Reproductive Biology Factors Hampering Lemon [Citrus limon (L.) Burm. f.] Genetic Improvement
Abstract
1. Introduction
2. Materials and Methods
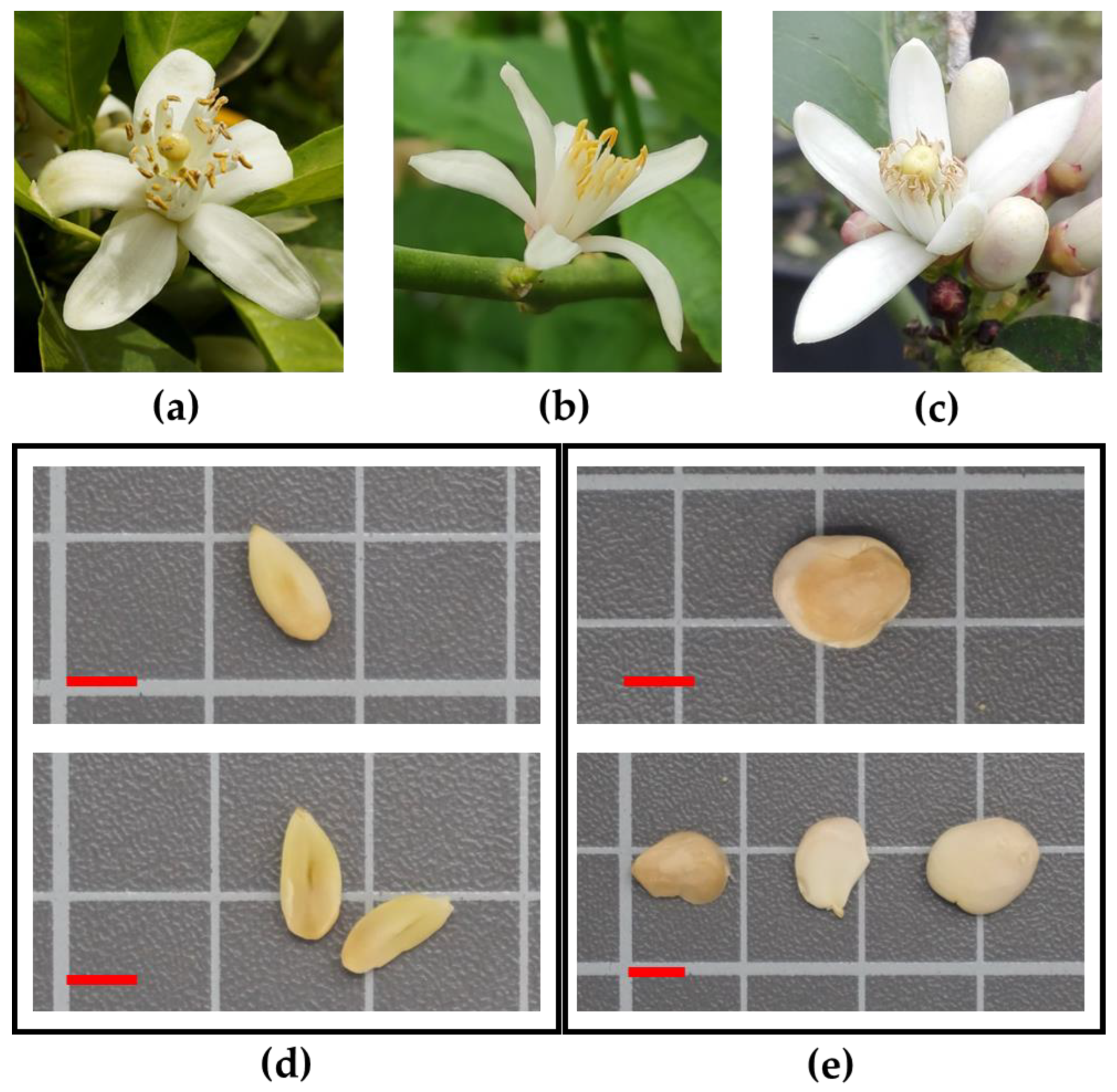
2.1. Plant Material
2.2. Flower and Seed Analysis
2.3. DNA Extraction
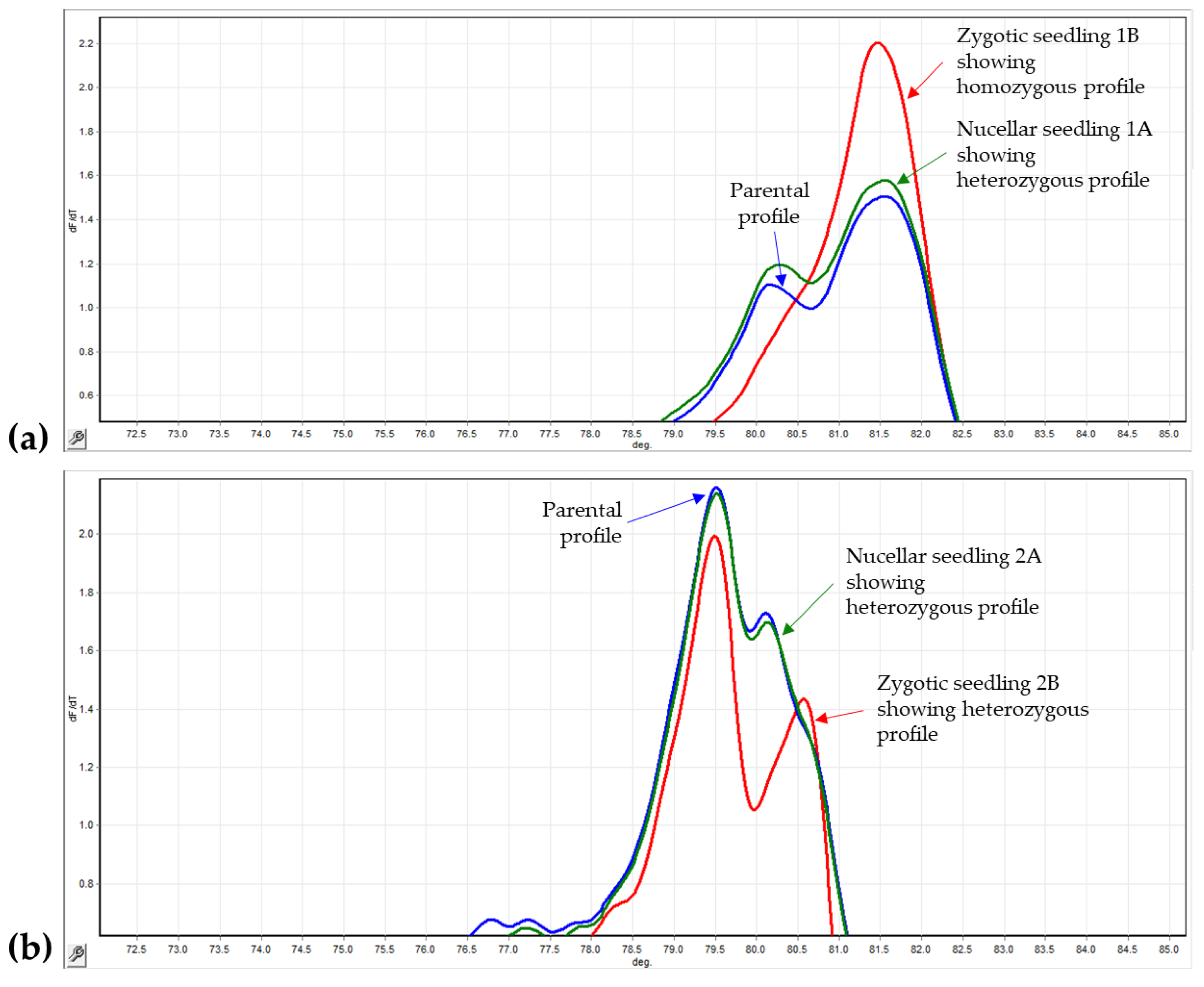
2.4. Identification of Nucellar and Zygotic Seedlings
2.5. Mono/Poly Embryonic Allelic Genotypes Discrimination by PCR Genotyping
2.6. Data Analysis
2.7. Sequencing
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Caruso, M.; Smith, M.W.; Froelicher, Y.; Russo, G.; Gmitter, F.G. Traditional Breeding. In The Genus Citrus; Woodhead Publishing: Sawston, UK, 2020. [Google Scholar]
- Singh, S.N.; Tomar, B.S. Bearing Habits of Kaghzi Lime. Indian Farming 1949, 10, 532–538. [Google Scholar]
- Frost, H.B. Citrus Industry; University of California Press: Berkeley, CA, USA, 1943; Volume 1. [Google Scholar]
- Minessy, F.A.; Schroeder, C.A. Pistil Development in Citrus. Bot. Gaz. 1956, 117, 343–347. [Google Scholar] [CrossRef]
- Wilms, H.J.; van Went, J.L.; Cresti, M.; Ciampolini, A. Structural Aspects Of Female Sterility In Citrus Limon. Acta Bot. Neerl. 1983, 32, 87–96. [Google Scholar] [CrossRef]
- Vardi, A.; Levin, I.; Carmi, N. Induction of Seedlessness in Citrus: From Classical Techniques to Emerging Biotechnological Approaches. J. Am. Soc. Hortic. Sci. 2008, 133, 117–126. [Google Scholar] [CrossRef]
- Guo, W.W.; Xie, K.D.; Wu, X.M.; Xie, Z.Z.; Xu, Q.; Deng, X.X. Ploidy Manipulation via Cell Engineering for Citrus Improvement Facilitated by Application of Molecular Markers. Acta. Hortic. 2018, 1203, 105–110. [Google Scholar] [CrossRef]
- Bennici, S.; Distefano, G.; las Casas, G.; di Guardo, M.; Lana, G.; Pacini, E.; la Malfa, S.; Gentile, A. Temperature Stress Interferes with Male Reproductive System Development in Clementine (Citrus Clementina Hort. Ex. Tan.). Ann. Appl. Biol. 2019, 175, 29–41. [Google Scholar] [CrossRef]
- Liang, M.; Cao, Z.; Zhu, A.; Liu, Y.; Tao, M.; Yang, H.; Xu, Q.; Wang, S.; Liu, J.; Li, Y.; et al. Evolution of Self-Compatibility by a Mutant Sm -RNase in Citrus. Nat. Plants 2020, 6, 131–142. [Google Scholar] [CrossRef]
- Wu, G.A.; Terol, J.; Ibanez, V.; López-García, A.; Pérez-Román, E.; Borredá, C.; Domingo, C.; Tadeo, F.R.; Carbonell-Caballero, J.; Alonso, R.; et al. Genomics of the Origin and Evolution of Citrus. Nature 2018, 554, 311–316. [Google Scholar] [CrossRef]
- Di Guardo, M.; Moretto, M.; Moser, M.; Catalano, C.; Troggio, M.; Deng, Z.; Cestaro, A.; Caruso, M.; Distefano, G.; la Malfa, S.; et al. Correction to: The Haplotype-resolved Reference Genome of Lemon (Citrus Limon L. Burm f.). Tree Genet. Genomes 2021, 17, 49. [Google Scholar] [CrossRef]
- Frost, H.B.; Soost, R.K. Seed Reproduction: Development of Gametes and Embryos. In The Citrus Industry; Reuther, W., Batchelor, L.D., Webber, H.J., Eds.; University of California Press: Berkeley, CA, USA, 1968; Volume 2, pp. 290–324. [Google Scholar]
- Filho, W.D.S.S.; Souza, U.; Ledo, C.A.D.S.; Santana, L.G.L.; Passos, O.S. Poliembrionia e Potencial de Obtenção de HÍbridos Em Citros. Rev. Bras. Frutic. 2014, 36, 950–956. [Google Scholar] [CrossRef]
- Shimada, T.; Endo, T.; Fujii, H.; Nakano, M.; Sugiyama, A.; Daido, G.; Ohta, S.; Yoshioka, T.; Omura, M. MITE Insertion-Dependent Expression of CitRKD1 with a RWP-RK Domain Regulates Somatic Embryogenesis in Citrus Nucellar Tissues. BMC Plant Biol. 2018, 18, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Emilia, F.; de Oliveira Duarte, V.; Dos, D.; Barros, R.; Girardi, E.A.; Dos, W.; Filho, S.S.; Passos, O.S. Poliembrionia E Atributos Morfológicos De Sementes De Porta-Enxertos De Citros 1. Rev. Bras. De Frutic. 2013, 35, 246–254. [Google Scholar]
- Bowman, K.D.; Gmitter, F.G.; Hu, X. Relationships of Seed Size and Shape with Polyembryony and the Zygotic or Nucellar Origin of Citrus spp. Seedlings. HortScience 1995, 30, 1279–1282. [Google Scholar] [CrossRef]
- Moore, G.A.; Castle, W.S. Morphological and Isozymic Analysis of Open-POllinated Citrus Rootstock Populations. J. Hered. 1988, 79, 59–63. [Google Scholar] [CrossRef]
- Anderson, C.M.; Castle, W.S.; Moore, G.A. Isozymic Identification of Zygotic Seedlings in Swingle Citrumelo Citrus Paradisi × Poncirus Trifoliata Nursery and Field Populations. J. Am. Soc. Hortic. Sci. 2019, 116, 322–326. [Google Scholar] [CrossRef]
- Garcìa, R.; Asìn, M.J.; Forner, J.; Carbonell, E.A. Genetic Analysis of Apomixis in Citrus and Poncirus by Molecular Markers. Theor. Appl. Genet. 1999, 99, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Scarano, M.T.; Tusa, N.; Abbate, L.; Lucretti, S.; Nardi, L.; Ferrante, S. Flow Cytometry, SSR and Modified AFLP Markers for the Identification of Zygotic Plantlets in Backcrosses between “Femminello” Lemon Cybrids (2n and 4n) and a Diploid Clone of “Femminello” Lemon (Citrus Limon L. Burm. F.) Tolerant to Mal Secco Disease. Plant Sci. 2003, 164, 1009–1017. [Google Scholar] [CrossRef]
- Kepiro, J.L.; Roose, M.L. AFLP Markers Closely Linked to a Major Gene Essential for Nucellar Embryony (Apomixis) in Citrus Maxima × Poncirus Trifoliata. Tree Genet. Genomes 2010, 6, 1–11. [Google Scholar] [CrossRef]
- Andrade-Rodríguez, M.; Villegas-Monter, A.; Carrillo-Castañeda, G.; García-Velázquez, A. Polyembryony and Identification of Volkamerian Lemon Zygotic and Nucellar Seedlings Using RAPD; Pesquisa Agropecuária Brasileira: Brasilia, Brazil, 2004. [Google Scholar]
- Nageswara Rao, M.; Soneji, J.R.; Chen, C.; Huang, S.; Gmitter, F.G. Characterization of Zygotic and Nucellar Seedlings from Sour Orange-like Citrus Rootstock Candidates Using RAPD and EST-SSR Markers. Tree Genet. Genomes 2008, 4, 113–124. [Google Scholar] [CrossRef]
- Jin, S.B.; Yun, S.H.; Park, J.H.; Park, S.M.; Koh, S.W.; Lee, D.H. Early Identification of Citrus Zygotic Seedlings Using Pollen-Specific Molecular Markers. Korean J. Hortic. Sci. Technol. 2015, 33, 598–604. [Google Scholar] [CrossRef]
- Nakano, M.; Shimizu, T.; Kuniga, T.; Nesumi, H.; Omura, M. Mapping and Haplotyping of the Flanking Region of the Polyembryony Locus in Citrus Unshiu Marcow. J. Jpn. Soc. Hortic. Sci. 2008, 77, 109–114. [Google Scholar] [CrossRef]
- de Oliveira, A.C.; Novac Garcia, A.; Cristofani, M.; Machado, M.A. Identification of Citrus Hybrids through the Combination of Leaf Apex Morphology and SSR Markers. Euphytica 2002, 128, 397–403. [Google Scholar] [CrossRef]
- Aleza, P.; Juárez, J.; Ollitrault, P.; Navarro, L. Polyembryony in Non-Apomictic Citrus Genotypes. Ann. Bot. 2010, 106, 533–545. [Google Scholar] [CrossRef] [PubMed]
- Yildiz, E.; Kaplankiran, M.; Hakan Demirkeser, T.; Uzun, A.; Toplu, C. Identification of Zygotic and Nucellar Individuals Produced from Several Citrus Crosses Using SSRs Markers. Not Bot Horti Agrobo 2013, 41, 478–484. [Google Scholar] [CrossRef][Green Version]
- Woo, J.K.; Park, Y.C.; Lee, J.W.; Yun, S.H.; Kim, M.; Park, S.; Lee, Y.; Song, K.J.; Kim, H.B. Evaluation of Polyembryony for Genetic Resources and Efficacy of Simple Sequence Repeat Markers for the Identification of Nucellar and Zygotic Embryo-Derived Individuals in Citrus. Appl. Biol. Chem. 2019, 62, 1–11. [Google Scholar] [CrossRef]
- Caruso, M.; Distefano, G.; Pietro Paolo, D.; la Malfa, S.; Russo, G.; Gentile, A.; Recupero, G.R. High Resolution Melting Analysis for Early Identification of Citrus Hybrids: A Reliable Tool to Overcome the Limitations of Morphological Markers and Assist Rootstock Breeding. Sci. Hortic. 2014, 180, 199–206. [Google Scholar] [CrossRef]
- Navarro-García, N.; García-Almodóvar, R.C.; Córdoba, F.; López-Pérez, A.J.; Jiménez-Alfaro, Y.; Pérez-Tornero, O. Identification of Zygotic and Nucellar Seedlings in Citrus Limon: The Search for Molecular Markers. Acta Hortic. 2019, 1230, 33–39. [Google Scholar] [CrossRef]
- Nakano, M.; Shimada, T.; Endo, T.; Fujii, H.; Nesumi, H.; Kita, M.; Ebina, M.; Shimizu, T.; Omura, M. Characterization of Genomic Sequence Showing Strong Association with Polyembryony among Diverse Citrus Species and Cultivars, and Its Synteny with Vitis and Populus. Plant Sci. 2012, 183, 131–142. [Google Scholar] [CrossRef]
- Wang, X.; Xu, Y.; Zhang, S.; Cao, L.; Huang, Y.; Cheng, J.; Wu, G.; Tian, S.; Chen, C.; Liu, Y.; et al. Genomic Analyses of Primitive, Wild and Cultivated Citrus Provide Insights into Asexual Reproduction. Nat. Genet. 2017, 49, 765–772. [Google Scholar] [CrossRef]
- Nigro, F.; Ippolito, A.; Salerno, M.G. Mal Secco Disease of Citrus: A Journey through a Century of Research. J. Plant Pathol. 2011, 93, 523–560. [Google Scholar] [CrossRef]
- Catalano, C.; di Guardo, M.; Distefano, G.; Caruso, M.; Nicolosi, E.; Deng, Z.; Gentile, A.; la Malfa, S.G. Biotechnological Approaches for Genetic Improvement of Lemon (Citrus Limon (l.) Burm. f.) against Mal Secco Disease. Plants 2021, 10, 1002. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Tornero, O.; Porras, I. Assessment of Polyembryony in Lemon: Rescue and in Vitro Culture of Immature Embryos. Plant Cell Tissue Organ. Cult. 2008, 93, 173–180. [Google Scholar] [CrossRef]
- Luro, F.L.; Costantino, G.; Terol, J.; Argout, X.; Allario, T.; Wincker, P.; Talon, M.; Ollitrault, P.; Morillon, R. Transferability of the EST-SSRs Developed on Nules Clementine (Citrus Clementina Hort Ex Tan) to Other Citrus Species and Their Effectiveness for Genetic Mapping. BMC Genom. 2008, 9, 1–13. [Google Scholar] [CrossRef]
- Distefano, G.; la Malfa, S.; Gentile, A.; Wu, S.B. EST-SNP Genotyping of Citrus Species Using High-Resolution Melting Curve Analysis. Tree Genet. Genomes 2013, 9, 1271–1281. [Google Scholar] [CrossRef]
- Distefano, G.; Caruso, M.; la Malfa, S.; Gentile, A.; Tribulato, E. Histological and Molecular Analysis of Pollen-Pistil Interaction in Clementine. Plant Cell Rep. 2009, 28, 1439–1451. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: http://www.R-project.org (accessed on 1 October 2022).
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H. A Statistical Framework for SNP Calling, Mutation Discovery, Association Mapping and Population Genetical Parameter Estimation from Sequencing Data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef]
- Dewi, P.S.; Wakana, A.; Tanimoto, Y.; Fujiwara, Y.; Sakai, K.; Kajiwara, K. Morphology of Sterile Anthers and Inheritance of Cytoplasmic-Genetic Male Sterility in Zygotic Seedlings of Polyembryonic Acid Citrus. J. Jpn. Soc. Hortic. Sci. 2013, 82, 203–214. [Google Scholar] [CrossRef]
- Curk, F.; Ollitrault, F.; Garcia-Lor, A.; Luro, F.; Navarro, L.; Ollitrault, P. Phylogenetic Origin of Limes and Lemons Revealed by Cytoplasmic and Nuclear Markers. Ann. Bot. 2016, 117, 565–583. [Google Scholar] [CrossRef]
- Hu, Z.; Tong, Z.; Wei, J.; Yi, H.; Deng, X. Mitochondrial gene expression in stamens is differentially regulated during male gametogenesis in Citrus unshiu. J. Hortic. Sci. Biotechnol. 2006, 81, 565–569. [Google Scholar] [CrossRef]
- Hu, Z.; Zhang, M.; Wen, Q.; Wei, J.; Yi, H.; Deng, X.; Xu, X. Abnormal microspore development leads to pollen abortion in a seedless mutant of ‘Ougan’ mandarin (Citrus suavissima Hort. ex Tanaka). J. Am. Soc. Hortic. Sci. 2007, 132, 777–782. [Google Scholar] [CrossRef]
- Zhang, S.; Shi, Q.; Albrecht, U.; Shatters, R.G., Jr.; Stange, R.; McCollum, G.; Zhang, S.; Fan, C.; Stover, E. Comparative transcriptome analysis during early fruit development between three seedy citrus genotypes and their seedless mutants. Hortic. Res. 2017, 4, 17041. [Google Scholar] [CrossRef]
- Zhang, C.; Yu, D.; Ke, F.; Zhu, M.; Xu, J.; Zhang, M. Seedless mutant ‘Wuzi Ougan’ (Citrus suavissima Hort. ex Tanaka ‘seedless’) and the wild type were compared by iTRAQ-based quantitative proteomics and integratedly analyzed with transcriptome to improve understanding of male sterility. BMC Genet. 2018, 19, 106. [Google Scholar] [CrossRef]
- Wang, R.; Fang, Y.N.; Wu, X.M.; Qing, M.; Li, C.C.; Xie, K.D.; Deng, X.X.; Guo, W.W. The miR399-CsUBC24 Module Regulates Reproductive Development and Male Fertility in Citrus. Plant Physiol. 2020, 183, 1681–1695. [Google Scholar] [CrossRef]
| Accession | Pistil Abortion (%) | Atrophic Anthers (%) | Number of Seeds per Fruit | Average Seed Weight (g) | Number of Embryos per Seed | Polyembryonic Seeds (%) | Germination (%) | Seeds Giving More than One Plantlet (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Average | SD | Average | SD | Average | SD | Average | SD | Average | SD | Average | SD | Average | SD | Average | SD | |
| Adamopoulos | 72.3 | 0.22 | 5.3 | 0.05 | 7.47 | 2.02 | 0.20 | 0.002 | 2.98 | 0.18 | 67.6 | 0.22 | 85.0 | 0.13 | 46.1 | 0.30 |
| Akragas | 16.0 | 0.07 | 5.5 | 0.06 | 2.50 | 2.12 | 0.15 | - | 2.43 | - | 22.0 | - | 70.0 | - | 14.3 | - |
| Cerza | 68.3 | 0.19 | 10.0 | 0.09 | - | - | NA | - | NA | - | NA | - | 60.0 | - | 0.0 | - |
| Chaparro | 53.7 | 0.19 | 10.3 | 0.17 | 14.23 | 2.25 | 0.15 | 0.0007 | 2.65 | 0.16 | 49.5 | 0.14 | 80.0 | 0.13 | 24.9 | 0.19 |
| Cirò | 80.0 | - | 20.0 | - | 1.05 | 0.07 | 0.18 | - | 2.00 | - | 9.0 | - | 25.0 | - | 0.0 | - |
| Erice | 17.7 | 0.02 | 7.1 | 0.03 | 3.45 | 1.48 | 0.15 | 0.03 | 2.62 | 0.13 | 57.9 | - | 67.5 | 0.11 | 20.8 | 0.18 |
| Femminello Adamo (seedless) | 22.4 | 0.26 | 76.0 | 0.40 | 1.00 | 0.00 | NA | - | NA | - | NA | - | NA | - | NA | - |
| Femminello apireno Continella m84 | NA | - | NA | - | 6.80 | - | 0.11 | - | 2.50 | - | 32.3 | - | 80.0 | - | 18.8 | - |
| Femminello Campisi (seedless) | 92.2 | 0.09 | 71.7 | 0.38 | NA | - | NA | - | NA | - | NA | - | 47.5 | 0.11 | 26.1 | 0.02 |
| Femminello Cucuzzaro | 18.5 | 0.08 | 2.5 | 0.02 | 17.54 | 4.37 | 0.15 | 0.03 | 2.82 | 0.34 | 37.6 | 0.04 | 66.7 | 0.08 | 29.9 | 0.06 |
| Femminello Dosaco m503 | 53.4 | 0.01 | 20.0 | 0.11 | 5.00 | 2.26 | 0.14 | 0.01 | 2.68 | 0.01 | 44.9 | 0.10 | 92.5 | 0.11 | 16.3 | 0.02 |
| Femminello Fior d’Arancio | 72.9 | 0.23 | 4.3 | 0.06 | 11.70 | 2.96 | 0.16 | 0.04 | 2.55 | 0.47 | 39.4 | 0.13 | 66.7 | 0.10 | 42.0 | 0.19 |
| Femminello Germanà (seedless) | 34.3 | 0.18 | 96.1 | 0.01 | 1.80 | 1.13 | 0.14 | - | 2.40 | - | 19.2 | - | NA | - | NA | - |
| Femminello Greco apireno | 72.1 | 0.13 | 6.3 | 0.05 | 5.77 | 4.99 | 0.15 | 0.00 | 3.03 | 0.04 | 44.8 | - | 70.0 | - | 85.7 | - |
| Femminello Pennisi Carrubbaro | 42.3 | 0.23 | 6.7 | 0.08 | 10.65 | 0.35 | 0.11 | 0.02 | 2.57 | 0.61 | 37.2 | 0.20 | 75.0 | 0.07 | 16.5 | 0.03 |
| Femminello S01 | NA | - | NA | - | 7.30 | - | NA | - | NA | - | 19.4 | - | NA | - | NA | - |
| Femminello S02 | NA | - | NA | - | 5.70 | - | NA | - | NA | - | 30.3 | - | NA | - | NA | - |
| Femminello Santa Teresa | 7.7 | 0.03 | 4.0 | 0.04 | 15.13 | 3.80 | 0.14 | 0.02 | 2.76 | 0.08 | 38.3 | 0.02 | 73.3 | 0.13 | 52.1 | 0.20 |
| Femminello Scandurra (seedless) | 89.8 | 0.09 | 48.3 | 0.43 | 1.00 | 0.00 | NA | - | NA | - | NA | - | NA | - | NA | - |
| Femminello Siracusano 2Kr | 78.0 | - | 12.0 | - | 5.97 | 1.46 | 0.13 | - | NA | - | 53.0 | 0.14 | 77.5 | 0.18 | 25.4 | 0.03 |
| Femminello Siracusano m296 | 48.9 | 0.25 | 5.6 | 0.08 | 20.33 | 3.04 | 0.13 | 0.03 | 3.42 | 0.68 | 51.5 | 0.06 | 80.0 | 0.13 | 14.0 | 0.05 |
| Femminello Zagara Bianca Fragalà | 62.9 | 0.22 | 10.1 | 0.07 | 7.53 | 1.18 | 0.17 | 0.02 | 2.73 | 0.32 | 39.3 | 0.12 | 78.3 | 0.20 | 20.2 | 0.06 |
| Fino Iniasel 49 | 66.0 | 0.17 | 40.0 | 0.24 | 13.00 | 0.71 | 0.13 | 0.05 | 2.64 | 0.52 | 45.8 | 0.13 | 77.5 | 0.11 | 22.5 | 0.01 |
| Fino Iniasel 95 | 58.3 | 0.15 | 35.7 | 0.31 | 11.03 | 4.66 | 0.14 | 0.004 | 2.50 | 0.06 | 34.8 | 0.02 | 91.7 | 0.06 | 12.5 | 0.06 |
| Incappucciato m504 | 17.0 | - | 0.0 | - | 10.30 | - | 0.10 | - | 2.33 | - | 26.0 | - | NA | - | NA | - |
| Interdonato | 46.0 | 0.17 | 11.5 | 0.06 | 7.95 | 1.91 | 0.16 | - | 2.33 | - | 13.5 | 0.09 | 65.0 | - | 23.1 | - |
| Kamarina | 26.3 | 0.14 | 96.1 | 0.05 | 2.62 | 2.38 | 0.11 | 0.01 | 2.33 | 0.24 | 16.3 | 0.06 | 100.0 | - | 15.0 | - |
| Lemox (seedless) | 63.0 | - | 12.0 | - | 1.00 | 0.00 | NA | - | NA | - | NA | - | NA | - | NA | - |
| Lisbon | 62.7 | 0.18 | 5.7 | 0.07 | 12.27 | 0.78 | 0.14 | 0.03 | 3.03 | 0.31 | 49.7 | 0.11 | 75.0 | 0.13 | 21.6 | 0.07 |
| Lunario | 75.0 | 0.17 | 5.7 | 0.06 | 2.78 | 0.74 | 0.11 | 0.02 | 2.17 | 0.23 | 8.7 | 0.02 | 50.0 | 0.22 | 12.0 | 0.07 |
| Messina old line | 70.5 | 0.37 | 52.5 | 0.60 | NA | - | 0.14 | - | NA | - | NA | - | 70.0 | - | 35.7 | - |
| Meyer ((C. maxima × C. reticulata) × C. medica hybrid) | 99.0 | - | 12.0 | - | 11.83 | 1.65 | 0.14 | 0.02 | 1.10 | 1.56 | 9.7 | 0.09 | 76.7 | 0.03 | 10.7 | 0.10 |
| Monachello Continella old line | 31.0 | - | 3.0 | - | 2.30 | - | 0.17 | - | 2.60 | - | 42.0 | - | NA | - | NA | - |
| Monachello nucellar line | 89.0 | - | 18.0 | - | 3.05 | 0.49 | NA | - | 2.27 | - | 25.0 | 0.11 | 67.5 | 0.04 | 3.6 | 0.05 |
| Ovale di Sorrento | 70.0 | 0.19 | 6.7 | 0.04 | 13.60 | 5.40 | 0.15 | 0.05 | 2.75 | 0.49 | 45.1 | 0.10 | 88.3 | 0.03 | 26.5 | 0.04 |
| Quattrocchi | 50.5 | 0.26 | 51.0 | 0.52 | 5.74 | 0.80 | 0.15 | - | 2.43 | - | 23.0 | - | NA | - | NA | - |
| Segesta (seedless) | 42.0 | - | 0.0 | - | 1.00 | 0.00 | NA | - | NA | - | NA | - | NA | - | NA | - |
| Selinunte | 52.9 | 0.15 | 7.7 | 0.07 | 2.80 | 1.13 | 0.13 | 0.03 | 2.88 | - | 38.0 | 0.11 | 72.5 | 0.04 | 34.5 | 0.02 |
| Sfusato Amalfitano | 47.3 | 0.23 | 4.7 | 0.07 | 12.17 | 3.68 | 0.21 | 0.03 | 3.15 | 0.21 | 43.1 | 0.09 | 88.3 | 0.03 | 11.5 | 0.06 |
| Verna | 70.3 | 0.14 | 6.0 | 0.08 | 6.70 | 3.90 | 0.15 | 0.03 | 2.85 | 0.05 | 32.2 | 0.05 | 78.3 | 0.20 | 17.2 | 0.01 |
| SSRs Marker | Primer Sequences | Size |
|---|---|---|
| INRA 1388 | F: AAAACAAAGCACCC AGATCG R: ACGGCAGCAACGAG ATAAGT | 139 |
| INRA 1210 | F: GCCAAAATGCATGT TCAAGA R: GTGCCAATGATGAT CACGTC | 175 |
| INRA 818 | F: GTAGATTCGTTCAA GGCCCA R: GTGAAGCTGGAAGA GATGGC | 134 |
| INRA 116 | F: GAATTGGGAGGACG AACTGA R: CGAGCCCTAGACAG AGATGG | 252 |
| INRA 338 | F: TTTCTAAAATTTCCT TCATGGC R: CAGGTGAAATCTCA TCGCCT | 204 |
| SNPs Marker | Base Variation | SNP Position | Primer Sequences | Size |
|---|---|---|---|---|
| U455 | C/T-C/T | 907–933 | F: ACTTCCGTGAGC CAGTGAAC R: GATAGGTAGCTT CTTGTCCTCAAA | 98 |
| U513 | A/T-C/G | 340–345 | F: AATAACGAATACGC ACACGGA R: CAGTGTCAGAAG CGAAAGATTG | 124 |
| U555 | C/T-A/G-A/G-C/T | 802–806–864–879 | F: GTCCCAATCCAA GTGGCTTA R: GGAGTCTGAGGT ATCCTTCATTAG | 124 |
| U10304 | G/T-C/G-G/TG/C-A/G | 368–390–400–411–426 | F: AGAAGAAGCATA CGGGCTCA R: GCTCAGTCCCT TTGAACCAA | 146 |
| U7190 | A/T-A/G-C/G-C/T-C/GC/T | 636–655–665–667–678–687 | F: GCTTTCATTTGG TTTGCTGC R: GGTGCCTATTTT GTCCCTGAT | 132 |
| U56 | A/G-AG | 514–531 | F: GCCACATCCC AGTTTAGCC R: ATATTCAGCG GAAAGCAAGG | 104 |
| Accessions | Seedlings | Origin | Number of Recombinant Markers | Accessions | Seedlings | Origin | Number of Recombinant Markers |
| ‘Femminello Siracusano m296’ | 1 | zygotic | 5 | ‘Chaparro’ | 1A | nucellar | 0 |
| 2 | zygotic | 6 | 1B | zygotic | 3 | ||
| 3 | zygotic | 2 | 2 | zygotic | 1 | ||
| 4 | zygotic | 7 | 3 | zygotic | 2 | ||
| 5A | zygotic | 2 | 4 | nucellar | 0 | ||
| 5B | nucellar | 0 | 5 | nucellar | 0 | ||
| 6A | nucellar | 0 | 6A | nucellar | 0 | ||
| 6B | nucellar | 0 | 6B | zygotic | 1 | ||
| 6C | zygotic | 6 | ‘Femminello Siracusano 2Kr’ | 1 | nucellar | 0 | |
| ‘Femminello Zagara Bianca’ | 1 | zygotic | 7 | 2A | zygotic | 1 | |
| 2 | zygotic | 4 | 2B | nucellar | 0 | ||
| 3 | zygotic | 3 | 3 | zygotic | 2 | ||
| 4 | zygotic | 3 | 4 | zygotic | 1 | ||
| 5 | zygotic | 8 | 5 | zygotic | 2 | ||
| ‘Lisbon’ | 1A | nucellar | 0 | ‘Fino Iniasel 95’ | 1 | zygotic | 3 |
| 1B | zygotic | 6 | 2 | zygotic | 3 | ||
| 2 | zygotic | 5 | 3 | nucellar | 0 | ||
| 3 | nucellar | 0 | 4 | zygotic | 2 | ||
| 4 | zygotic | 5 | 5A | zygotic | 1 | ||
| 5 | nucellar | 0 | 5B | nucellar | 0 | ||
| ‘Verna’ | 1 | zygotic | 3 | ‘Adamopoulos’ | 1 | nucellar | 0 |
| 2 | zygotic | 3 | 2A | nucellar | 0 | ||
| 4 | zygotic | 9 | 2B | nucellar | 0 | ||
| 5 | zygotic | 8 | 3 | zygotic | 2 | ||
| 6A | zygotic | 9 | 4A | nucellar | 0 | ||
| 6B | nucellar | 0 | 4B | nucellar | 0 | ||
| ‘Femminello Santa Teresa’ | 1 | nucellar | 0 | 5 | nucellar | 0 | |
| 2A | nucellar | 0 | 6 | zygotic | 5 | ||
| 2B | zygotic | 4 | |||||
| 3 | zygotic | 5 | |||||
| 4 | zygotic | 6 | |||||
| 5 | nucellar | 0 |
| Accession | Polyembryony | Allelic Constitution of MITE Gene | |
|---|---|---|---|
| 1.3 kbp Band | 0.7 kbp Band | ||
| Pummelo (C. maxima) | monoembryonic | - | + |
| Sour orange (C. aurantium) | polyembryonic | + | + |
| Adamopoulos | polyembryonic | + | + |
| Akragas | polyembryonic | + | + |
| Chaparro | polyembryonic | + | + |
| Femminello Cucuzzaro | polyembryonic | + | + |
| Femminello Dosaco m503 | polyembryonic | + | + |
| Femminello Fior d’Arancio | polyembryonic | + | + |
| Femminello Greco apireno | polyembryonic | + | + |
| Femminello Pennisi Carrubbaro | polyembryonic | + | + |
| Femminello Santa Teresa | polyembryonic | + | + |
| Femminello Siracusano 2Kr | polyembryonic | + | + |
| Femminello Siracusano m296 | polyembryonic | + | + |
| Femminello Zagara Bianca Fragalà | polyembryonic | + | + |
| Fino Iniasel 49 | polyembryonic | + | + |
| Fino Iniasel 95 | polyembryonic | + | + |
| Kamarina | polyembryonic | + | + |
| Lisbon | polyembryonic | + | + |
| Lunario | polyembryonic | + | + |
| Meyer | monoembryonic | - | + |
| Monachello Continella old line | polyembryonic | + | + |
| Monachello nucellar line | polyembryonic | + | + |
| Ovale di Sorrento | polyembryonic | + | + |
| Quattrocchi | polyembryonic | + | + |
| Segesta (seedless) | polyembryonic | + | + |
| Selinunte | polyembryonic | + | + |
| Sfusato Amalfitano | polyembryonic | + | + |
| Verna | polyembryonic | + | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Catalano, C.; Las Casas, G.; Giuffrida, A.; Ferlito, F.; Di Guardo, M.; Continella, A.; Bennici, S.; La Malfa, S.; Gentile, A.; Distefano, G. Reproductive Biology Factors Hampering Lemon [Citrus limon (L.) Burm. f.] Genetic Improvement. Agriculture 2022, 12, 2020. https://doi.org/10.3390/agriculture12122020
Catalano C, Las Casas G, Giuffrida A, Ferlito F, Di Guardo M, Continella A, Bennici S, La Malfa S, Gentile A, Distefano G. Reproductive Biology Factors Hampering Lemon [Citrus limon (L.) Burm. f.] Genetic Improvement. Agriculture. 2022; 12(12):2020. https://doi.org/10.3390/agriculture12122020
Chicago/Turabian StyleCatalano, Chiara, Giuseppina Las Casas, Alessio Giuffrida, Filippo Ferlito, Mario Di Guardo, Alberto Continella, Stefania Bennici, Stefano La Malfa, Alessandra Gentile, and Gaetano Distefano. 2022. "Reproductive Biology Factors Hampering Lemon [Citrus limon (L.) Burm. f.] Genetic Improvement" Agriculture 12, no. 12: 2020. https://doi.org/10.3390/agriculture12122020
APA StyleCatalano, C., Las Casas, G., Giuffrida, A., Ferlito, F., Di Guardo, M., Continella, A., Bennici, S., La Malfa, S., Gentile, A., & Distefano, G. (2022). Reproductive Biology Factors Hampering Lemon [Citrus limon (L.) Burm. f.] Genetic Improvement. Agriculture, 12(12), 2020. https://doi.org/10.3390/agriculture12122020












