Effects of Inhibiting the Expression of Chitin Synthase Gene SfCHSB on the Metabolism of Trehalose and Chitin in Spodoptera frugiperda Larvae
Abstract
:1. Introduction
2. Material and Methods
2.1. Test Insect
2.2. Total RNA Extraction and cDNA First Strand Synthesis
2.3. Synthesis of Double-Stranded RNA (dsRNA)
2.4. dsRNA Injection
2.5. Collection and Observation of Spodoptera Frugiperda
2.6. Real-Time Fluorescence Quantitative PCR (qRT-PCR)
2.7. Carbohydrates Content and Trehalase Activity Tests
2.8. Chitin Content and Chitinase Activity Tests
2.9. Statistical Analysis
3. Results
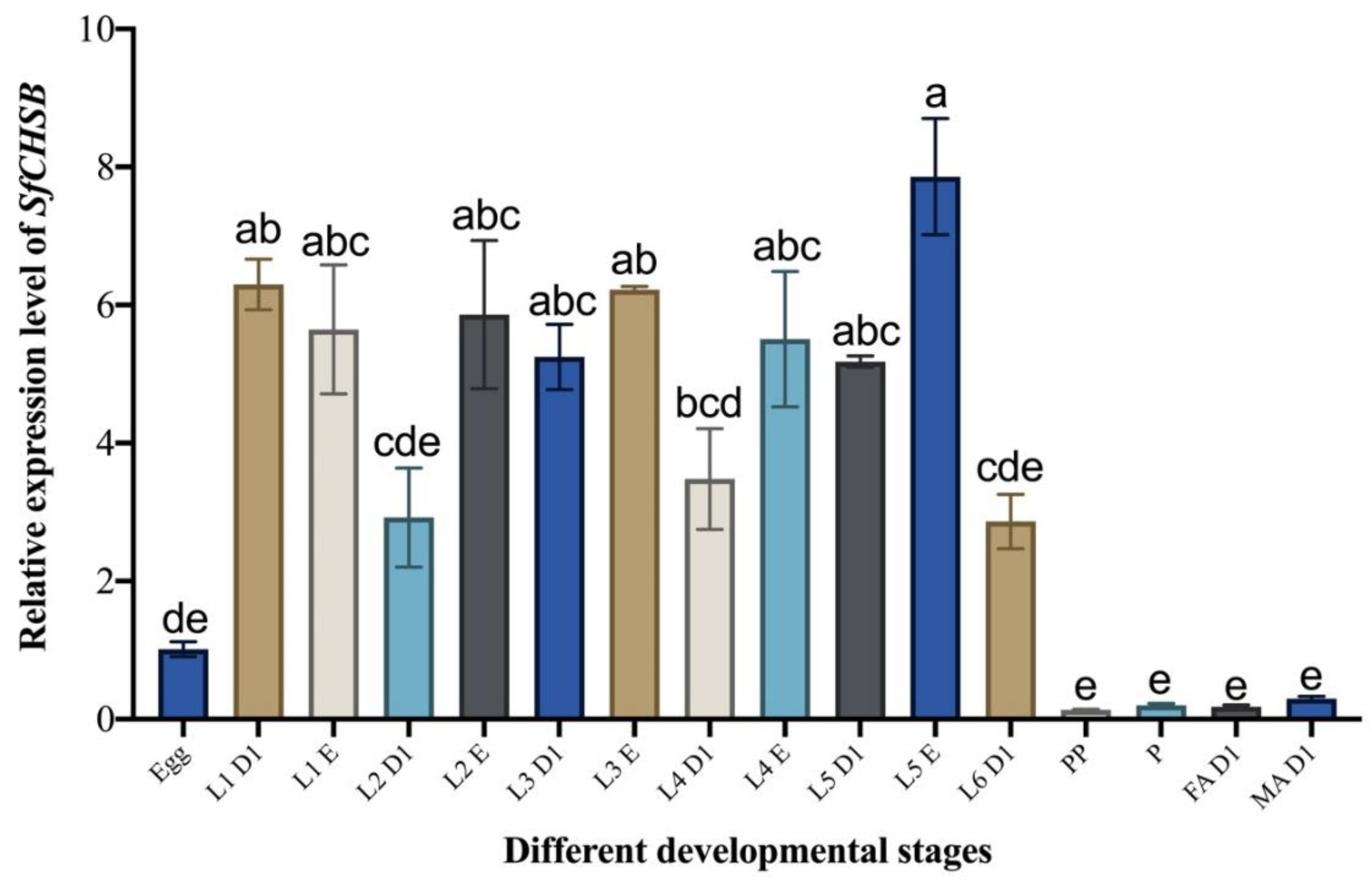
3.1. Developmental Expression Pattern of SfCHSB
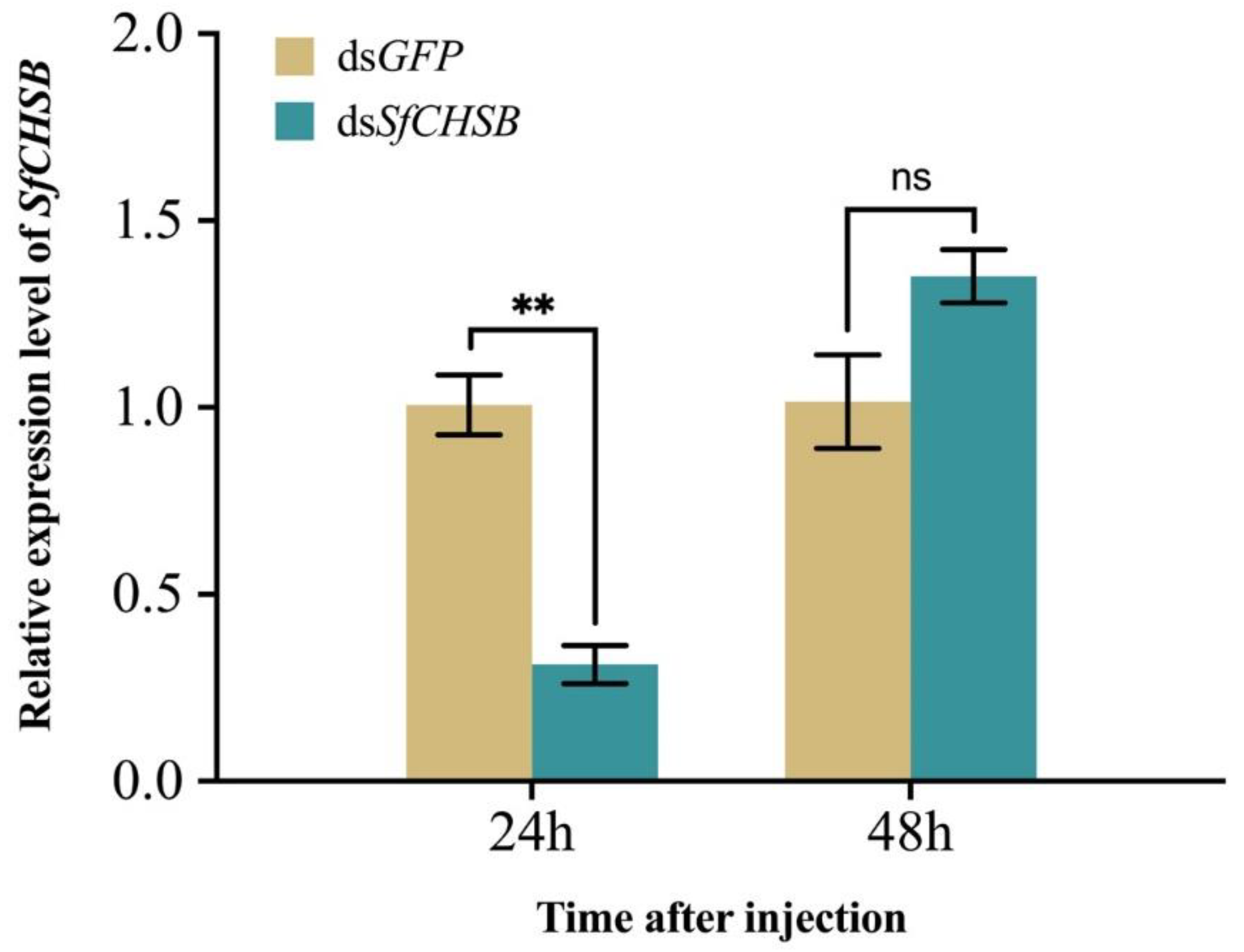
3.2. Expression of SfCHSB after RNAi
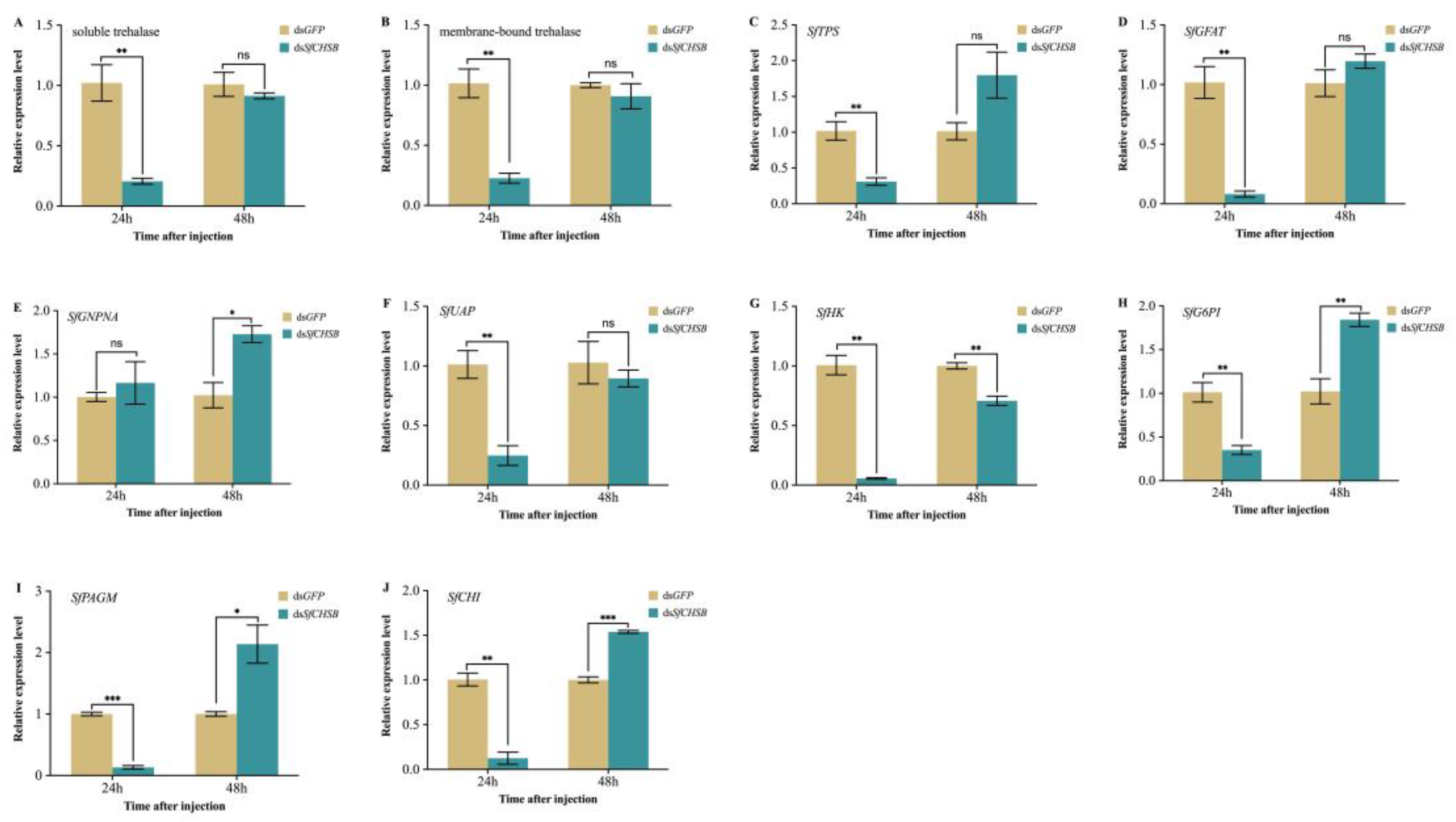
3.3. Expression of Trehalose and Chitin Metabolism-Related Genes after RNAi
3.4. Trehalase and Chitinase Activities after RNAi
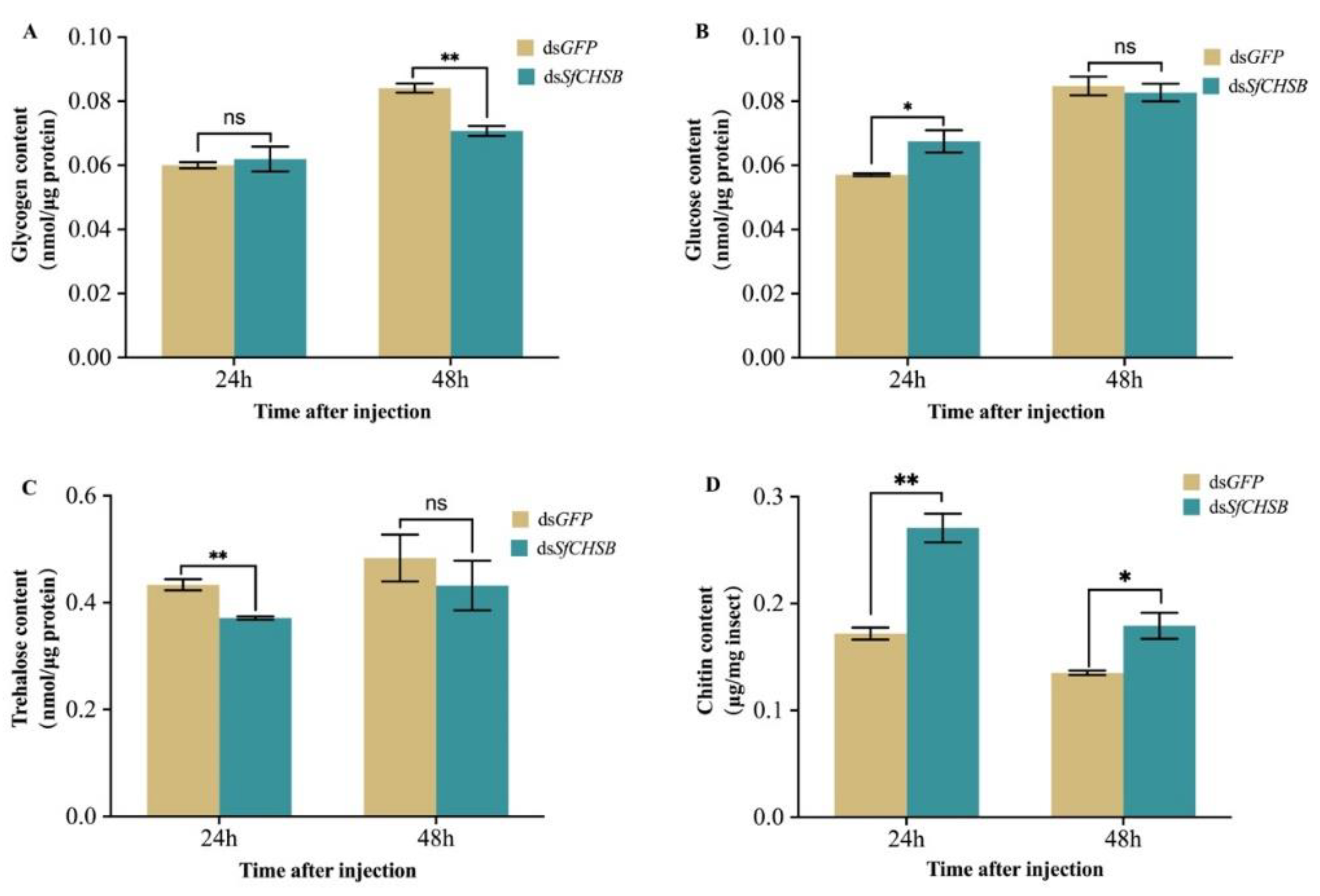
3.5. Carbohydrate and Chitin Contents after RNAi
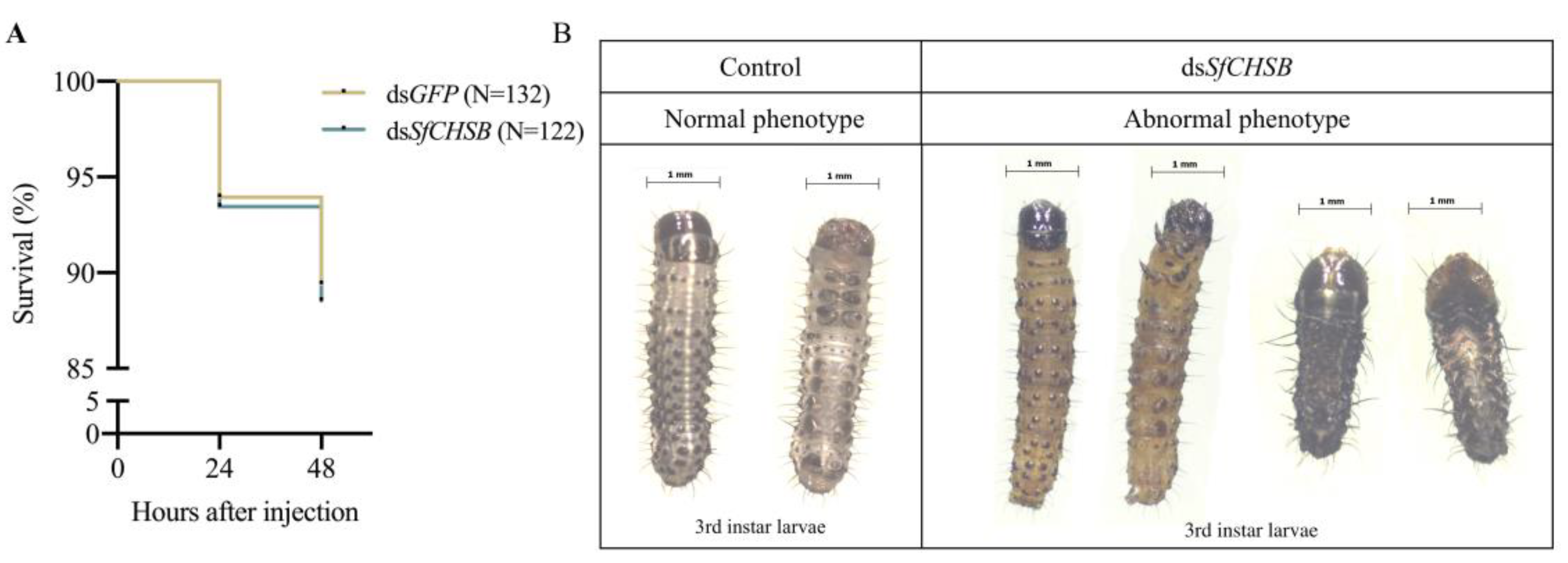
3.6. Survival and Abnormal Phenotype
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Kenis, M.; Benelli, G.; Biondi, A.; Calatayud, P.; Day, R.; Desneux, N.; Harrison, R.; Kriticos, D.; Rwomushana, I.; van den Berg, J.; et al. Invasiveness, biology, ecology, and management of the fall armyworm, Spodoptera frugiperda. Entomol. Gen. 2022. [Google Scholar] [CrossRef]
- Early, R.; Gonzalez-Moreno, P.; Murphy, S.T.; Day, R. Forecasting the global extent of invasion of the cereal pest Spodoptera frugiperda, the fall armyworm. NeoBiota 2018, 40, 25–50. [Google Scholar] [CrossRef] [Green Version]
- Westbrook, J.K.; Fleischer, S.J.; Jairam, S.; Meagher, R.L.; Nagoshi, R.N. Multigenerational migration of fall armyworm, a pest insect. Ecosphere 2019, 10, e02919. [Google Scholar] [CrossRef] [Green Version]
- Goergen, G.; Kumar, P.L.; Sankung, S.B.; Togola, A.; Tamò, M. First report of outbreaks of the fall armyworm Spodoptera frugiperda (J E Smith) (Lepidoptera, Noctuidae), a new alien invasive pest in West and Central Africa. PLoS ONE 2016, 11, e0165632. [Google Scholar] [CrossRef] [Green Version]
- Otim, M.H.; Tay, W.T.; Walsh, T.K.; Kanyesigye, D.; Adumo, S.; Abongosi, J.; Ochen, S.; Sserumaga, J.; Alibu, S.; Abalo, G.; et al. Detection of sister-species in invasive populations of the fall armyworm Spodoptera frugiperda (Lepidoptera: Noctuidae) from Uganda. PLoS ONE 2018, 13, e0194571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brévault, T.; Ndiaye, A.; Badiane, D.; Bal, A.B.; Sembène, M.; Silvie, P.; Haran, J. First records of the fall armyworm, Spodoptera frugiperda (Lepidoptera: Noctuidae), in Senegal. Entomol. Gen. 2018, 37, 129–142. [Google Scholar] [CrossRef]
- Centre Agriculture Bioscience International (CABI). Datasheet Spodoptera Frugiperda (Fall armyworm). Invasive Species Compendium. 2019. Available online: https://www.cabi.org/isc/datasheet/29810#94987198-9f50-4173-8bbd30bd93840e73?tdsourcetag=s_pcqq_aiomsg (accessed on 2 September 2022).
- Jiang, Y.Y.; Zhang, Y.Y.; Zhou, X.Y.; Hong, X.Y.; Chen, L. Population genetics reveal multiple independent invasions of Spodoptera frugiperda (Lepidoptera: Noctuidae) in China. Bull. Entomol. Res. 2022, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.X.; Ren, Q.L.; Wang, W.; Ma, Z.; Zhang, R.Z. A bet-hedging strategy rather than just a classic fast life-history strategy exhibited by invasive fall armyworm. Entomol. Gen. 2021, 41, 337–344. [Google Scholar] [CrossRef]
- Wang, P.; He, P.-C.; Hu, L.; Chi, X.-L.; Keller, M.A.; Chu, D. Host selection and adaptation of the invasive pest Spodoptera frugiperda to indica and japonica rice cultivars. Entomol. Gen. 2022, 42, 403–411. [Google Scholar] [CrossRef]
- Guedes, R.N.C.; Roditakis, E.; Campos, M.R.; Haddi, K.; Bielza, P.; Siqueira, H.A.A.; Tsagkarakou, A.; Vontas, J.; Nauen, R. Insecticide resistance in the tomato pinworm Tuta absoluta: Patterns, spread, mechanisms, management and outlook. J. Pest Sci. 2019, 92, 1329–1342. [Google Scholar] [CrossRef]
- Wang, X.; Xu, X.; Ullah, F.; Ding, Q.; Gao, X.; Desneux, N.; Song, D. Comparison of full-length transcriptomes of different imidacloprid-resistant strains of Rhopalosiphum padi (L.). Entomol. Gen. 2021, 41, 289–304. [Google Scholar] [CrossRef]
- Pires Paula, D.; Lozano, R.E.; Menger, J.P.; Andow, D.A.; Koch, R.L. Identification of point mutations related to pyrethroid resistance in voltage-gated sodium channel genes in Aphis glycines. Entomol. Gen. 2021, 41, 243–255. [Google Scholar] [CrossRef]
- Damalas, C.A.; Ilias, G.E. Pesticide exposure, safety issues, and risk assessment indicators. Int. J. Environ. Res. Public Health 2011, 8, 402–419. [Google Scholar] [CrossRef] [PubMed]
- Desneux, N.; Decourtye, A.; Delpuech, J.M. The sublethal effects of pesticides on beneficial arthropods. Annu. Rev. Entomol. 2007, 52, 81–106. [Google Scholar] [CrossRef]
- Chapman, R.F. The Insects: Structure and Function; Cambridge University Press: Cambridge, UK, 2013. [Google Scholar]
- Moussian, B. Chitin: Structure, chemistry and biology. Adv. Exp. Med. Biol. 2019, 1142, 5–18. [Google Scholar] [PubMed]
- Candy, D.J.; Kilby, B.A. Studies on chitin synthesis in the desert locust. J. Exp. Biol. 1962, 39, 129–140. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, J.; Zhu, K.Y. Chitin in Arthropods: Biosynthesis, modification, and metabolism. Adv. Exp. Med. Biol. 2019, 1142, 169–207. [Google Scholar] [PubMed]
- Ou, J.; Deng, H.M.; Zheng, S.C.; Huang, L.H.; Feng, Q.L.; Liu, L. Transcriptomic analysis of developmental features of Bombyx mori wing disc during metamorphosis. BMC Genom. 2014, 15, 820. [Google Scholar] [CrossRef] [Green Version]
- Zhuo, W.; Fang, Y.; Kong, L.; Li, X.; Sima, Y.; Xu, S. Chitin synthase A: A novel epidermal development regulation gene in the larvae of Bombyx mori. Mol. Biol. Rep. 2014, 41, 4177–4186. [Google Scholar] [CrossRef]
- Li, T.; Chen, J.; Fan, X.; Chen, W.; Zhang, W. MicroRNA and dsRNA targeting chitin synthase A reveal a great potential for pest management of the hemipteran insect Nilaparvata lugens. Pest Manag. Sci. 2017, 73, 1529–1537. [Google Scholar] [CrossRef]
- Lu, Z.J.; Huang, Y.L.; Yu, H.Z.; Li, N.Y.; Xie, Y.X.; Zhang, Q.; Zeng, X.D.; Hu, H.; Huang, A.J.; Yi, L.; et al. Silencing of the chitin synthase gene is lethal to the Asian Citrus Psyllid, Diaphorina citri. Int. J. Mol. Sci. 2019, 20, 3734. [Google Scholar] [CrossRef] [Green Version]
- Ullah, F.; Gul, H.; Wang, X.; Ding, Q.; Said, F.; Gao, X.; Desneux, N.; Song, D. RNAi-mediated knockdown of chitin synthase 1 (CHS1) gene causes mortality and decreased longevity and fecundity in Aphis gossypii. Insects 2019, 11, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Xu, Y.; Yin, Q.; Zhang, H.; Yin, H.; Sun, Y.; Ma, L.; Zhou, D.; Shen, B. Physiological characterization of chitin synthase A responsible for the biosynthesis of cuticle chitin in Culex pipiens pallens (Diptera: Culicidae). Parasit Vectors 2021, 14, 234. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.H.; Mu, L.L.; Jin, L.; Anjum, A.A.; Li, G.Q. RNAi for chitin synthase 1 rather than 2 causes growth delay and molting defect in Henosepilachna vigintioctopunctata. Pestic. Biochem. Physiol. 2021, 178, 104934. [Google Scholar] [CrossRef]
- Kelkenberg, M.; Odman-Naresh, J.; Muthukrishnan, S.; Merzendorfer, H. Chitin is a necessary component to maintain the barrier function of the peritrophic matrix in the insect midgut. Insect Biochem. Mol. Biol. 2015, 56, 21–28. [Google Scholar] [CrossRef]
- Wu, J.J.; Chen, Z.C.; Wang, Y.W.; Fu, K.Y.; Guo, W.C.; Li, G.Q. Silencing chitin deacetylase 2 impairs larval-pupal and pupal-adult molts in Leptinotarsa decemlineata. Insect Mol. Biol. 2019, 28, 52–64. [Google Scholar] [CrossRef] [Green Version]
- Bolognesi, R.; Arakane, Y.; Muthukrishnan, S.; Kramer, K.J.; Terra, W.R.; Ferreira, C. Sequences of cDNAs and expression of genes encoding chitin synthase and chitinase in the midgut of Spodoptera frugiperda. Insect Biochem. Mol. Biol. 2005, 35, 1249–1259. [Google Scholar] [CrossRef]
- Arakane, Y.; Hogenkamp, D.G.; Zhu, Y.C.; Kramer, K.J.; Specht, C.A.; Beeman, R.W.; Kanost, M.R.; Muthukrishnan, S. Characterization of two chitin synthase genes of the red flour beetle, Tribolium castaneum, and alternate exon usage in one of the genes during development. Insect Biochem. Mol. Biol. 2004, 34, 291–304. [Google Scholar] [CrossRef]
- Hogenkamp, D.G.; Arakane, Y.; Zimoch, L.; Merzendorfer, H.; Kramer, K.J.; Beeman, R.W.; Kanost, M.R.; Specht, C.A.; Muthukrishnan, S. Chitin synthase genes in Manduca sexta: Characterization of a gut-specific transcript and differential tissue expression of alternately spliced mRNAs during development. Insect Biochem. Mol. Biol. 2005, 35, 529–540. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.S.; Shi, M.R.; Xu, J.; Liu, J.H.; Ye, H. Interference Efficiency and effects of bacterium-mediated RNAi in the fall armyworm (Lepidoptera: Noctuidae). J. Insect Sci. 2021, 21, 8. [Google Scholar] [CrossRef]
- Pinto, J.R.L.; Torres, A.F.; Truzi, C.C.; Vieira, N.F.; Vacari, A.M.; De Bortoli, S.A. Artificial corn-based diet for rearing Spodoptera frugiperda (Lepidoptera: Noctuidae). J. Insect Sci. 2019, 19, 2. [Google Scholar] [CrossRef]
- Gurusamy, D.; Mogilicherla, K.; Shukla, J.N.; Palli, S.R. Lipids help double-stranded RNA in endosomal escape and improve RNA interference in the fall armyworm, Spodoptera frugiperda. Arch. Insect Biochem. Physiol. 2020, 104, e21678. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Qiu, L.Y.; Yang, H.L.; Wang, H.J.; Zhou, M.; Wang, S.G.; Tang, B. Study on the effect of wing bud chitin metabolism and its developmental network genes in the Brown Planthopper, Nilaparvata lugens, by knockdown of TRE gene. Front. Physiol. 2017, 8, 750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arakane, Y.; Muthukrishnan, S.; Kramer, K.J.; Specht, C.A.; Tomoyasu, Y.; Lorenzen, M.D.; Kanost, M.; Beeman, R.W. The Tribolium chitin synthase genes TcCHS1 and TcCHS2 are specialized for synthesis of epidermal cuticle and midgut peritrophic matrix. Insect Mol. Biol. 2005, 14, 453–463. [Google Scholar] [CrossRef]
- Kumar, N.S.; Tang, B.; Chen, X.; Tian, H.; Zhang, W. Molecular cloning, expression pattern and comparative analysis of chitin synthase gene B in Spodoptera exigua. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2008, 149, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, G.H.; Smartt, C.T.; Kiley, L.M.; Christensen, B.M. Cloning and characterization of a chitin synthase cDNA from the mosquito Aedes aegypti. Insect Biochem. Mol. Biol. 2000, 30, 1213–1222. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, H.; Li, S.; Zhu, K.Y.; Ma, E.; Zhang, J. Characterization of a midgut-specific chitin synthase gene (LmCHS2) responsible for biosynthesis of chitin of peritrophic matrix in Locusta migratoria. Insect Biochem. Mol. Biol. 2012, 42, 902–910. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Feng, Z.J.; Chen, Z.S.; Wang, X.X.; Cong, H.S.; Fan, Y.L.; Liu, T.X. Connection between cuticular hydrocarbons and melanization in Harmonia axyridis revealed by RNAi-mediated silencing of the CYP4G79. Entomol. Gen. 2021, 41, 83–96. [Google Scholar] [CrossRef]
- Li, Y.; Gao, H.; Zhang, Y.W.; Lin, X.D. Role of the transcription factor Taiman in moulting and ovarian development of Nilaparvata lugens. Entomol. Gen. 2021, 41, 169–177. [Google Scholar] [CrossRef]
- Nandety, R.S.; Kuo, Y.W.; Nouri, S.; Falk, B.W. Emerging strategies for RNA interference (RNAi) applications in insects. Bioengineered 2015, 6, 8–19. [Google Scholar] [CrossRef] [Green Version]
- Ben-Amar, A.; Daldoul, S.; Reustle, G.M.; Krczal, G.; Mliki, A. Reverse genetics and high throughput sequencing methodologies for plant functional genomics. Curr. Genom. 2016, 17, 460–475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, T.; Nunes, M.A.; España, M.U.; Namin, H.H.; Jin, P.; Bensoussan, N.; Zhurov, V.; Rahman, T.; De Clercq, R.; Hilson, P.; et al. RNAi-based reverse genetics in the chelicerate model Tetranychus urticae: A comparative analysis of five methods for gene silencing. PLoS ONE 2017, 12, e0180654. [Google Scholar] [CrossRef]
- Zhu, K.Y.; Palli, S.R. Mechanisms, applications, and challenges of insect RNA interference. Annu. Rev. Entomol. 2020, 65, 293–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wynant, N.; Santos, D.; Verdonck, R.; Spit, J.; Van Wielendaele, P.; Vanden Broeck, J. Identification, functional characterization and phylogenetic analysis of double stranded RNA degrading enzymes present in the gut of the desert locust. Schistocerca gregaria. Insect Biochem. Mol. Biol. 2014, 46, 1–8. [Google Scholar] [CrossRef]
- Song, H.; Zhang, J.; Li, D.; Cooper, A.; Silver, K.; Li, T.; Liu, X.; Ma, E.; Zhu, K.Y.; Zhang, J. A double-stranded RNA degrading enzyme reduces the efficiency of oral RNA interference in migratory locust. Insect Biochem. Mol. Biol. 2017, 86, 68–80. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Peng, Y.; Pu, J.; Fu, W.; Wang, J.; Han, Z. Variation in RNAi efficacy among insect species is attributable to dsRNA degradation in vivo. Insect Biochem. Mol. Biol. 2016, 77, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Singh, I.K.; Singh, S.; Mogilicherla, K.; Shukla, J.N.; Palli, S.R. Comparative analysis of double-stranded RNA degradation and processing in insects. Sci. Rep. 2017, 7, 17059. [Google Scholar] [CrossRef] [Green Version]
- Pan, B.Y.; Li, G.Y.; Wu, Y.; Zhou, Z.S.; Zhou, M.; Li, C. Glucose utilization in the regulation of chitin synthesis in Brown Planthopper. J. Insect Sci. 2019, 19, 3. [Google Scholar] [CrossRef]
- Xu, C.D.; Liu, Y.K.; Qiu, L.Y.; Wang, S.S.; Pan, B.Y.; Li, Y.; Wang, S.G.; Tang, B. GFAT and PFK genes show contrasting regulation of chitin metabolism in Nilaparvata lugens. Sci. Rep. 2021, 11, 5246. [Google Scholar] [CrossRef]
- Kato, N.; Mueller, C.R.; Wessely, V.; Lan, Q.; Christensen, B.M. Aedes aegypti phosphohexomutases and uridine diphosphate-hexose pyrophosphorylases: Comparison of primary sequences, substrate specificities and temporal transcription. Insect Mol. Biol. 2005, 14, 615–624. [Google Scholar] [CrossRef] [PubMed]
- Arakane, Y.; Baguinon, M.C.; Jasrapuria, S.; Chaudhari, S.; Doyungan, A.; Kramer, K.J.; Muthukrishnan, S.; Beeman, R.W. Both UDP N-acetylglucosamine pyrophosphorylases of Tribolium castaneum are critical for molting, survival and fecundity. Insect Biochem. Mol. Biol. 2011, 41, 42–50. [Google Scholar] [CrossRef]
- Wang, Z.; Long, G.Y.; Zhou, C.; Jin, D.C.; Yang, H.; Yang, W.J. Molecular characterization of UDP-N-Acetylglucosamine pyrophosphorylase and its role in the growth and development of the white-backed planthopper Sogatella furcifera (Hemiptera: Delphacidae). Genes 2022, 13, 1340. [Google Scholar] [CrossRef] [PubMed]
- Shakeel, M.; Du, J.; Li, S.W.; Zhou, Y.J.; Sarwar, N.; Guo, X. Glucosamine-6-phosphate N-acetyltransferase gene silencing by parental RNA interference in rice leaf folder, Cnaphalocrocis medinalis (Lepidoptera: Pyralidae). Sci. Rep. 2022, 12, 2141. [Google Scholar] [CrossRef]
- Shen, X.N.; Ji, S.X.; Liu, W.X.; Guo, J.Y.; Lü, Z.C.; Wan, F.H. Molecular characteristics of three cold resistance genes and their roles in temperature stress response in two Bemisia tabaci cryptic species. Entomol. Gen. 2021, 41, 317–328. [Google Scholar] [CrossRef]
- Wang, S.S.; Chen, X.; Li, Y.; Pan, B.Y.; Wang, S.G.; Dai, H.J.; Wang, S.; Tang, B. Effects of changing temperature on the physiological and biochemical properties of Harmonia axyridis larvae. Entomol. Gen. 2020, 40, 229–241. [Google Scholar] [CrossRef]
- Luo, Y.J.; Chen, Y.; Wang, X.J.; Wang, S.T.; Yang, Y.Y.; Xu, H.X.; Qu, C.; Wu, Y.; Li, C.; Wang, S.G.; et al. Validamycin affects the development and chitin metabolism in Spodoptera frugiperda by inhibiting trehalase activity. Entomol. Gen. 2022. [Google Scholar] [CrossRef]
- Yu, H.Z.; Huang, Y.L.; Lu, Z.J.; Zhang, Q.; Su, H.N.; Du, Y.M.; Yi, L.; Zhong, B.L.; Chen, C.X. Inhibition of trehalase affects the trehalose and chitin metabolism pathways in Diaphorina citri (Hemiptera: Psyllidae). Insect Sci. 2021, 28, 718–734. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Yang, M.; Shen, Q.; Liu, X.; Shi, Z.; Wang, S.; Tang, B. Functional characterization of three trehalase genes regulating the chitin metabolism pathway in rice brown planthopper using RNA interference. Sci. Rep. 2016, 6, 27841. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Gou, Y.; Guo, S.; Zhou, J.J.; Liu, C. RNA interference of trehalose-6-phosphate synthase and trehalase genes regulates chitin metabolism in two color morphs of Acyrthosiphon pisum Harris. Sci. Rep. 2021, 11, 948. [Google Scholar] [CrossRef]
- Zhou, M.; Shen, Q.; Wang, S.; Li, G.; Wu, Y.; Xu, C.; Tang, B.; Li, C. Regulatory function of the trehalose-6-phosphate synthase gene TPS3 on chitin metabolism in brown planthopper, Nilaparvata lugens. Insect Mol. Biol. 2022, 31, 241–250. [Google Scholar] [CrossRef]
- Merzendorfer, H.; Zimoch, L. Chitin metabolism in insects: Structure, function and regulation of chitin synthases and chitinases. J. Exp. Biol. 2003, 206, 4393–4412. [Google Scholar] [PubMed] [Green Version]
- Hegedus, D.D.; Toprak, U.; Erlandson, M. Peritrophic matrix formation. J. Insect Physiol. 2019, 117, 103898. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhu, K.Y. Biochemical characterization of chitin synthase activity and inhibition in the African malaria mosquito, Anopheles gambiae. Insect Sci. 2013, 20, 158–166. [Google Scholar] [CrossRef] [Green Version]
- Kato, N.; Mueller, C.R.; Fuchs, J.F.; Wessely, V.; Lan, Q.; Christensen, B.M. Regulatory mechanisms of chitin biosynthesis and roles of chitin in peritrophic matrix formation in the midgut of adult Aedes aegypti. Insect Biochem. Mol. Biol. 2006, 36, 1–9. [Google Scholar] [CrossRef]
- Macedo, L.L.; de Souza, A.; Junior, J.D.; Coelho, R.R.; Fonseca, F.C.A.; Firmino, A.A.P.; Silva, M.C.M.; Fragoso, R.R.; Albuquerque, E.V.S.; Silva, M.S.; et al. Knocking down chitin synthase 2 by RNAi is lethal to the cotton boll weevil. Biotechnol. Res. Innov. 2017, 1, 72–86. [Google Scholar] [CrossRef]
- Bolognesi, R.; Ribeiro, A.F.; Terra, W.R.; Ferreira, C. The peritrophic membrane of Spodoptera frugiperda: Secretion of peritrophins and role in immobilization and recycling digestive enzymes. Arch. Insect Biochem. Physiol. 2001, 47, 62–75. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Granados, R.R. Molecular structure of the peritrophic membrane (PM): Identification of potential PM target sites for insect control. Arch. Insect Biochem. Physiol. 2001, 47, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Wang, B.; Lei, G.; Chen, G.; Liu, D. Advances in nanocarriers to improve the stability of dsRNA in the environment. Front. Bioeng. Biotechnol. 2022, 10, 974646. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; Shi, Y.; Wu, J.N.; Li, H.; Smagghe, G.; Liu, T.X. CRISPR/Cas9 in lepidopteran insects: Progress, application and prospects. J. Insect Physiol. 2021, 135, 104325. [Google Scholar] [CrossRef]
- Kunte, N.; McGraw, E.; Bell, S.; Held, D.; Avila, L.A. Prospects, challenges and current status of RNAi through insect feeding. Pest Manag. Sci. 2020, 76, 26–41. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, X.; Wang, S.; Yu, Y.; Cheng, Y.; Hu, C.; Zhou, M.; Li, C.; Tang, B.; Wu, Y. Effects of Inhibiting the Expression of Chitin Synthase Gene SfCHSB on the Metabolism of Trehalose and Chitin in Spodoptera frugiperda Larvae. Agriculture 2022, 12, 2019. https://doi.org/10.3390/agriculture12122019
Liu X, Wang S, Yu Y, Cheng Y, Hu C, Zhou M, Li C, Tang B, Wu Y. Effects of Inhibiting the Expression of Chitin Synthase Gene SfCHSB on the Metabolism of Trehalose and Chitin in Spodoptera frugiperda Larvae. Agriculture. 2022; 12(12):2019. https://doi.org/10.3390/agriculture12122019
Chicago/Turabian StyleLiu, Xiangyu, Shasha Wang, Yuanyi Yu, Yisha Cheng, Chaoxing Hu, Min Zhou, Can Li, Bin Tang, and Yan Wu. 2022. "Effects of Inhibiting the Expression of Chitin Synthase Gene SfCHSB on the Metabolism of Trehalose and Chitin in Spodoptera frugiperda Larvae" Agriculture 12, no. 12: 2019. https://doi.org/10.3390/agriculture12122019
APA StyleLiu, X., Wang, S., Yu, Y., Cheng, Y., Hu, C., Zhou, M., Li, C., Tang, B., & Wu, Y. (2022). Effects of Inhibiting the Expression of Chitin Synthase Gene SfCHSB on the Metabolism of Trehalose and Chitin in Spodoptera frugiperda Larvae. Agriculture, 12(12), 2019. https://doi.org/10.3390/agriculture12122019





