Abstract
Micronutrients, such as zinc (Zn), are essential for the growth and development of a wide range of crops. To overcome Zn deficiency in the soil, Zn-solubilizing bacteria (ZSB) have recently been employed. In the present study, samples from the rice fields in the state of Selangor, Malaysia, were collected to isolate, characterize, and identify the ZSB. A total of 88 strains were isolated, and only 9 strains were able to solubilize the insoluble Zn on zinc oxide (ZnO)-, zinc carbonate (ZnCO3)-, and zinc phosphate (Zn3(PO4)2)-amended Tris-minimal media agar and broth assays. The highest Zn solubilization (20.99%) was measured for the TM23 isolate when exposed to Zn3(PO4)2-modified media culture, whereas ZnCO3 showed the lowest (3.35%) Zn solubilization by ZSB. In addition, nine isolated ZSB also exhibited plant-growth-promoting (PGP) traits, including nitrogen fixation ability, siderophore production, indole acetic acid production (35.28–65.48 mL−1), phosphate solubilization (27.69–77.38%), enzyme hydrolysis, and production of organic acids. Most of the isolated strains (88) were Gram-negative, except for TM54, which was Gram-positive. The four potential ZSB isolates based on 16RS rDNA sequence analysis were identified as Serratia sp. and Acinetobacter sp. Hence, this study’s findings suggest that these isolates could be prospective candidates to overcome Zn deficiencies and reduce the consumption of chemical fertilizers in agricultural areas.
1. Introduction
Rice is the most widely consumed grain, and nearly 50% of the world’s population relies on it as their primary food. Rice production in Asia accounts for over 90% of the world’s total rice production, and Asia’s growing population has increased rice production to meet future demand [1,2,3]. According to [3], about 2.5 servings of white rice are consumed daily by adults in Malaysia. To increase rice production which meets the high demand, chemical fertilizers are typically used in rice fields to fulfil the needs for vital macronutrients (N, P, and K) and micronutrients (B, Zn, Fe, Cu, Mn, and Mo). Among the micronutrients, Zn is a vital element in several metabolic pathways in plants for improved development and nutrition [4,5]. According to Gontia-Mishra et al. [2], Zn plays a significant role in various enzymatic processes, such as glucose metabolism, cellular membrane integrity maintenance, protein synthesis, and auxin synthesis. It is also vital in regulating gene expression, which is required for plants to tolerate abiotic stresses [2]. Zn deficiency can impede plant growth by reducing the development of flowers and fruits, lowering carbohydrate levels, and reducing phytohormone synthesis. These Zn deficiency symptoms can result in reduced crop yields and decreased nutritional quality [6]. Rice crops have been identified as one of the crops that are vulnerable to Zn deficiency. Zn, nitrogen, and phosphorus deficiencies are reported more in flooded rice production systems [7].
Zn deficiency is influenced by environmental and soil conditions such as pH, temperature, salinity, organic matter, phosphorus, and iron availability [8]. One of the fertilizers used in agricultural areas is zinc (ZnSO4). Zinc sulfate is supplied in fertilization schemes for Malaysian rice farmers and can be called a mixture fertilizer. It is recommended to apply about 5–10 kg/ha [9]. However, various forms of insoluble Zn are formed after applying ZnSO4 to the soil, such as zinc oxide (ZnO), zinc hydroxide (Zn(OH)2), zinc carbonate (ZnCO3), zinc phosphate (Zn3(PO4)2), and zinc (ZnS), depending on the chemical reactions in the soil [6,8]. This insoluble Zn cannot be assimilated by crops, resulting in Zn deficiency. To combat Zn deficiency, more Zn fertilizers are applied to crops. However, this technique is costly and can be harmful to both human health and the natural environment [10,11]. Thus, eco-friendly and cost-effective agro-technologies are needed to increase crop yield and reduce Zn deficiency [11].
Recently, numerous reports have been published on the use of soil bacteria in transforming insoluble Zn into plant-accessible, soluble forms to eradicate Zn deficiency [1,2,6,10,12,13,14]. Bacterial genera such as Pseudomonas, Bacillus, Acinetobacter, Azotobacter, Azospirillum, Gluconacetobacter, Burkholderia, and Thiobacillus have shown their ability to solubilize Zn [14]. In addition, these ZSB have also demonstrated the ability to improve crop quality via PGP traits such as producing various phytohormones and solubilizing nutrients (e.g., P and K), synthesizing exopolysaccharides and siderophores, and reducing environmental stresses [15]. Pseudomonas pseudoalcaligenes and Bacillus pumilus are the examples of bacteria that have been proven to improve some growth parameters for rice, and they have a higher potential compared to other bacteria [16]. These bacteria have also been found to be efficient in terms of zinc mineralization capability.
Despite rice being Malaysia’s most popular grain, studies on zinc-solubilizing bacteria (ZSB) have not been conducted extensively in Malaysian rice fields. The high humidity of Malaysia’s tropical climate ensures the presence of various microorganisms in the soil of wetland rice areas due to active rice cultivation activity. There are numerous findings on the potential usage of zinc-solubilizing bacteria outside Malaysia [15,16,17]. However, there is limited information about ZSB compared to the other plant-growth-promoting bacteria—such as nitrogen-fixing bacteria and phosphate-solubilizing bacteria—that have been isolated from Malaysian soil. Due to a lack of knowledge and literature on eco-friendly methods of zinc substitution, farmers employ biofertilizers sparingly in the Malaysian rice fields [18]. Understanding of the performance and the dynamic, diverse, and cooperative functions of ZSB, along with their relative abundance in the soil, is necessary, as is their identification and characterization [13,14]. It is hypothesized that there exists indigenous zinc-solubilizing bacteria in the Malaysian wetland soil areas, and these microbes have potential as plant-growth-promoting bacteria due to their beneficial traits, such as nitrogen fixation, phosphate solubilization, IAA production, and siderophore production. Furthermore, ZSB can be identified using 16S rRNA molecular identification to determine the exact name of the strain. Thus, the present study was undertaken to isolate, characterize, and identify the ZSB in the wetland rice fields and evaluate them for beneficial traits for plant growth promotion.
2. Materials and Methods
2.1. Study Area and Sample Collection
Rice fields in the state of Selangor, Malaysia, were selected as the study area. The average annual rainfall in this region is around 2000 mm; the average monthly temperature is 28 °C; and the average monthly humidity is 77%. Rice is grown twice per year, during the dry or off season (January–June) and the rainy or primary season (July–December) [19]. Figure 1 depicts the sampling points for sample collection. Different types of soil (i.e., Bernam, organic clay and muck (OCM), Jawa, and Sedu) were collected from six sampling points. Table 1 shows the detailed information, and Figure 1 illustrates the locations of the sample collection. The collected samples were transported to the microbiology laboratory in sterilized polyethylene bags and a 4 °C cool box for further analysis.
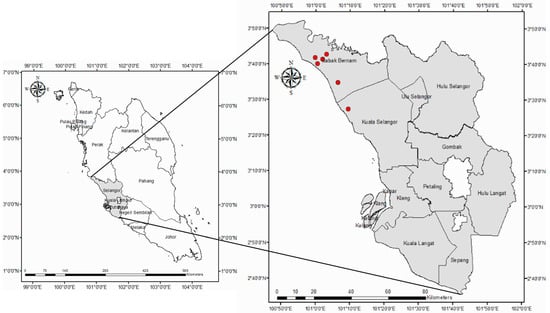
Figure 1.
Locations of sample collection from wetland rice fields in the state of Selangor, Peninsular Malaysia.

Table 1.
Detailed information on soil sample collection.
2.2. Isolation of Zn-Solubilizing Bacteria
Total plate counts (TPCs) were used to isolate bacteria from the rice’s non-rhizosphere, rhizosphere, and endophytes using Zn-modified media plates. Non-rhizosphere bacterial isolation was performed by adding 10 g of soil sample to 90 mL of distilled water and homogenizing the mixture. Then, 0.1 mL was isolated and gently swirled on a nutrient agar (NA) plate. Rice roots were used to isolate bacteria from the rhizosphere. The roots were initially cut and gently washed with sterile water to eliminate the excess soil while retaining the soil attached to the roots. For the endophytes, 1 g of roots was washed and then sterilized for 5 minutes in 70% ethanol. Following that, it was treated for 1 minute with 3% Clorox. The roots were pulverized with a sterile mortar and pestle after being repeatedly washed with sterile water. The efficacy of surface sterilization was next assessed on the roots. A 10-fold series of dilutions up to 10−10 was prepared. Then, the non-rhizosphere, rhizosphere, and endosphere bacterial populations were determined using the drop plate count method.
2.3. Screening for Zn Solubilization Activity
The isolates were evaluated for their ability to solubilize Zn via two methods: qualitative (plate assay) and quantitative (broth assay).
2.3.1. Qualitative Analysis Using the Plate Assay
All isolates were inoculated into liquid mineral salts medium (MSM) (g/L) as described by Saravanan et al. [17] (containing dextrose: 10.0; (NH4)2SO4: 1.0; KCl: 0.2; K2HPO4: 0.1; MgSO4:0.2; pH: 7.0; insoluble Zn compounds (i.e., ZnO, ZnCO3, and Zn3(PO4)2): 0.1%; and agar: 15.0 g) and then autoclaved at 121 °C for 20 min. The bacteria were inoculated (3 µL) on these media. They were then incubated for 48 h at 28 °C. The diameters of the colonies and the clearance zones around the colonies were measured. The isolate with the highest Zn solubilization efficiency (SE)% was an effective Zn solubilizer [12]. The Zn SE of the isolates based on the agar plate assay was determined using the following equation [20]:
2.3.2. Quantitative Analysis Using the Broth Assay
The quantitative Zn solubilization ability of the isolates was evaluated in 150 mL conical flasks containing 50 mL of liquid mineral salts medium in the presence of 0.1% of each insoluble Zn compound (i.e., ZnO, ZnCO3, and Zn3(PO4)2). The isolates were inoculated and cultured for 24 h in nutrient broth (NB). Following that, the broth was then inoculated with 10 µL of overnight-grown bacterial inoculum and incubated for 48 h at 28 °C in an incubator shaker under shaking conditions (160 rpm). After incubation, 1 mL of the sample was diluted with 99 mL of sterile distilled water. It was then filtered with 0.22 µm filter paper. The soluble Zn in each sample was then measured using an atomic absorbance spectrophotometer (AAS). The following equation was used to calculate the Zn solubilization percentage [20]:
2.4. Biochemical Characterization of Zn-Solubilizing Bacteria
2.4.1. Determination of Nitrogen Fixation Activity
A preliminary screening was conducted to determine the bacteria’s ability to grow in nitrogen-deprived environments by incubating the isolates on N-free solid malate medium for 24 h at 30 °C. As described by Bhakat et al. [14], nitrogen-fixing bacteria altered the medium’s color from green to blue by elevating its pH.
2.4.2. Determination of Siderophore Production
On Chrome Azurol S (CAS) agar, the bacteria’s ability to produce siderophore was assessed. Each bacterial strain was spot-injected on CAS agar and incubated for 72 h at 33 °C. The formation of an orange halo zone around the bacterial colony indicated a positive result, as the siderophores removed the Fe from the Fe–CAS dye complex in the media [21].
2.4.3. Determination of Indole Acetic Acid (IAA) Production
The fully grown bacterial cultures were inoculated in 100 mL of NB, and the mixture was then shaken vigorously for 24 h using a rotary shaker. Then, 1 mL of each bacterial culture was transferred into a new 100 mL NB with the addition of 5 mL of L-tryptophan as an IAA precursor. Concurrently, non-inoculated control sets were prepared. The 1.5 mL bacterial cultures were transferred to Eppendorf tubes and centrifuged for 7 min at 7000 rpm [22]. The IAA concentration was determined using a spectrophotometer with a wavelength of 535 nm, and the IAA concentration of each isolate was compared to the IAA standard curve.
2.4.4. Determination of Phosphate Solubilization Ability
The isolates were cultured for 24 h at 33 °C using NB to determine their phosphate solubilization ability. Then, they were inoculated on the National Botanical Research Institute’s (Uttar Pradesh, India) phosphate growth medium (NBRIP) for 48 h. The halo zone formation indicated the phosphate solubilization ability [2,20].
2.4.5. Determination of Hydrolyzing Enzyme Ability (Cellulose Degradation)
Hydrolyzing enzyme ability was tested on carboxymethyl cellulose (CMC) agar using a modified method from [23]. A cork borer (diameter: 0.5 cm) was used to cut a hole in the centercentre of the agar plate. After 24 h of incubation, the isolates were injected into the hole using a sterile syringe. The plates were then incubated for 48 h. After incubation, the agar was inundated with Congo red. The unstained color indicated CMC breakdown caused by the bacteria’s hydrolyzing enzyme ability.
2.4.6. Determination of Organic Acid Production
For the bacterial organic acid production test, bacterial cultures were inoculated in minimal salts medium for 2–3 days at 30 °C. The appearance of a pink color in the medium indicated the formation of organic acids using methyl red as an indicator.
2.5. Gram Staining
After 48 hours, the bacterial cultures were analyzed for Gram staining as described by Buchanan and Gibbons [24]. Heat-fixing bacteria were used to create Gram-stained slides. The stained smears from the slides were observed using oil emulsion at 10–40× magnification and a light microscope to observe the reaction. A Gram-positive strain appeared bluish or purple, whereas a Gram-negative strain appeared reddish.
2.6. Identification of Zn-Solubilizing Bacteria
Four selected isolates (TM56, TM23, TM13, and TM9) were genetically identified using molecular procedures that involved DNA extraction and polymerase chain reaction (PCR) amplification [25]. Genomic DNA was extracted from 24 h presumptive bacterial cultures using the HiYield Genomic DNA Mini Kit (Cat no. YGB 100/YGB 300) from Real Biotech Corporation (Taipei, Taiwan). The bacterial species were identified using multiplex PCR with two primers: 27F and 1492R. The bacterial cells (1 × 109 CFU) were transferred to 1.5 mL microcentrifuge tubes. The samples were then centrifuged at high speed (13,000 rpm). Then, the tubes were filled with 200 µL of GT buffer. The cells were then resuspended by vortexing. The samples were incubated in a water bath at 70 °C for 10–15 minutes or until the sample’s lysate became clear. During the incubation period, the tubes were inverted every 3 minutes. The elution buffer (200 µL per sample) was preheated at 70 °C for DNA elution. After completing all the processes, the samples were allowed to stand for 3–5 minutes to allow the buffer to be absorbed by the matrix. After that, the materials were centrifuged to extract the pure DNA. The 16S rDNA was amplified from the extracted DNA samples using PCR (MJ Mini Personal Thermal Cycler, Bio-Rad, Hercules, CA, USA) using the universal forward (27F, 5’-AGAGTTTGATCMTGGCTGAG-3’) and reverse (1429R, 5’-TACGGYTACCTTGTTACGACTT-3’) primers (Integrated DNA Technologies, Coralville, IA, USA). The mixture was mixed with 5 µL of sample DNA in a 0.2 mL PCR tube and run using the cycling steps described in Table 2.

Table 2.
Polymerase chain reaction (PCR) cycling steps [25].
To visualize the DNA bands, the amplified DNA was electrophoresed in a 1% agarose gel electrophoresis. Therefore, the DNA bands needed to be 1500 bands long to be sequenced. The sequences were analysed and aligned using BioEdit 7.2.5 software (Irvine, CA, USA). The aligned partial 16S rRNA sequence of the bacterial strains (1425 bp) was compared with the genes from Basic Local Alignment Search Tool (BLAST), NCBI GenBank (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 6 July 2022, and EzBioCloud (https://www.ezbiocloud.net/, accessed on 6 July 2022). The multiple alignment and phylogenetic tree were constructed by using the Molecular Evolutionary Genetics Analysis program (MEGA, version 10.1.6, Irvine, CA, USA). The phylogenetic tree was constructed using a neighbour-joining algorithm and 1000 bootstraps of replicates.
2.7. Data Analysis
The SAS software program (version 9.4; SAS Institute., Cary, NC, USA) was used to analyse all the data. The normal distribution analysis showed that our data were normally distributed at p > 0.05, and the null hypothesis of normal data distribution was accepted. The triplicate data were analysed for each experiment using one-way ANOVA, and significant differences between means were compared using Tukey’s post hoc HSD test at 5% probability, using SPSS software (version 28.0.0.0, IBM, Austin, TX, USA).
3. Results
3.1. Soil Characteristics
The chemical properties of soil in the sampling sites are elaborated in Table 3. Three rice varieties were collected: MR 220-CL2, MR 253, and MR 219. The pH of all the soil samples studied ranged from 4.50 to 6.38. The soil’s organic carbon (OC) percentage ranged from 2.35% to 15.45%. The soil nitrogen (N) content was lower, ranging from 0.19% to 0.65%. The available phosphorus in the soils ranged from 39.40 to 68.50 mg/kg (P1). The sampling sites with the highest exchangeable magnesium and calcium values were P1 (60.10 mg/kg) and P5 (25.46 mg/kg), respectively. Site P1 showed the highest Fe, Mn, and Cu contents, with 217.65, 6.56, and 0.33 mg/kg, respectively.

Table 3.
Chemical properties of the sampling sites in the wetland fields of Selangor, Malaysia.
3.2. Isolation of Zn-Solubilizing Bacteria
The maximum bacterial abundance in the rice fields was observed in the rhizosphere, while the minimum was from endosphere-isolated bacterial samples. The highest rhizosphere population (6.57 × 106) was found in the P2 (Jawa 1) site. However, the highest non-rhizosphere (5.78 × 105) and endosphere populations (2.29 × 103) were reported in the P4 (Jawa 3) sampling location (Table 4).

Table 4.
Bacterial populations (CFU g−1) in the non-rhizosphere, rhizosphere, and endosphere of wetland rice from different locations and soil series.
3.3. Screening for Zn Solubilization Activity
3.3.1. Qualitative Analysis Using the Plate Assay
A total of 88 bacterial isolates were isolated on various liquid mineral salts media, among which 9 isolates (TM9, TM13, TM23, TM29, TM54, TM56, TM61, TM65, and TM67) were able to solubilize Zn on liquid mineral salts media plates, showing a clear halo zone (Figure 2). The isolate TM61 showed the highest Zn solubilization efficiency (SE) for ZnO, at 631%, followed by TM56 (533%). For ZnCO3, the highest Zn SE was found in TM67, with 612%, followed by TM61 with 567% (Figure 3). However, for Zn3(PO4)2, the highest SE was recorded for TM67 (540%), followed by TM65 (530%).

Figure 2.
Zn solubilization by bacterial isolates showing halo zone formation on mineral salts agar medium.
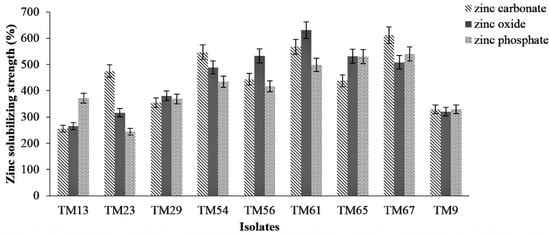
Figure 3.
Zn solubilization efficiency (SE) of isolates on various mineral salts agar media.
3.3.2. Quantitative Analysis of Zn Solubilization Using Various Mineral Salts Broth Assays
The ability of the ZSB to solubilize 0.1% of the various forms of insoluble Zn (i.e., ZnO, ZnCO3, and Zn3(PO4)2) was quantitatively determined while growing in liquid mineral salts broth media, and the results are shown in Table 5. For ZnO, TM56 had the highest solubilization ability (20.3%), while in ZnCO3-modified broth culture, TM29 had the highest solubilization (13.98%) compared to the other isolates. However, TM23 demonstrated the highest (20.99%) Zn3(PO4)2 solubilization ability. Most of the isolates could solubilize Zn in all modified broth assays, and the lowest Zn solubility was observed in ZnCO3 media.

Table 5.
Zn solubilization by ZSB in various insoluble Zn mineral-supplemented salts after 48 h of incubation.
3.4. Biochemical Characterization of Zn-Solubilizing Bacteria
All nine ZSBs were evaluated further for their biochemical characteristics (Table 6). Regarding IAA production, TM23 produced the maximum IAA (65.48 mg/L), followed by TM56 (59.72 mg/L). The results indicated that all ZSBs could solubilize phosphate, and the percentage of phosphate solubilization ranged from 27.69 to 77.38%. The maximum phosphate solubilization was shown by the isolate TM56 (77.38%), followed by TM61 (51.93%). Additionally, all the ZSB tested positive for nitrogen fixation ability, which resulted in the medium changing color from green to blue. The production of siderophores was determined based on the color change in the CAS medium from blue to orange. All isolated ZSB except for TM23 were positive for siderophore production. For cellulose and organic acid production tests, all the tested strains showed positive results except for TM61, which showed negative results for both tests. Only one isolate (TM54) was Gram-positive for the Gram staining test, while the others were Gram-negative.

Table 6.
Biochemical properties of Zn-solubilizing bacteria.
3.5. Identification of Zn-Solubilizing Bacteria (ZSB)
Based on the 16RS rDNA sequence analysis, the BLAST comparison searches against the NCBI nucleotide database revealed similarity to known rice-plant-associated bacteria, such as Serratia nematodiphilia strain DZ0503SBS1 (NCBI accession no.: 044385.1), with 98% similarity for TM56 (Figure 4a); and Acinetobacter pitti strain ATCC 9004 (NCBI accession no.: 117621.1), with similarity of up to 99% for TM 13 and TM9 (Figure 4b). The isolate TM23 was also identified as Serratia sp. and Acinetobacter sp. (Figure 4).
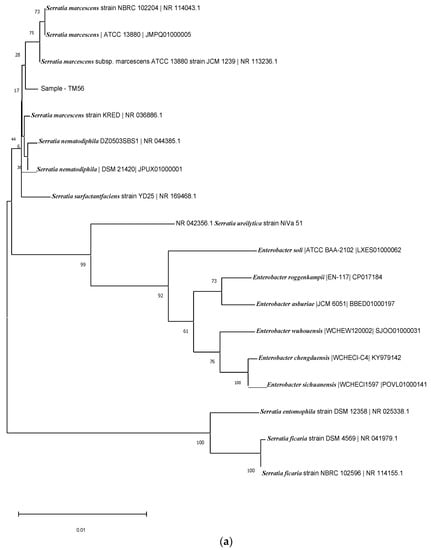
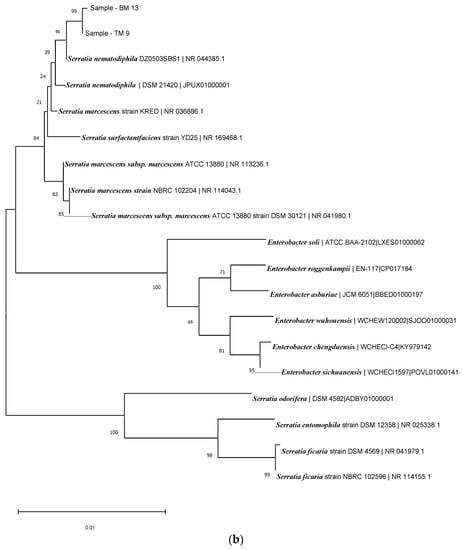
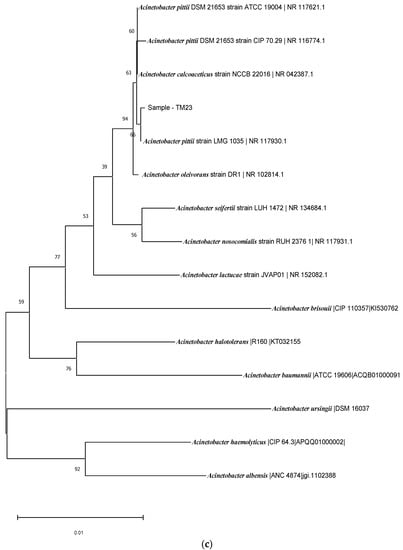
Figure 4.
(a) Phylogenetic tree of isolate TM56. (b) Phylogenetic tree of isolates TM13 and TM9. (c) Phylogenetic tree of isolate TM23.
4. Discussion
The ZSB were successfully isolated from different types of soil in wetland rice fields in the state of Selangor, Malaysia. The similar results are also found in [15,16,17], where bacteria were isolated from mineral soil, agricultural soil, and organic soil, which also represented both rhizosphere and non-rhizosphere bacterial isolates. The chemical properties of the soil types in this research were also evaluated. The results showed that the soil chemical properties varied significantly among the sampling areas. It was revealed that soils collected in the sampling areas were acidic and had significant soil nutrient levels—particularly of P and Fe. The higher nutrient levels in the soils can be attributed to increased bioactivity due to their involvement in the rate-limiting processes of soil organic matter decomposition and nutrient transformation in soils [12].
Zn is an essential micronutrient that is required by plants, humans, and microorganisms for performing a wide range of physiological and metabolic activities [4,6]. According to [26], flooded rice soils are more susceptible to Zn deficiency. High bacterial abundance was observed in all collected soils except for Sedu, which had the lowest bacterial abundance. However, there were no significant differences in the ZSB populations across all soil types. This was expected, because the farmers in the area were using the same agricultural practices, including the same types and amounts of fertilizers, along with the environmental factors that influence the bacterial populations. In addition, a range of factors—including crop types, soil properties, and nutrient levels—was also found to affect the diversity and distribution of bacteria in the rhizosphere, endosphere, and non-rhizosphere [27,28,29,30,31].
The plate assay method (qualitative analysis) is commonly used to identify microorganisms with mineral solubilization ability [12]. The isolated bacteria from the rice fields that produced a halo or clear zone in the surrounding medium with inorganic Zn was considered to be potential ZSB. However, only 9 out of the 88 isolated bacteria produced a halo zone on Tris-minimal media supplemented with three insoluble Zn compounds (i.e., Zn oxide, Zn carbonate, and Zn phosphate) at a 0.1% concentration for each compound. Similar findings were also reported in that the bacteria were not major contributors to the high solubilization index, which relied on a few factors that affect zinc solubilization, including bacterial growth rates [8,16]. Bacillus thuringiensis has lower efficiency in solubilizing zinc oxide and zinc phosphate compared to zinc carbonate [16]. In contrast, Bacillus aryabhattai showed a contradicting finding as a previous study discovered it to achieve high solubilization of zinc phosphate compared to zinc carbonate and less efficiency in solubilizing zinc oxide [24]. Generally, the bacterial isolates’ processes of solubilizing insoluble Zn could be due to proton extrusion and the formation of microbial-derived organic acids, resulting in Zn solubilization and, thus, affecting the bioaccumulation and bioavailability of Zn [12,24]. Based on the Zn solubilization efficiency (SE) results, no single isolate demonstrated uniform efficiency on all three insoluble Zn compounds. ZnO had the highest Zn SE in this investigation, followed by ZnCO3 and Zn3(PO4)2. The findings of this study also showed that the efficiency of solubilizing Zn in various Zn compounds varied between strains, which is consistent with the results of other studies [12,32].
All nine isolates in this study could produce the PGP traits to enhance plant growth—such as nitrogen fixation, phosphate solubilization, potassium solubilization, production of cellulose-hydrolyzing enzymes, siderophore production, and organic acid production. Similar studies have shown the necessity of PGP traits when using bacterial isolates to improve the development and growth of plants such as black pepper, rice, and chilies [2,4,33]. One of the important PGP traits is the ability to produce IAA, which promotes the elongation of cells and boosts the growth of root hairs and lateral roots in plants, resulting in the plants having access to sufficient water and nutrients [33,34]. Most of the isolated ZSB in this research were able to produce siderophores. The same results were obtained by Wang et al. [35], where zinc solubilization characteristics were shown by efficient siderophore-producing soil bacteria isolated from Indian soil. Siderophores are chemical molecules with a low molecular weight and high affinity and selectivity for iron (Fe). Siderophores boost plants’ development by increasing Fe uptake and crop yield [35]. In addition, the ZSB isolates could hydrolyze enzymes, fix nitrogen, and solubilize phosphate. The ability of bacteria to solubilize phosphate may improve phosphate uptake by plants, resulting in improvements in vegetative and biological parameters [32].
Furthermore, almost all the ZSB isolates showed positive results for organic acid production. The ability of ZSB to solubilize Zn could be a result of the production of organic acids. The formation of organic acids by bacteria decreases the pH value of the rhizosphere and leads to an increase in solubilized Zn [35]. Similar findings were also reported by Gontia Mishra [2]—that Zn solubilization may occur due to the microbes in soil producing various organic acids, such as gluconic acid, 2-ketogluconic acid, 5-ketogluconic acid, or pentanoic acid [2]. The bacterial production of organic acids and proton extrusion are important mechanisms affecting the solubilization of metal compounds through the excretion of other metabolites and siderophores, contributing to the solubilization process [12,36]. The solubilization of ZnO and Zn3(PO4)2 was accompanied by a rise in the medium’s H+ concentration, which was most likely caused by the synthesis of 2-ketogluconic acid [37].
Numerous crop species’ rhizospheres exhibit a stronger connection with Gram-negative rhizobacteria than with Gram-positive rhizobacteria [14,15]. The ZSB strains were morphologically and biochemically examined, finding that eight isolates were Gram-negative rods, while only one isolate (TM54) was Gram-positive. This finding was corroborated by Gupta et al. [15] who discovered the dominance of Gram-negative bacteria over Gram-positive bacteria in the rhizosphere. The rhizodeposition promotes and facilitates the movement of Gram-negative bacteria, while Gram-positive bacteria are inhibited by it. Additionally, root exudates attract Gram-negative bacteria, increasing their population around the roots and promoting plant growth. In contrast, Gram-positive bacteria are aerobic, and their population declines due to the lack of oxygen surrounding the roots [15].
The selected ZSB isolates in this study were identified as Serratia nematodiphila and Acinetobacter pitti. According to previous studies, most of the ZSB belong to Pseudomonas, Bacillus, Enterobacter, Xanthomonas, Stenotrophomonas, and Acinetobacter [8,11]. Serratia sp. is a rod-shaped, Gram-negative, facultative anaerobic bacterium of the Enterobacteriaceae family [38,39]. Serratia sp. has been isolated from various sources, including animals, soil, and in plants as endophytes. Serratia nematodiphilaa improves the health and growth of its host plant and alleviates abiotic impacts through multiple mechanisms, including the synthesis of numerous bioactive substances and the enhancement of nutrient intakes in host plants [33,39]. According to Khai et al. [38], Serratia nematodiphila has been utilized as a biostimulant, biofertilizer, and bioprotectant to inhibit the adverse effects of low temperature on pepper plants. Serratia sp. is proven to promote growth in various plant species, including black pepper, coconut palms, wheat, and corn [39]. For Acinetobacter pitti, similar findings to Acinetobacter sp. have been reported by several studies [29]. A study conducted by Gandhi and Muralidharan [4] isolated 143 ZSB from the rice rhizosphere and identified that the most efficient ZSB (i.e., AGM3 and AGM9) belonged to the Acinetobacter sp. based on biochemical and molecular characterization. According to He and Wan [40], Acinetobacter pitti significantly promoted soybean growth, as evidenced by improved vegetative growth properties, by producing small molecular organic acids such as IAA, oxalic acid, gluconic acid, and citric acid.
Implementing the ZSB may increase the availability of Zn in soil, avoiding crop loss and, as a result, improving the nutritional quality of rice [2]. This study demonstrates that Serratia nematodiphila and Acinetobacter pitti can potentially overcome Zn deficiency in rice plants. Due to their PGP traits, they may qualify as efficient strains for field application as biofertilizers.
5. Conclusions
In conclusion, a total of 88 strains were isolated, and only 9 strains were able to solubilize insoluble Zn on zinc oxide (ZnO)-, zinc carbonate (ZnCO3)-, and zinc phosphate (Zn3(PO4)2)-amended Tris-minimal media agar and broth assays. The highest Zn solubilization was shown by the TM23 isolate when exposed to Zn3(PO4)2-modified medium culture, whereas ZnCO3 showed the lowest Zn solubilization by ZSB. In addition, nine isolated ZSB also exhibited plant-growth-promoting traits, including nitrogen fixation ability, siderophore production, indole acetic acid production, phosphate solubilization, enzyme hydrolysis, and production of organic acids. The four potential ZSB isolates based on 16RS rDNA sequence analysis were identified as Serratia sp. and Acinetobacter sp. Zn release by promising strains in soil per se may be included as a future line of work. Moreover, further validation in actual field conditions for soil Zn solubilization by the promising isolates is imperative.
Author Contributions
N.M.I.O.: Conceptualization, formal analysis, investigation, writing original draft. R.O.: Supervision. A.T.K.Z.: Methodology. A.S.S.: Writing-editing and review. N.B.K.Z.: Funding acquisition. N.A.S.: Investigation. Q.A.P.: Validation. All authors have read and agreed to the published version of the manuscript.
Funding
The research was funded by Pembiayaan Yuran Artikel Berindeks (PYPA), Tabung Dana Kecemerlangan Pendidikan (DKP), Universiti Teknologi Mara (UiTM) Malaysia.
Institutional Review Board Statement
Ethical review and approval were waived for this study this study did not involve a prospective evaluation, did not involve laboratory animals and humans and only involved non-invasive procedures.
Informed Consent Statement
Not applicable for studies not involving human.
Data Availability Statement
The data are available upon reasonable request to the corresponding author.
Acknowledgments
The authors would like to thank Universiti Teknologi Mara (UiTM), Faculty of Plantation and Agrotechnology, Universiti Teknologi Mara (UiTM), Jasin Campus, Melaka and the Faculty of Agriculture, University Putra Malaysia, Serdang, Selangor, for their support.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Rajamoorthy, Y.; Rahim, K.A.; Munusamy, S. Rice Industry in Malaysia: Challenges, Policies, and Implications. Procedia Econom. Bus. Adm. 2015, 31, 861–867. [Google Scholar] [CrossRef]
- Gontia-Mishra, I.; Sapre, S.; Tiwari, S. Zinc Solubilizing Bacteria from the Rhizosphere of Rice as Prospective Modulator of Zinc Biofortilation in Rice. Rhizosphere 2017, 3, 185–190. [Google Scholar] [CrossRef]
- Radin Firdaus, R.B.; Tan, M.L.; Rahmat, S.R.; Gunaratne, M.S. Paddy, Rice, and Food Security in Malaysia: A Review of Climate Change Impacts. Cogent Soc. Sci. 2020, 6, 1818373. [Google Scholar] [CrossRef]
- Gandhi, A.; Muralidharan, G. Assessment of Zinc Solubilizing Potentiality of Acinetobacter Sp. Isolated from Rice Rhizosphere. Eur. J. Soil Biol. 2016, 76, 1–8. [Google Scholar] [CrossRef]
- Othman, N.M.I.; Othman, R.; Saud, H.M.; Wahab, P.E.M. Effects of Root Colonization by Zinc-Solubilizing Bacteria on Rice Plant (Oryza Sativa MR219) Growth. Agric. Nat. Resour. 2017, 51, 532–537. [Google Scholar] [CrossRef]
- Mumtaz, M.Z.; Ahmad, M.; Jamil, M.; Hussain, T. Zinc Solubilizing Bacillus Spp. Potential Candidates for Biofortification in Maize. Microbiol. Res. 2017, 202, 51–60. [Google Scholar] [CrossRef]
- Impa, S.M.; Morete, M.J.; Ismail, A.M.; Schulin, R.; Johnson-Beebout, S.E. Zn Uptake, Translocation and Grain Zn Loading in Rice (Oryza Sativa L.) Genotypes Selected For Zn Deficiency Tolerance And High Grain Zn. J. Exp. Bot. 2013, 64, 2739–2751. [Google Scholar] [CrossRef]
- Khanghahi, M.Y.; Ricciuti, P.; Allegretta, I.; Terzano, R.; Crecchio, C. Solubilization Of Insoluble Zinc Compounds by Zinc Solubilizing Bacteria (ZSB) and Optimization of Their Growth Conditions. Environ. Sci. Pollut. Res. 2018, 25, 25862–25868. [Google Scholar] [CrossRef]
- Fageria, N.K.; Dos Santos, A.B.; Cobucci, T. Zinc Nutrition of Lowland Rice. Commun. Soil Sci. Plant Anal. 2011, 42, 1719–1727. [Google Scholar] [CrossRef]
- Zaheer, A.; Malik, A.; Sher, A.; Qaisrani, M.M.; Mehmood, A.; Khan, S.U.; Ashraf, M.; Mirza, Z.; Karim, S.; Rasool, M. Isolation, Characterization, and Effect of Phosphate-Zinc-Solubilizing Bacterial Strains on Chickpea (Cicerarietinum L.) Growth. Saudi J. Biol. Sci. 2019, 26, 1061–1067. [Google Scholar] [CrossRef]
- Khanghahi, M.Y.; Strafella, S.; Allegretta, I.; Crecchio, C. Isolation of Bacteria with Potential Plant-Promoting Traits and Optimization of Their Growth Conditions. Curr. Microbiol. 2021, 78, 464–478. [Google Scholar] [CrossRef]
- Khande, R.; Sharma, S.K.; Ramesh, A.; Sharma, M.P. Zinc Solubilizing Bacillus Strains that Modulate Growth, Yield and Zinc Biofortification of Soybean and Wheat. Rhizosphere 2017, 4, 126–138. [Google Scholar] [CrossRef]
- Shaikh, S.; Saraf, M. Biofortification of Triticumaestivum through the Inoculation of Zinc Solubilizing Plant Growth Promoting Rhizobacteria in Field Experiment. Biocatal. Agric. Biotechnol. 2017, 9, 120–126. [Google Scholar] [CrossRef]
- Bhakat, K.; Chakraborty, A.; Islam, E. Characterization of Zinc Solubilization Potential of Arsenic Tolerance Burkholderia Spp. Isolated From Rice Rhizospheric Soil. World J. Microbiol. Biotechnol. 2021, 37, 39. [Google Scholar] [CrossRef]
- Gupta, R.; Kumari, A.; Sharma, S.; Alzahrani, O.M.; Noureldeen, A. Identification, Characterization and Optimization Of Phosphate Solubilizing Rhizobacteria (PSRB) from Rice Rhizosphere. Saudi J. Biol. Sci. 2022, 29, 35–42. [Google Scholar] [CrossRef]
- Jha, Y. The Importance of Zinc-Mobilizing Rhizosphere Bacteria to the Enhancement of Physiology and Growth Parameters for Paddy under Salt-Stress Conditions. Jordan J. Biol. Sci. 2019, 12, 167–173. [Google Scholar]
- Saravanan, V.S.; Kalaiarasan, P.; Madhaiyan, M.; Thangaraju, M. Solubilization of Insoluble Zinc Compounds by Gluconacetobacterdiazotrophicus and the Detrimental Action of Zinc Ion (Zn2+) and Zinc Chelates on Root Knot Nematode Meloidogyne Incognita. Lett. Appl. Microbiol. 2007, 44, 235–241. [Google Scholar] [CrossRef]
- Liew, Y.A.; Omar, S.R.S.; Husni, M.H.A.; Abidin, M.A.Z.; Abdullah, N.A.P. Effects of Micronutrient Fertilizers on the Production of MR 219 Rice (Oryza Sativa L.). Malays. J. Soil Sci. 2010, 14, 71–82. [Google Scholar]
- Ismail, H.; Kamal, M.R.; Abdullah, A.F.; Mohd, M.S.F. Climate-Smart Agro-Hydrological Model for a Large-Scale Rice Irrigation Scheme in Malaysia. Appl. Sci. 2020, 10, 3906. [Google Scholar] [CrossRef]
- Nguyen, C.; Yan, W.; Le Tacon, F.; Lapeyrie, F. Genetic Variability of Phosphate Solubilizing Activity by Monocaryotic and Dicaryotic Mycelia of the Ectomycorrhizal Fungus Laccaria Bicolor (Maire) P.D. Orton. Plant Soil 1992, 143, 193–199. [Google Scholar] [CrossRef]
- Yasmin, R.; Hussain, S.; Rasool, M.H.; Siddique, M.H.; Muzammil, S. Isolation, Characterization of Zn Solubilizing Bacterium (Pseudomonas Protegens RY2) and Its Contribution in Growth of Chickpea (Cicerarietinum L) as Deciphered by Improved Growth Parameters and Zn Content. Dose Response 2021, 19, 1–12. [Google Scholar] [CrossRef]
- Gordon, A.S.; Weber, R.P. Colorimetric Estimation of Indoleacetic Acid. Plant Physiol. 1951, 26, 192–195. [Google Scholar] [CrossRef]
- Udayashankar, A.C.; Chandra Nayaka, S.; Reddy, M.; Srinivas, C. Plant Growth-Promoting Rhizobacteria Mediate Induced Systemic Resistance in Rice Against Bacterial Leaf Blight Caused by Xanthomonasoryzae pv. Oryzae. Biol. Control 2011, 59, 114–122. [Google Scholar]
- Buchanan, R.E.; Gibbons, N.E. Bergey’s Manual of Determinative Bacteriology, 8th ed.; Williams & Wilkins Co.: Baltimore, MD, USA, 1974; p. 1246. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. Molecular Evolutionary Genetics Analysis. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Nitu, R.; Rajinder, K.; Sukhminderjit, K. Zinc Solubizing Bacteria to Augment Soil Fertility–A Comprehensive Review. Int. J. Agric. Vet. Sci. 2020, 8, 38–44. [Google Scholar]
- Schreiter, S.; Sandmann, M.; Smalla, K.; Grosch, R. Soil Type Dependent Rhizosphere Competence and Biocontrol of Two Bacterial Inoculant Strains and Their Effects on the Rhizosphere Microbial Community on Field-Grown Lettuce. PLoS ONE 2014, 9, e103726. [Google Scholar] [CrossRef]
- Sugiyama, A.; Yazaki, K. Flavonoids in Plant Rhizospheres: Secretion, Fate and Their Effects on Biological Communication. Plant Biotech. 2014, 31, 431–443. [Google Scholar] [CrossRef]
- Vaid, S.K.; Kumar, B.; Sharma, A.; Shukla, A.K.; Srivastava, P.C. Effect of Zinc Solubilizing Bacteria on Growth Promotion and Zinc Nutrition of Rice. J. Plant Nutr. Soil Sci. 2014, 14, 889–910. [Google Scholar]
- Hameed, A.; Yeh, M.W.; Hsieh, Y.T.; Chung, W.C.; Lo, C.T.; Young, L.S. Diversity And Functional Characterization of Bacterial Endophytes Dwelling in Various Rice (Oryza Sativa L.) Tissues, and Their Seed Borne Dissemination into Rhizosphere under Genotobiotic P-Stress. Plant Soil 2015, 394, 177–197. [Google Scholar] [CrossRef]
- Ramesh, A.; Sharma, S.K.; Sharma, M.P.; Yadav, N.; Joshi, O.P. Inoculation of Zinc Solubilizing Bacillus Aryabhattai Strains for Improved Growth, Mobilization and Biofortification of Zinc in Soybean and Wheat Cultivated in Vertisols of Central India. Appl. Soil Ecol. 2014, 73, 87–96. [Google Scholar] [CrossRef]
- Bhatt, K.; Maheshwari, D.K. Zinc Solubilizing Bacteria (Bacillus Megaterium) with Multifarious Plant Growth Promoting Activities Alleviates Growth in Capsicum Annuum L. Biotech 2020, 10, 36. [Google Scholar] [CrossRef]
- Dastager, S.G.; Deepa, C.K.; Pandey, A. Potential Plant Growth-Promoting Activity of Serratianematodiphila NII-0928 on Black Pepper (Piper Nigrum L.). World J. Microbiol. Biotechnol. 2011, 27, 259–265. [Google Scholar] [CrossRef]
- Eshaghi, E.; Nosrati, R.; Owlia, P.; Malboobi, M.A.; Ghaseminejad, P.; Ganjali, M.R. Zinc Solubilization Characteristics of Efficient Siderophore-Producing Soil Bacteria. Iran J. Microbiol. 2019, 11, 419–430. [Google Scholar] [CrossRef]
- Wang, J.; Li, R.; Zhang, H.; Wei, G.; Li, Z. Beneficial Bacteria Active Nutrients and Promote Wheat Growth under Conditions of Reduced Fertilizer Application. BMC Microbiol. 2020, 20, 38. [Google Scholar] [CrossRef]
- Sayer, J.A.; Gadd, G.M. Binding of Cobalt and Zinc by Organic Acids and Culture Filtrates of Aspergillusniger Grown in the Absence or Presence of Insoluble Cobalt or Zinc Phosphate. Mycol. Res. 2001, 105, 1261–1267. [Google Scholar] [CrossRef]
- Fasim, F.; Ahmed, N.; Parsons, R.; Gadd, G.M. Solubilization of Zinc Salts by a Bacterium Isolated from the Air Environment of a Tannery. FEMS Microbiol. Lett. 2002, 213, 1–6. [Google Scholar] [CrossRef]
- Khoa, N.Ð.; Giàu, N.Ð.N.; Tuàn, T.Q. Effects of Serratianematodiphila CT-78 on Rice Bacterial Leaf Blight Caused by Xanthomonasoryzae pv. Oryzae. Biol. Control 2016, 103, 1–10. [Google Scholar] [CrossRef]
- Martínez, O.A.; Encina, C.; Tomckowiack, C.; Droppelmann, F.; Jara, R.; Maldonado, C.; Muñoz, O.; García-Fraile, P.; Rivas, R. Serratia Strains Isolated from the Rhizosphere of Raulí (Nothofagus Alpine) in Volcanic Soils Harbor PGPR Mechanisms and Promote Raulí Plantlet Growth. J. Plant. Nutr. Soil Sci. 2018, 18, 804–819. [Google Scholar]
- He, D.; Wan, W. Phosphate-Solubilizng Bacterium Acinetobacterpitti Gp-1 Affects Rhizosphere Bacterial Community to Alleviate Soil Phosphorus Limitation for Growth of Soybean (Glycine Max). Front. Microbiol. 2021, 12, 737116. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).