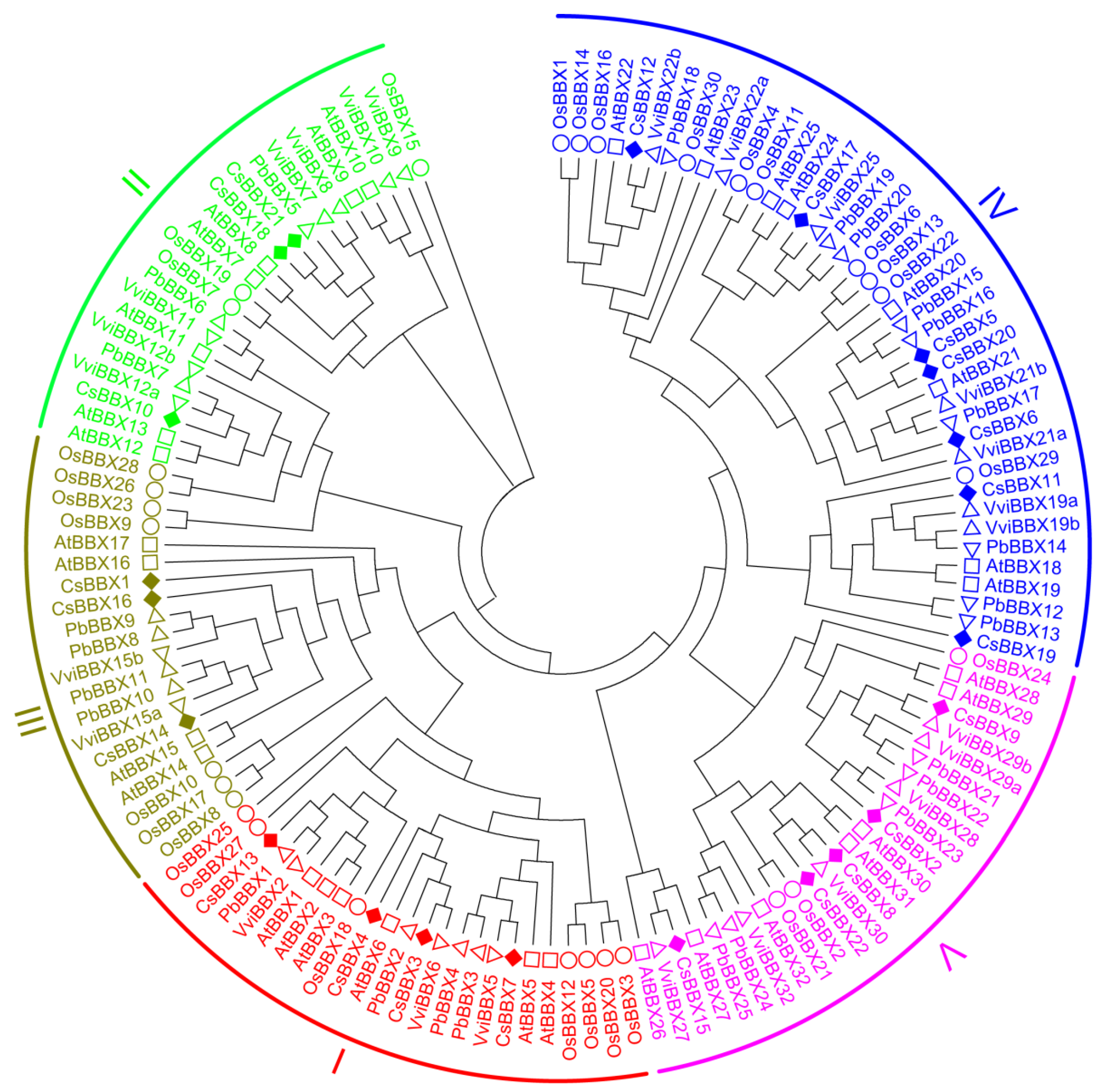
3.2. Phylogenetic Analysis of BBX Family Members
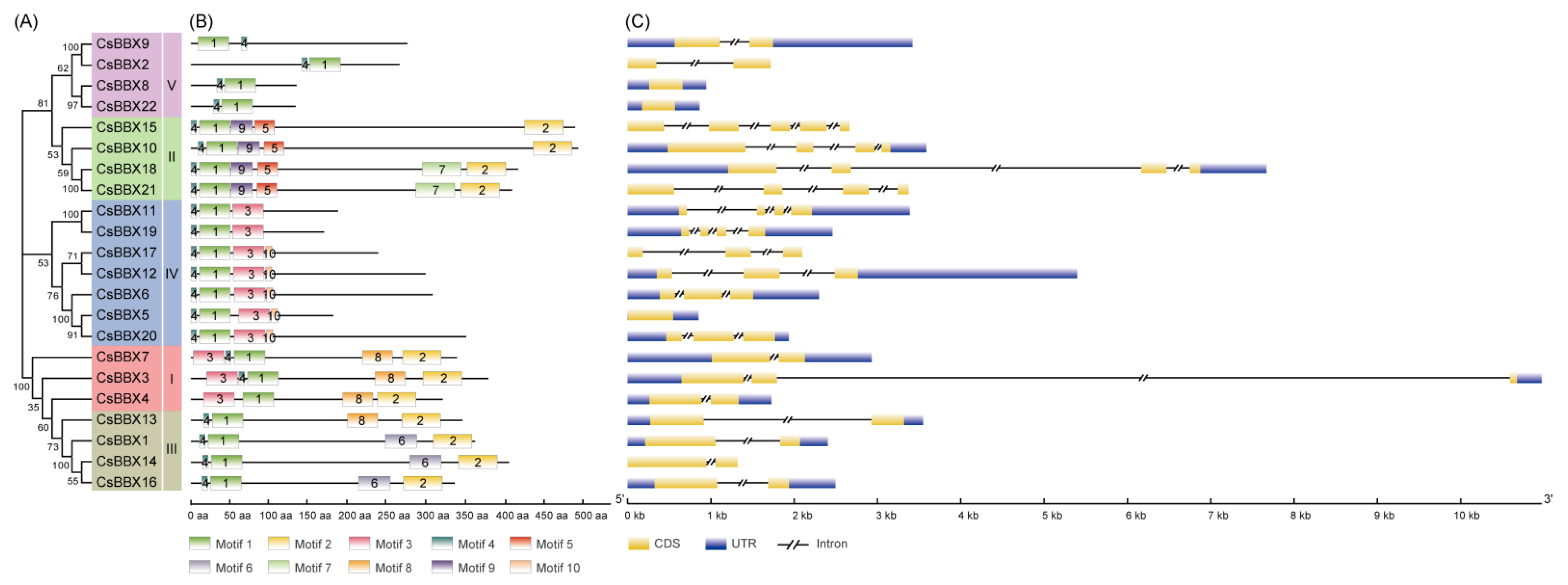
3.3. Protein Motif and Gene Structure Analysis of BBX Members in Cucumber
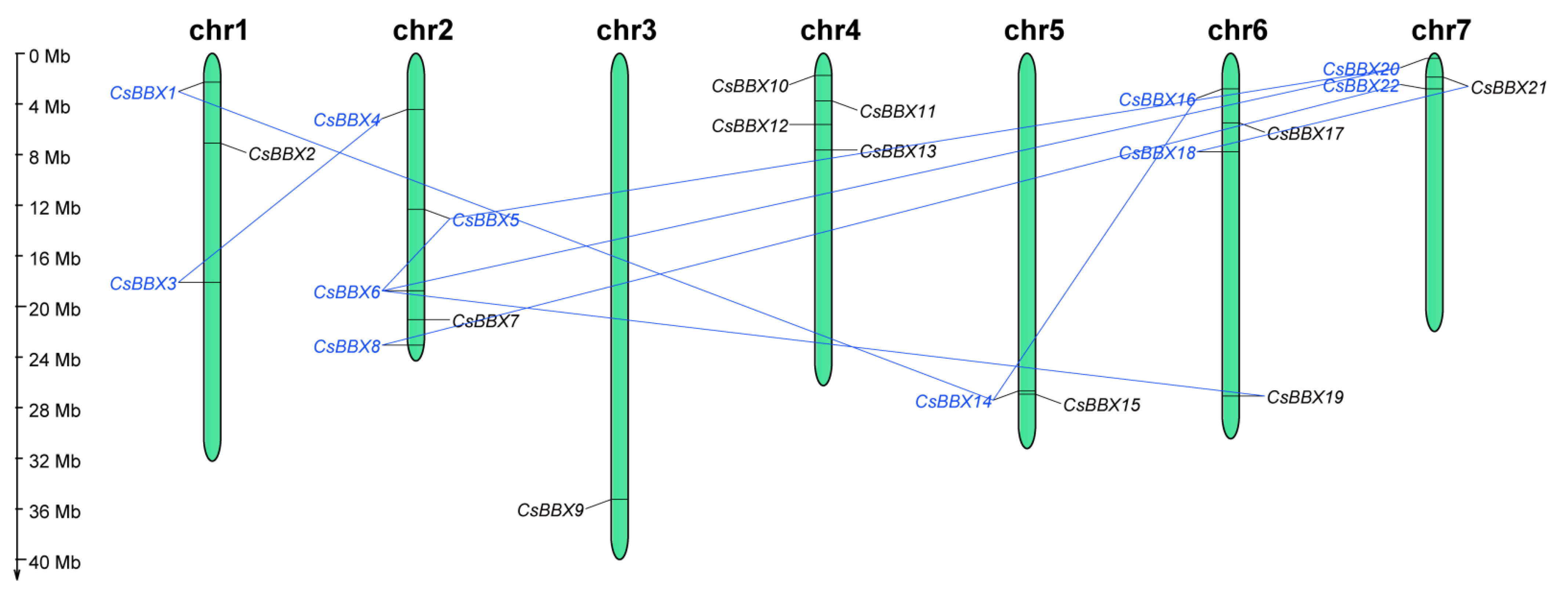
3.4. Chromosomal Location and Gene Duplication of the CsBBX Genes
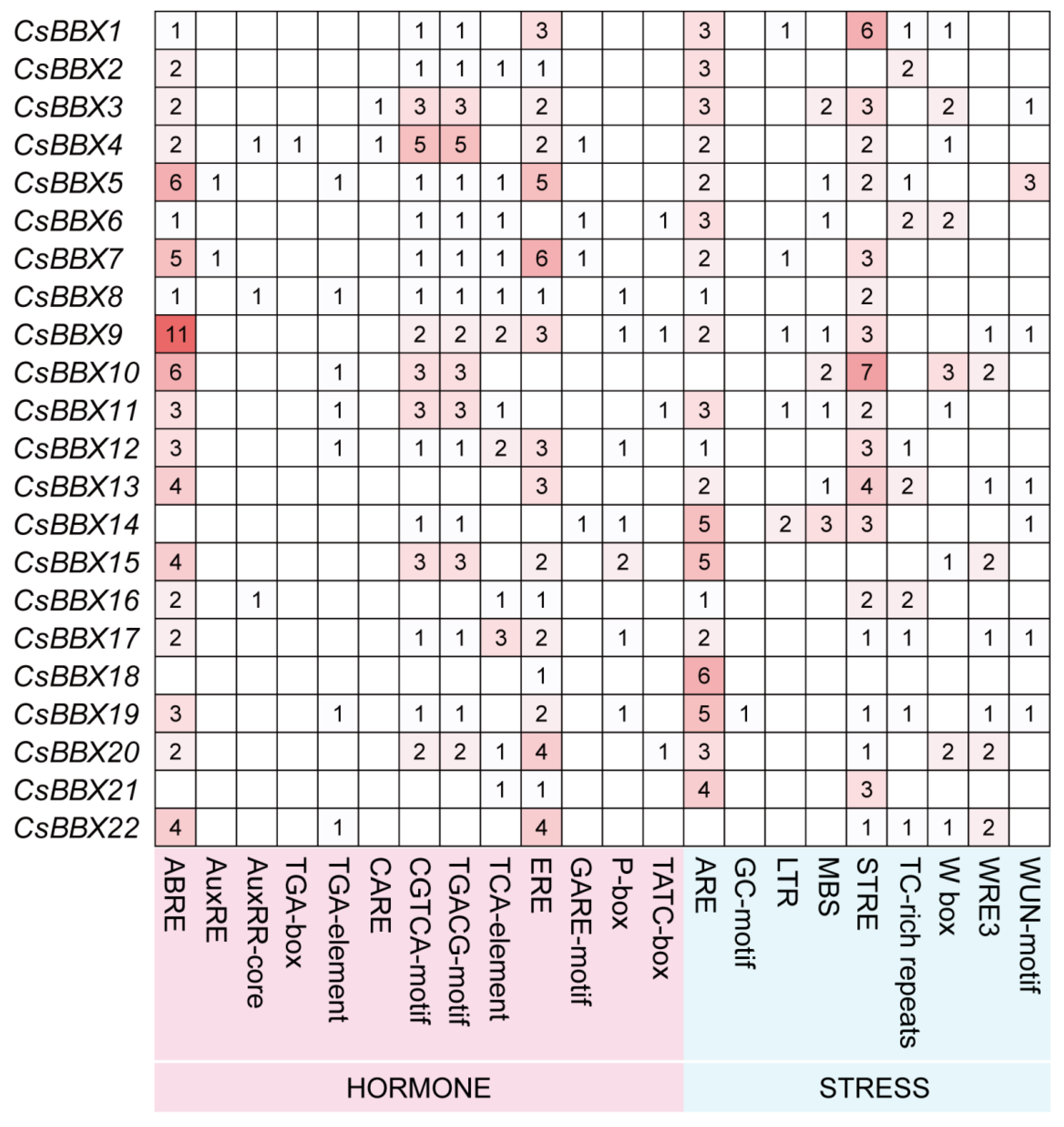
3.5. Cis-Acting Elements in the Promoter Regions of the CsBBX Genes
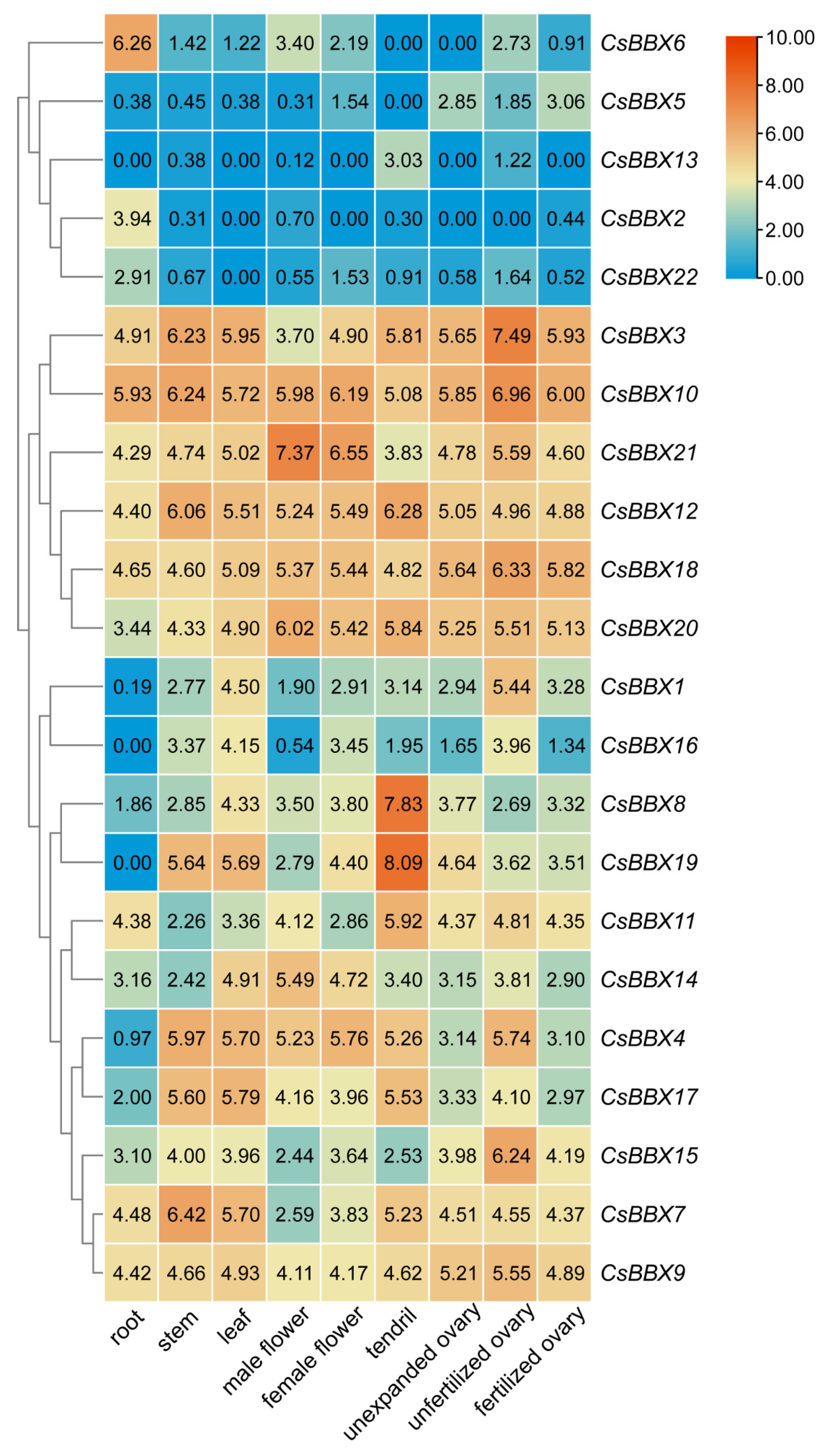
3.6. Expression Patterns of CsBBX Genes in Different Tissues of Cucumber
3.7. Expression Patterns of CsBBX Genes under Multiple Abiotic Stresses
3.8. Expression Patterns of CsBBX Genes in Response to Biotic Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gangappa, S.N.; Botto, J.F. The BBX family of plant transcription factors. Trends Plant Sci. 2014, 19, 460–470. [Google Scholar] [CrossRef] [PubMed]
- Crocco, C.D.; Botto, J.F. BBX proteins in green plants: Insights into their evolution, structure, feature and functional diversification. Gene 2013, 531, 44–52. [Google Scholar] [CrossRef]
- Khanna, R.; Kronmiller, B.; Maszle, D.R.; Coupland, G.; Holm, M.; Mizuno, T.; Wu, S.H. The Arabidopsis B-box zinc finger family. Plant Cell 2009, 21, 3416–3420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graeff, M.; Straub, D.; Eguen, T.; Dolde, U.; Rodrigues, V.; Brandt, R.; Wenkel, S. MicroProtein-mediated recruitment of CONSTANS into a TOPLESS trimeric complex represses flowering in Arabidopsis. PLoS Genet. 2016, 12, e1005959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, L.; Wang, S.; Song, Z.T.; Jiang, Y.; Han, J.J.; Lu, S.J.; Li, L.; Liu, J.X. Two B-Box domain proteins, BBX18 and BBX23, interact with ELF3 and regulate thermomorphogenesis in Arabidopsis. Cell Rep. 2018, 25, 1718–1728.e1714. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Zhao, X.; Aiwaili, P.; Mu, X.; Zhao, M.; Zhao, J.; Cheng, L.; Ma, C.; Gao, J.; Hong, B. A zinc finger protein BBX19 interacts with ABF3 to affect drought tolerance negatively in chrysanthemum. Plant J. 2020, 103, 1783–1795. [Google Scholar] [CrossRef]
- Bursch, K.; Toledo-Ortiz, G.; Pireyre, M.; Lohr, M.; Braatz, C.; Johansson, H. Identification of BBX proteins as rate-limiting cofactors of HY5. Nat. Plants 2020, 6, 921–928. [Google Scholar] [CrossRef]
- Song, Z.; Yan, T.; Liu, J.; Bian, Y.; Heng, Y.; Lin, F.; Jiang, Y.; Wang Deng, X.; Xu, D. BBX28/BBX29, HY5 and BBX30/31 form a feedback loop to fine-tune photomorphogenic development. Plant J. 2020, 104, 377–390. [Google Scholar] [CrossRef]
- Gangappa, S.N.; Crocco, C.D.; Johansson, H.; Datta, S.; Hettiarachchi, C.; Holm, M.; Botto, J.F. The Arabidopsis B-BOX protein BBX25 interacts with HY5, negatively regulating BBX22 expression to suppress seedling photomorphogenesis. Plant Cell 2013, 25, 1243–1257. [Google Scholar] [CrossRef] [Green Version]
- Shen, C.; Liu, H.; Guan, Z.; Yan, J.; Zheng, T.; Yan, W.; Wu, C.; Zhang, Q.; Yin, P.; Xing, Y. Structural insight into DNA recognition by CCT/NF-YB/YC complexes in plant photoperiodic flowering. Plant Cell 2020, 32, 3469–3484. [Google Scholar] [CrossRef]
- Lv, X.; Zeng, X.; Hu, H.; Chen, L.; Zhang, F.; Liu, R.; Liu, Y.; Zhou, X.; Wang, C.; Wu, Z.; et al. Structural insights into the multivalent binding of the Arabidopsis FLOWERING LOCUS T promoter by the CO-NF-Y master transcription factor complex. Plant Cell 2021, 33, 1182–1195. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Lyu, Z.; Liu, H.; Zhang, G.; He, C.; Zhang, J. Insights into the evolutionary origin and expansion of the BBX gene family. Plant Biotechnol. Rep. 2022, 16, 205–214. [Google Scholar] [CrossRef]
- Yadukrishnan, P.; Job, N.; Johansson, H.; Datta, S. Opposite roles of group IV BBX proteins: Exploring missing links between structural and functional diversity. Plant Signal. Behav. 2018, 13, e1462641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yadav, A.; Ravindran, N.; Singh, D.; Rahul, P.V.; Datta, S. Role of Arabidopsis BBX proteins in light signaling. J. Plant Biochem. Biotechnol. 2020, 29, 623–635. [Google Scholar] [CrossRef]
- Bai, S.; Tao, R.; Yin, L.; Ni, J.; Yang, Q.; Yan, X.; Yang, F.; Guo, X.; Li, H.; Teng, Y. Two B-box proteins, PpBBX18 and PpBBX21, antagonistically regulate anthocyanin biosynthesis via competitive association with Pyrus pyrifolia ELONGATED HYPOCOTYL 5 in the peel of pear fruit. Plant J. 2019, 100, 1208–1223. [Google Scholar] [CrossRef]
- Bai, S.; Tao, R.; Tang, Y.; Yin, L.; Ma, Y.; Ni, J.; Yan, X.; Yang, Q.; Wu, Z.; Zeng, Y.; et al. BBX16, a B-box protein, positively regulates light-induced anthocyanin accumulation by activating MYB10 in red pear. Plant Biotechnol. J. 2019, 17, 1985–1997. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Zhu, Z.Z.; Qu, D.; Wang, B.C.; Hao, N.N.; Yang, Y.Z.; Yang, H.J.; Zhao, Z.Y. MdBBX21, a B-Box protein, positively regulates light-induced anthocyanin accumulation in apple peel. Front. Plant Sci. 2021, 12, 774446. [Google Scholar] [CrossRef]
- Zhang, B.; Yang, H.J.; Qu, D.; Zhu, Z.Z.; Yang, Y.Z.; Zhao, Z.Y. The MdBBX22-miR858-MdMYB9/11/12 module regulates proanthocyanidin biosynthesis in apple peel. Plant Biotechnol. J. 2022. [Google Scholar] [CrossRef]
- An, J.P.; Wang, X.F.; Zhang, X.W.; Bi, S.Q.; You, C.X.; Hao, Y.J. MdBBX22 regulates UV-B-induced anthocyanin biosynthesis through regulating the function of MdHY5 and is targeted by MdBT2 for 26S proteasome-mediated degradation. Plant Biotechnol. J. 2019, 17, 2231–2233. [Google Scholar] [CrossRef]
- Bai, M.; Sun, J.; Liu, J.; Ren, H.; Wang, K.; Wang, Y.; Wang, C.; Dehesh, K. The B-box protein BBX19 suppresses seed germination via induction of ABI5. Plant J. 2019, 99, 1192–1202. [Google Scholar] [CrossRef]
- Crocco, C.D.; Locascio, A.; Escudero, C.M.; Alabadí, D.; Blázquez, M.A.; Botto, J.F. The transcriptional regulator BBX24 impairs DELLA activity to promote shade avoidance in Arabidopsis thaliana. Nat. Commun. 2015, 6, 6202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Chen, H.; Ping, Q.; Zhang, Z.; Guan, Z.; Fang, W.; Chen, S.; Chen, F.; Jiang, J.; Zhang, F. The heterologous expression of CmBBX22 delays leaf senescence and improves drought tolerance in Arabidopsis. Plant Cell Rep. 2019, 38, 15–24. [Google Scholar] [CrossRef] [PubMed]
- An, J.P.; Zhang, C.L.; Li, H.L.; Wang, G.L.; You, C.X. Apple SINA E3 ligase MdSINA3 negatively mediates JA-triggered leaf senescence by ubiquitinating and degrading the MdBBX37 protein. Plant J. 2022. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Yu, Y.; Liu, M.; Song, Y.; Li, H.; Sun, J.; Wang, Q.; Xie, Q.; Wang, L.; Xu, X. BBX19 fine-tunes the circadian rhythm by interacting with PSEUDO-RESPONSE REGULATOR proteins to facilitate their repressive effect on morning-phased clock genes. Plant Cell 2021, 33, 2602–2617. [Google Scholar] [CrossRef] [PubMed]
- Hwang, G.; Park, J.; Kim, S.; Park, J.; Seo, D.; Oh, E. Overexpression of BBX18 promotes thermomorphogenesis through the PRR5-PIF4 pathway. Front. Plant Sci. 2021, 12, 782352. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Wang, Q.; Li, W.; Hu, T.; Wang, Q.; Yin, Y.; Liu, X.; He, S.; Zhang, M.; Liang, Y.; et al. Overexpression of SlBBX17 affects plant growth and enhances heat tolerance in tomato. Int. J. Biol. Macromol. 2022, 206, 799–811. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Lu, Y.; Zhou, Z.; Wang, X.; Ge, H.; Sun, Q. B-box containing protein 1 from Malus domestica (MdBBX1) is involved in the abiotic stress response. PeerJ 2022, 10, e12852. [Google Scholar] [CrossRef]
- Liu, X.; Li, R.; Dai, Y.; Yuan, L.; Sun, Q.; Zhang, S.; Wang, X. A B-box zinc finger protein, MdBBX10, enhanced salt and drought stresses tolerance in Arabidopsis. Plant Mol. Biol. 2019, 99, 437–447. [Google Scholar] [CrossRef]
- Zhou, Y.; Ge, L.; Li, G.; He, P.; Yang, Y.; Liu, S. In silico identification and expression analysis of Rare Cold Inducible 2 (RCI2) gene family in cucumber. J. Plant Biochem. Biot. 2020, 29, 56–66. [Google Scholar] [CrossRef]
- Lai, W.; Zhu, C.; Yang, S.; Hu, Z.; Liu, S.; Zhou, Y. Comprehensive identification of the VQ family genes in cucumber and their roles in response to abiotic and biotic stresses. Sci. Hortic. 2022, 295, 110874. [Google Scholar] [CrossRef]
- Huang, J.; Zhao, X.; Weng, X.; Wang, L.; Xie, W. The rice B-box zinc finger gene family: Genomic identification, characterization, expression profiling and diurnal analysis. PLoS ONE 2012, 7, e48242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Talar, U.; Kiełbowicz-Matuk, A.; Czarnecka, J.; Rorat, T. Genome-wide survey of B-box proteins in potato (Solanum tuberosum)—Identification, characterization and expression patterns during diurnal cycle, etiolation and de-etiolation. PLoS ONE 2017, 12, e0177471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Li, R.; Dai, Y.; Chen, X.; Wang, X. Genome-wide identification and expression analysis of the B-box gene family in the apple (Malus domestica Borkh.) genome. Mol. Genet. Genom. 2018, 293, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Meng, D.; Han, Y.; Chen, T.; Jiao, C.; Chen, Y.; Jin, Q.; Cai, Y. Comparative analysis of B-BOX genes and their expression pattern analysis under various treatments in Dendrobium officinale. BMC Plant Biol. 2019, 19, 245. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Chen, J.; Huang, B.; Huang, Z.; Zhang, Z. The BBX gene family in Moso bamboo (Phyllostachys edulis): Identification, characterization and expression profiles. BMC Genom. 2021, 22, 533. [Google Scholar] [CrossRef]
- Liu, W.; Tang, R.; Zhang, Y.; Liu, X.; Gao, Y.; Dai, Z.; Li, S.; Wu, B.; Wang, L. Genome-wide identification of B-box proteins and VvBBX44 involved in light-induced anthocyanin biosynthesis in grape (Vitis vinifera L.). Planta 2021, 253, 114. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, L.; Ji, M.; Wu, Y.; Zhang, S.; Zhu, Y.; Yao, J.; Li, Z.; Gao, H.; Wang, X. Genome-wide identification and expression analysis of the B-box transcription factor gene family in grapevine (Vitis vinifera L.). BMC Genom. 2021, 22, 221. [Google Scholar] [CrossRef]
- Wei, H.; Wang, P.; Chen, J.; Li, C.; Wang, Y.; Yuan, Y.; Fang, J.; Leng, X. Genome-wide identification and analysis of B-BOX gene family in grapevine reveal its potential functions in berry development. BMC Plant Biol. 2020, 20, 72. [Google Scholar] [CrossRef]
- Wang, J.; Yang, G.; Chen, Y.; Dai, Y.; Yuan, Q.; Shan, Q.; Pan, L.; Dai, L.; Zou, X.; Liu, F.; et al. Genome-wide characterization and anthocyanin-related expression analysis of the B-BOX gene family in Capsicum annuum L. Front. Genet. 2022, 13, 847328. [Google Scholar] [CrossRef]
- Ma, J.; Dai, J.X.; Liu, X.W.; Lin, D. Genome-wide and expression analysis of B-box gene family in pepper. BMC Genom. 2021, 22, 883. [Google Scholar] [CrossRef]
- Obel, H.O.; Cheng, C.; Li, Y.; Tian, Z.; Njogu, M.K.; Li, J.; Lou, Q.; Yu, X.; Yang, Z.; Ogweno, J.O.; et al. Genome-wide identification of the B-Box gene family and expression analysis suggests their potential role in photoperiod-mediated β-carotene accumulation in the endocarp of cucumber (Cucumis sativus L.) fruit. Genes 2022, 13, 658. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, X.; Han, J.; Lu, W.; Ren, Z. Genome-wide analysis of the WRKY gene family in the cucumber genome and transcriptome-wide identification of WRKY transcription factors that respond to biotic and abiotic stresses. BMC Plant Biol. 2020, 20, 443. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.X.; Yang, L.; Liu, N.; Yang, J.; Zhou, X.K.; Xia, Y.C.; He, Y.; He, Y.Q.; Gong, H.J.; Ma, D.F.; et al. Genome-wide identification, structure characterization, and expression pattern profiling of aquaporin gene family in cucumber. BMC Plant Biol. 2019, 19, 345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Yin, J.; Liang, Y.; Liu, J.; Jia, J.; Huo, H.; Wu, Z.; Yang, R.; Gong, H. Transcriptomic dynamics provide an insight into the mechanism for silicon-mediated alleviation of salt stress in cucumber plants. Ecotoxicol. Environ. Saf. 2019, 174, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Xu, X.; Shi, Y.; Qi, X.; Chen, X. Elucidation of the molecular responses of a cucumber segment substitution line carrying Pm5.1 and its recurrent parent triggered by powdery mildew by comparative transcriptome profiling. BMC Genom. 2017, 18, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burkhardt, A.; Day, B. Transcriptome and small RNAome dynamics during a resistant and susceptible interaction between cucumber and downy mildew. Plant Genome 2016, 9, 1–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Cheng, C.; Zhang, K.; Tian, Z.; Xu, J.; Yang, S.; Lou, Q.; Li, J.; Chen, J.F. Comparative transcriptomics reveals suppressed expression of genes related to auxin and the cell cycle contributes to the resistance of cucumber against Meloidogyne incognita. BMC Genom. 2018, 19, 583. [Google Scholar] [CrossRef]
- Zhou, Y.; Ouyang, L.; Zhou, D.; Cai, Y.; He, H. Superoxide dismutase family genes in watermelon and their responses to different abiotic stresses. Front. Agric. Sci. Eng. 2021, 8, 645–658. [Google Scholar] [CrossRef]
- Cao, Y.; Han, Y.; Meng, D.; Li, D.; Jiao, C.; Jin, Q.; Lin, Y.; Cai, Y. B-BOX genes: Genome-wide identification, evolution and their contribution to pollen growth in pear (Pyrus bretschneideri Rehd.). BMC Plant Biol. 2017, 17, 156. [Google Scholar] [CrossRef] [Green Version]
- Lyu, G.; Li, D.; Li, S. Bioinformatics analysis of BBX family genes and its response to UV-B in Arabidopsis thaliana. Plant Signal. Behav. 2020, 15, 1782647. [Google Scholar] [CrossRef]
- Bu, X.; Wang, X.; Yan, J.; Zhang, Y.; Zhou, S.; Sun, X.; Yang, Y.; Ahammed, G.J.; Liu, Y.; Qi, M.; et al. Genome-wide characterization of B-Box gene family and its roles in responses to light quality and cold stress in tomato. Front. Plant Sci. 2021, 12, 698525. [Google Scholar] [CrossRef] [PubMed]
- Shalmani, A.; Jing, X.Q.; Shi, Y.; Muhammad, I.; Zhou, M.R.; Wei, X.Y.; Chen, Q.Q.; Li, W.Q.; Liu, W.T.; Chen, K.M. Characterization of B-BOX gene family and their expression profiles under hormonal, abiotic and metal stresses in Poaceae plants. BMC Genom. 2019, 20, 27. [Google Scholar] [CrossRef] [PubMed]
- Wen, S.; Zhang, Y.; Deng, Y.; Chen, G.; Yu, Y.; Wei, Q. Genomic identification and expression analysis of the BBX transcription factor gene family in Petunia hybrida. Mol. Biol. Rep. 2020, 47, 6027–6041. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Jiang, W.; Yin, J.; Wang, S.; Fang, Z.; Ma, D.; Gao, D. Genome-wide mining of wheat B-BOX zinc finger (BBX) gene family provides new insights into light stress responses. Crop Pasture Sci. 2021, 72, 17–37. [Google Scholar] [CrossRef]
- Chu, Z.; Wang, X.; Li, Y.; Yu, H.; Li, J.; Lu, Y.; Li, H.; Ouyang, B. Genomic organization, phylogenetic and expression analysis of the B-BOX gene family in tomato. Front. Plant Sci. 2016, 7, 1552. [Google Scholar] [CrossRef] [Green Version]
- Feng, Z.; Li, M.; Li, Y.; Yang, X.; Wei, H.; Fu, X.; Ma, L.; Lu, J.; Wang, H.; Yu, S. Comprehensive identification and expression analysis of B-Box genes in cotton. BMC Genom. 2021, 22, 439. [Google Scholar] [CrossRef]
- Singh, S.; Chhapekar, S.S.; Ma, Y.; Rameneni, J.J.; Oh, S.H.; Kim, J.; Lim, Y.P.; Choi, S.R. Genome-wide identification, evolution, and comparative analysis of B-Box genes in Brassica rapa, B. oleracea, and B. napus and their expression profiling in b. rapa in response to multiple hormones and abiotic stresses. Int. J. Mol. Sci. 2021, 22, 10367. [Google Scholar] [CrossRef]
- Wu, H.; Wang, X.; Cao, Y.; Zhang, H.; Hua, R.; Liu, H.; Sui, S. CpBBX19, a B-box transcription factor gene of Chimonanthus praecox, improves salt and drought tolerance in Arabidopsis. Genes 2021, 12, 1456. [Google Scholar] [CrossRef]
- An, J.P.; Wang, X.F.; Zhang, X.W.; You, C.X.; Hao, Y.J. Apple B-box protein BBX37 regulates jasmonic acid mediated cold tolerance through the JAZ-BBX37-ICE1-CBF pathway and undergoes MIEL1-mediated ubiquitination and degradation. New Phytol. 2021, 229, 2707–2729. [Google Scholar] [CrossRef]
- Liu, H.; Dong, S.; Sun, D.; Liu, W.; Gu, F.; Liu, Y.; Guo, T.; Wang, H.; Wang, J.; Chen, Z. CONSTANS-Like 9 (OsCOL9) interacts with Receptor for Activated C-Kinase 1 (OsRACK1) to regulate blast resistance through salicylic acid and ethylene signaling pathways. PLoS ONE 2016, 11, e0166249. [Google Scholar] [CrossRef]
- Hou, W.; Ren, L.; Zhang, Y.; Sun, H.; Shi, T.; Gu, Y.; Wang, A.; Ma, D.; Li, Z.; Zhang, L. Characterization of BBX family genes and their expression profiles under various stresses in the sweet potato wild ancestor Ipomoea trifida. Sci. Hortic. 2021, 288, 110374. [Google Scholar] [CrossRef]

| Gene | Accession No. (v3) | Chromosome: Location | Protein | Group | |||||
|---|---|---|---|---|---|---|---|---|---|
| CDS/bp | AA | pI | MW/kDa | GRAVY | Subcellular Localization | ||||
| CsBBX1 | CsaV3_1G003800 | chr1: 2,364,830–2,367,233 | 1083 | 360 | 5.25 | 41.63 | −0.949 | Nucleus | III |
| CsBBX2 | CsaV3_1G011690 | chr1: 7,244,708–7,246,426 | 795 | 264 | 5.29 | 29.49 | −0.316 | Nucleus | V |
| CsBBX3 | CsaV3_1G031230 | chr1: 18,437,778–18,448,748 | 1134 | 377 | 5.85 | 41.24 | −0.308 | Nucleus | I |
| CsBBX4 | CsaV3_2G008220 | chr2: 4,503,501–4,505,228 | 960 | 319 | 8.20 | 35.16 | −0.395 | Nucleus | I |
| CsBBX5 | CsaV3_2G015160 | chr2: 12,644,521–12,645,374 | 543 | 180 | 6.64 | 19.76 | −0.301 | Nucleus | IV |
| CsBBX6 | CsaV3_2G029230 | chr2: 19,152,113–19,154,409 | 921 | 306 | 6.65 | 32.67 | −0.221 | Nucleus | IV |
| CsBBX7 | CsaV3_2G032640 | chr2: 21,557,067–21,559,996 | 1014 | 337 | 6.08 | 36.93 | −0.505 | Nucleus | I |
| CsBBX8 | CsaV3_2G035230 | chr2: 23,569,718–23,570,660 | 402 | 133 | 8.40 | 14.66 | −0.041 | Nucleus | V |
| CsBBX9 | CsaV3_3G044220 | chr3: 36,085,222–36,088,641 | 825 | 274 | 4.31 | 30.15 | −0.829 | Nucleus | V |
| CsBBX10 | CsaV3_4G002870 | chr4: 1,790,472–1,794,059 | 1476 | 491 | 5.50 | 54.36 | −0.545 | Nucleus | II |
| CsBBX11 | CsaV3_4G005810 | chr4: 3,819,528–3,822,914 | 561 | 186 | 6.44 | 20.76 | −0.605 | Nucleus | IV |
| CsBBX12 | CsaV3_4G008210 | chr4: 5,753,161–5,758,556 | 894 | 297 | 5.31 | 32.30 | −0.384 | Nucleus | IV |
| CsBBX13 | CsaV3_4G009980 | chr4: 7,751,656–7,755,202 | 1035 | 344 | 5.24 | 38.47 | −0.772 | Nucleus | III |
| CsBBX14 | CsaV3_5G034320 | chr5: 27,321,989–27,323,306 | 1212 | 403 | 5.55 | 45.47 | −0.710 | Nucleus | III |
| CsBBX15 | CsaV3_5G034710 | chr5: 27,519,572–27,522,236 | 1464 | 487 | 6.81 | 54.84 | −0.614 | Nucleus | II |
| CsBBX16 | CsaV3_6G003540 | chr6: 2,826,773–2,829,268 | 1005 | 334 | 8.33 | 38.40 | −0.823 | Nucleus | III |
| CsBBX17 | CsaV3_6G006790 | chr6: 5,555,910–5,558,007 | 714 | 237 | 4.89 | 26.09 | −0.285 | Nucleus | IV |
| CsBBX18 | CsaV3_6G009750 | chr6: 7,889,202–7,896,870 | 1248 | 415 | 5.14 | 45.47 | −0.481 | Nucleus | II |
| CsBBX19 | CsaV3_6G046900 | chr6: 27,677,337–27,679,797 | 507 | 168 | 6.59 | 18.86 | −0.567 | Nucleus | IV |
| CsBBX20 | CsaV3_7G000270 | chr7: 395,757–397,689 | 1050 | 349 | 5.91 | 39.33 | −0.359 | Nucleus | IV |
| CsBBX21 | CsaV3_7G002350 | chr7: 1,863,318–1,866,691 | 1224 | 407 | 5.42 | 44.56 | −0.520 | Nucleus | II |
| CsBBX22 | CsaV3_7G003780 | chr7: 2,810,583–2,811,446 | 399 | 132 | 6.40 | 14.63 | −0.242 | Nucleus | V |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, C.; Xiao, L.; Hu, Y.; Liu, L.; Liu, H.; Hu, Z.; Liu, S.; Zhou, Y. Genome-Wide Survey and Expression Analysis of B-Box Family Genes in Cucumber Reveal Their Potential Roles in Response to Diverse Abiotic and Biotic Stresses. Agriculture 2022, 12, 827. https://doi.org/10.3390/agriculture12060827
Zhu C, Xiao L, Hu Y, Liu L, Liu H, Hu Z, Liu S, Zhou Y. Genome-Wide Survey and Expression Analysis of B-Box Family Genes in Cucumber Reveal Their Potential Roles in Response to Diverse Abiotic and Biotic Stresses. Agriculture. 2022; 12(6):827. https://doi.org/10.3390/agriculture12060827
Chicago/Turabian StyleZhu, Chuxia, Lingdi Xiao, Yaqi Hu, Liu Liu, Haoju Liu, Zhaoyang Hu, Shiqiang Liu, and Yong Zhou. 2022. "Genome-Wide Survey and Expression Analysis of B-Box Family Genes in Cucumber Reveal Their Potential Roles in Response to Diverse Abiotic and Biotic Stresses" Agriculture 12, no. 6: 827. https://doi.org/10.3390/agriculture12060827
APA StyleZhu, C., Xiao, L., Hu, Y., Liu, L., Liu, H., Hu, Z., Liu, S., & Zhou, Y. (2022). Genome-Wide Survey and Expression Analysis of B-Box Family Genes in Cucumber Reveal Their Potential Roles in Response to Diverse Abiotic and Biotic Stresses. Agriculture, 12(6), 827. https://doi.org/10.3390/agriculture12060827






