Root System Architecture and Omics Approaches for Belowground Abiotic Stress Tolerance in Plants
Abstract
1. Introduction
2. Phenomics Underlying Abiotic Stress Tolerance in Root
2.1. Growth and Development of Root
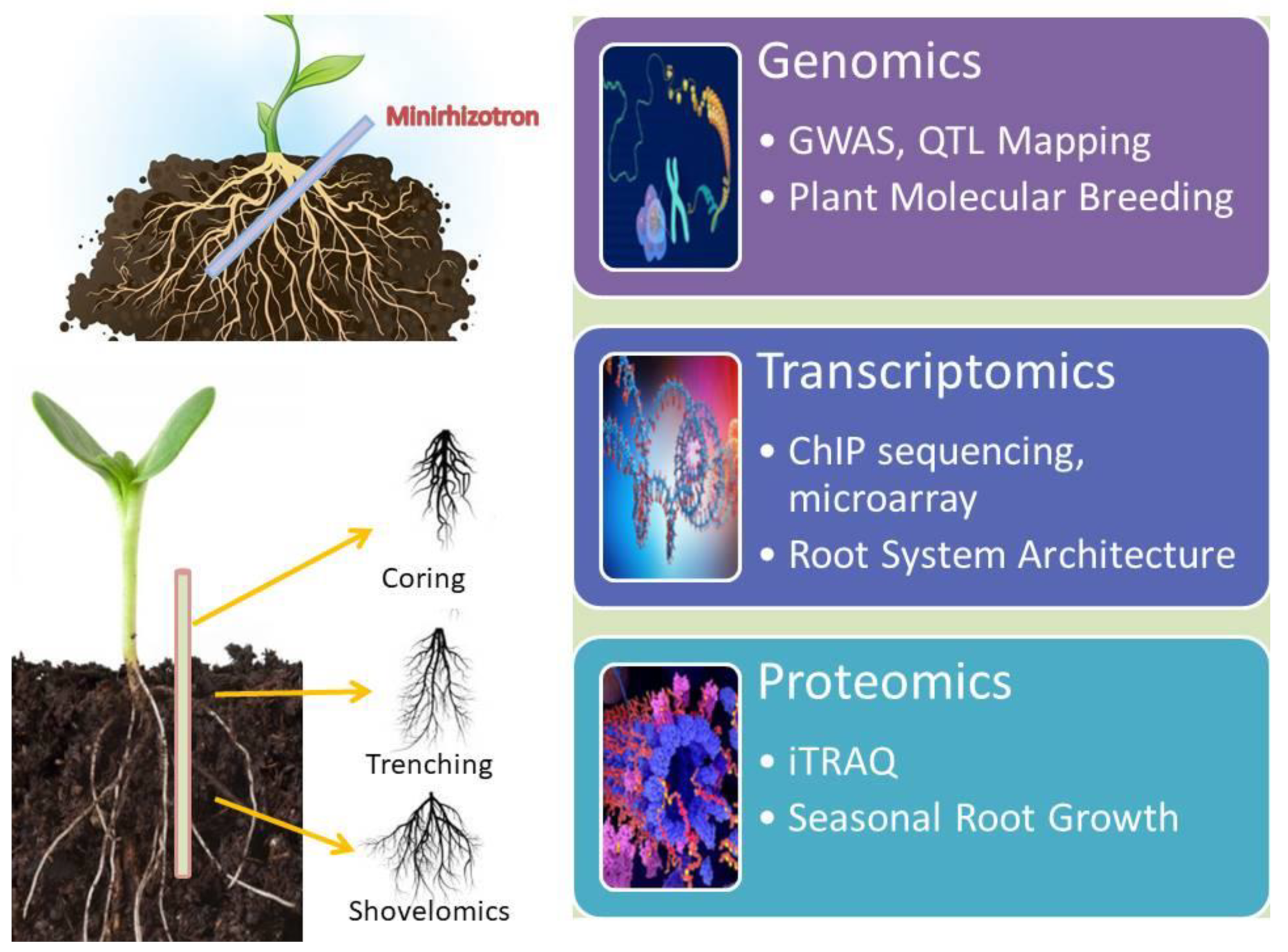
2.2. Root Phenotyping in Field/Laboratory
2.3. Phenotyping for Root Traits
3. Root Genomics Underlying Abiotic Stress Tolerance
3.1. Nutrient Stress
3.2. Drought Stress
4. Transcriptomics: A Key to Understand Abiotic Stress Tolerance in Root
4.1. Root Architectural Traits
4.2. Drought Stress
4.3. Salinity Stress
4.4. Temperature Stress
4.5. Nutrient Stress
4.6. Heavy Metal Stress
4.7. Applications of Transcriptomics in Roots
5. Proteomics of Roots in Response to Abiotic Stress
5.1. Temperature Stress
5.2. Drought Stress
5.3. Salinity Stress
5.4. Flooding Stress
6. Metabolomics of Roots in Response to Abiotic Stress
6.1. Metabolites in Root Development
6.2. Abiotic Stress Tolerance
7. Conclusions and Future Prospects
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bita, C.; Gerats, T. Plant tolerance to high temperature in a changing environment: Scientific fundamentals and production of heat stress tolerant crops. Front. Plant Sci. 2013, 4, 273. [Google Scholar] [CrossRef]
- Kitomi, Y.; Itoh, J.; Uga, Y. Genetic mechanisms involved in the formation of root system architecture. In Rice Genomics, Genetics and Breeding; Sasaki, T., Ashikari, M., Eds.; Springer: Singapore, 2018; pp. 241–274. [Google Scholar]
- Lin, P.; Wu, L.; Wei, D.; Chen, H.; Zhou, M.; Yao, X. Promoter analysis of cold-responsive (COR) gene from Capsella bursa-pastoris and expression character in response to low temperature. Int. J. Agri. Biol. 2016, 18, 346–352. [Google Scholar]
- Tyagi, W.; Rai, M. Root transcriptomes of two acidic soil adapted Indica rice genotypes suggest diverse and complex mechanism of low phosphorus tolerance. Protoplasma 2017, 254, 725–736. [Google Scholar] [CrossRef] [PubMed]
- Sebastian, J.; Yee, M.C.; Goudinho Viana, W.; Rellan-Alvarez, R.; Feldman, M.; Priest, H.D. Grasses suppress shoot-borne roots to conserve water during drought. Proc. Natl. Acad. Sci. USA 2016, 113, 8861–8866. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Du, Y.; Wang, J.; Wu, A.; Qiao, S.; Xu, B.; Zhang, S.; Siddique, K.H.M.; Chen, Y. Moderate drought stress affected root growth and grain yield in old, modern and newly released cultivars of winter wheat. Front. Plant Sci. 2017, 8, 672. [Google Scholar] [CrossRef] [PubMed]
- Burton, A.L.; Lynch, J.P.; Brown, K.M. Spatial distribution and phenotypic variation in root cortical aerenchyma of maize (Zea mays L.). Plant Soil 2013, 367, 263–274. [Google Scholar] [CrossRef]
- Gowda, V.R.; Henry, A.; Yamauchi, A.; Shashidhar, H.E.; Serraj, R. Root biology and genetic improvement for drought avoidance in rice. Field Crops Res. 2011, 122, 1–13. [Google Scholar] [CrossRef]
- Joshi, R.; Kumar, P. Lysigenous aerenchyma formation involves non-apoptotic programmed cell death in rice roots. Physiol. Mol. Biol. Plants 2012, 18, 1–9. [Google Scholar] [CrossRef]
- Manzano, C.; Pallero-Baena, M.; Casimiro, I.; De Rybel, B.; Orman-Ligeza, B.; Van Isterdael, G. The emerging role of reactive oxygen species signalling during lateral root development. Plant Physiol. 2014, 165, 1105–1119. [Google Scholar] [CrossRef]
- Joshi, R.; Shukla, A.; Mani, S.C.; Kumar, P. Hypoxia induced non-apoptotic cellular changes during aerenchyma formation in rice (Oryza sativa L.) roots. Physiol. Mol. Biol. Plants 2010, 16, 99–106. [Google Scholar] [CrossRef][Green Version]
- Lynch, J.P. Steep, cheap and deep: An ideotype to optimize water and N acquisition by maize root systems. Ann. Bot. 2013, 112, 347–357. [Google Scholar] [CrossRef]
- Kadam, N.N.; Yin, X.; Bindraban, P.S.; Struik, P.C.; Jagadish, K.S. Does morphological and anatomical plasticity during the vegetative stage make wheat more tolerant of water deficit stress than rice? Plant Physiol. 2015, 167, 1389–1401. [Google Scholar] [CrossRef]
- Naz, A.A.; Arifuzzaman, M.; Muzammil, S.; Pillen, K.; Léon, J. Wild barley introgression lines revealed novel QTL alleles for root and related shoot traits in the cultivated barley (Hordeum vulgare L.). BMC Genet. 2014, 15, 107. [Google Scholar] [CrossRef]
- Li, Y.; Wu, X.; Xu, W.; Sun, Y.; Wang, Y.; Li, G.; Xu, P. High-Throughput physiology-based stress response phenotyping: Advantages, applications and prospective in horticultural plants. Hort. Plant J. 2021, 7, 181–187. [Google Scholar] [CrossRef]
- Jeong, J.S.; Kim, Y.S.; Baek, K.H.; Jung, H.; Ha, S.H.; Choi, Y.D. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 2010, 153, 185–197. [Google Scholar] [CrossRef]
- Fenta, B.A.; Beebe, S.E.; Kunert, K.J.; Burridge, J.D.; Barlow, K.M.; Lynch, P.J. Field phenotyping of soybean roots for drought stress tolerance. Agronomy 2014, 4, 418–435. [Google Scholar] [CrossRef]
- Christopher, J.; Christopher, M.; Jennings, R.; Jones, S.; Fletcher, S.; Borrell, A.; Manschadi, A.M.; Jordan, D.; Mace, E.; Hammer, G. QTL for root angle and number in a population developed from bread wheat (Triticum aestivum) with contrasting adaptation to water-limited environments. Theor. Appl. Genet. 2013, 126, 1563–1574. [Google Scholar] [CrossRef]
- Byrt, C.S.; Munns, R.; Burton, R.A.; Gilliham, M.; Wege, S. Root cell wall solutions for crop plants in saline soils. Plant Sci. 2018, 269, 47–55. [Google Scholar] [CrossRef]
- Ding, Z.; Fu, L.; Tie, W.; Yan, Y.; Wu, C.; Dai, J.; Hu, W. Highly dynamic, coordinated, and stage-specific profiles are revealed by a multi-omics integrative analysis during tuberous root development in cassava. J. Expt. Bot. 2020, 71, 7003–7017. [Google Scholar] [CrossRef]
- Uga, Y.; Sugimoto, K.; Ogawa, S.; Rane, J.; Ishitani, M.; Hara, N.; Kitomi, Y.; Inukai, Y.; Ono, K.; Kanno, N.; et al. Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 2013, 45, 1097–1102. [Google Scholar] [CrossRef]
- Seidel, S.J.; Gaiser, T.; Srivastava, A.K.; Leitner, D.; Schmittmann, O.; Athmann, M.; Schnepf, A. Simulating Root Growth as a Function of Soil Strength and Yield with a Field-Scale Crop Model Coupled With a 3D Architectural Root Model. Front. Plant Sci. 2022, 13, 865188. [Google Scholar] [CrossRef] [PubMed]
- Kitomi, Y.; Kanno, N.; Kawai, S.; Mizubayashi, T.; Fukuoka, S.; Uga, Y. QTLs underlying natural variation of root growth angle among rice cultivars with functional allele of DEEPER ROOTING 1. Rice 2015, 8, 16. [Google Scholar] [CrossRef] [PubMed]
- Mamedov, A.I.; Husiyev, E.K. Allometric Model for Predicting Root Biomass of Field Crops in the Salt-Affected Clay Soil: Novel Approach. Environ. Sci. Proc. 2022, 16, 11. [Google Scholar]
- Jin, W.; Aufrecht, J.; Patino-Ramirez, F. Modeling root system growth around obstacles. Sci. Rep. 2020, 10, 15868. [Google Scholar] [CrossRef]
- Svane, S.F.; Jensen, C.S.; Thorup-Kristensen, K. Construction of a Large-Scale Semi-Field Facility to Study Genotypic Differences in Deep Root Growth and Resources Acquisition. Plant Methods 2019, 15, 26. [Google Scholar] [CrossRef]
- Amtmann, A.; Bennett, M.J.; Henry, A. Root phenotypes for the future. Plant Cell Env. 2022, 45, 595–601. [Google Scholar] [CrossRef]
- Atkinson, J.A.; Wingen, L.U.; Griffiths, M.; Pound, M.P.; Gaju, O.; Foulkes, M.J.; Le Gouis, J.; Griffiths, S.; Bennett, M.J.; King, J.; et al. Phenotyping pipeline reveals major seedling root growth QTL in hexaploid wheat. J. Expt. Bot. 2015, 66, 2283–2292. [Google Scholar] [CrossRef]
- Luo, D.; Zhou, Q.; Wu, Y.; Chai, X.; Liu, W.; Wang, Y. Full-length transcript sequencing and comparative transcriptomic analysis to evaluate the contribution of osmotic and ionic stress components towards salinity tolerance in the roots of cultivated alfalfa (Medicago sativa L.). BMC Plant Biol. 2019, 19, 32. [Google Scholar] [CrossRef]
- Liao, H.; Yan, X.; Rubio, G.; Beebe, S.E.; Blair, M.W.; Lynch, J.P. Genetic mapping of basal root gravitropism and phosphorus acquisition efficiency in common bean. Funct. Plant Biol. 2004, 31, 959–970. [Google Scholar] [CrossRef]
- Zhu, J.M.; Brown, K.M.; Lynch, J.P. Root cortical aerenchyma improves the drought tolerance of maize (Zea mays L.). Plant Cell Env. 2010, 33, 740–749. [Google Scholar]
- Jeudy, C.; Adrian, M.; Baussard, C.; Bernard, C.; Bernaud, E.; Bourion, V.; Busset, H.; Cabrera-Bosquet, L.; Cointault, F.; Han, S.; et al. RhizoTubes as a new tool for high throughput imaging of plant root development and architecture: Test, comparison with pot grown plants and validation. Plant Methods 2016, 12, 31. [Google Scholar] [CrossRef]
- Rutkoski, J.; Poland, J.; Mondal, S.; Autrique, E.; Pérez, L.G.; Crossa, J.; Singh, R. Canopy temperature and vegetation indices from high-throughput phenotyping improve accuracy of pedigree and genomic selection for grain yield in wheat. G3: Genes Genomes Genet. 2016, 6, 2799–2808. [Google Scholar] [CrossRef]
- Li, X.; Ingvordsen, C.H.; Weiss, M.; Rebetzke, G.J.; Condon, A.G.; James, R.A.; Richards, R.A. Deeper roots associated with cooler canopies, higher normalized difference vegetation index, and greater yield in three wheat populations grown on stored soil water. J, Exp. Bot. 2019, 70, 4963–4974. [Google Scholar] [CrossRef]
- Clarke, C.; Lukac, M.; Gregory, P.; Gooding, M. Associating remotely sensed canopy traits with deep rooting in wheat. Asp. Appl. Biol. 2017, 135, 1–10. [Google Scholar]
- Deery, D.M.; Rebetzke, G.J.; Jimenez-Berni, J.A.; James, R.A.; Condon, A.; Bovill, W.D.; Hutchinson, P.; Scarrow, J.; Davy, R.; Furbank, R.T. Methodology for high-throughput field phenotyping of canopy temperature using airborne thermography. Front. Plant Sci. 2016, 7, 1808. [Google Scholar] [CrossRef]
- Horton, M.W.; Hancock, A.M.; Huang, Y.S.; Toomajian, C.; Atwell, S.; Auton, A.; Bergelson, J. Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat. Genet. 2012, 44, 212–216. [Google Scholar] [CrossRef]
- Kang, H.M.; Zaitlen, N.A.; Wade, C.M.; Kirby, A.; Heckerman, D. Efficient control of population structure in model organism association mapping. Genetics 2008, 178, 1709–1723. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Alanís, D.; Yong-Villalobos, L.; Jiménez-Sandoval, P.; Alatorre-Cobos, F.; Oropeza-Aburto, A.; Mora-Macías, J.; Herrera-Estrella, L. Phosphate starvation-dependent iron mobilization induces CLE14 expression to trigger root meristem differentiation through CLV2/PEPR2 signaling. Dev. Cell 2017, 41, 555–570.e3. [Google Scholar] [CrossRef]
- Bouain, N.; Korte, A.; Satbhai, S.B.; Nam, H.I.; Rhee, S.Y.; Busch, W.; Rouached, H. Systems genomics approaches provide new insights into Arabidopsis thaliana root growth regulation under combinatorial mineral nutrient limitation. PLoS Genet. 2019, 15, e1008392. [Google Scholar] [CrossRef]
- Kitomi, Y.; Nakao, E.; Kawai, S.; Kanno, N.; Ando, T.; Fukuoka, S.; Irie, K.; Uga, Y. Fine mapping of QUICK ROOTING 1 and 2, quantitative trait loci increasing root length in rice. G3 2018, 8, 727–735. [Google Scholar] [CrossRef]
- Pandit, E.; Panda, R.K.; Sahoo, A.; Pani, D.R.; Pradhan, S.K. Genetic relationship and structure analysis of root growth angle for improvement of drought avoidance in early and mid-early maturing rice genotypes. Rice Sci. 2020, 27, 124–132. [Google Scholar] [CrossRef]
- Kumar, J.; Sen Gupta, D.; Djalovic, I.; Kumar, S.; Siddique, K.H. Root-omics for drought tolerance in cool-season grain legumes. Physiol. Plant. 2021, 172, 629–644. [Google Scholar] [CrossRef]
- Dwivedi, P.; Ramawat, N.; Dhawan, G.; Gopala Krishnan, S.; Vinod, K.K.; Singh, M.P.; Singh, A.K. Drought tolerant near isogenic lines (NILs) of Pusa 44 developed through marker assisted introgression of qDTY2. 1 and qDTY3. 1 enhances yield under reproductive stage drought stress. Agriculture 2021, 11, 64. [Google Scholar] [CrossRef]
- Fan, S.; Han, N.; Wu, H.; Jia, J.; Guo, J. Plasma membrane intrinsic protein SlPIP1; 7 promotes root growth and enhances drought stress tolerance in transgenic tomato (Solanum lycopersicum) plants. Plant Breed. 2021, 140, 1102–1114. [Google Scholar] [CrossRef]
- Azeem, F.; Bilal, A.; Rana, M.A.; Muhammad, A.A.; Habibullah, N.; Sabir, H.; Muhammad, A. Drought affects aquaporins gene expression in important pulse legume chickpea (Cicer arietinum L.). Pak. J. Bot. 2019, 51, 81–88. [Google Scholar] [CrossRef]
- Abbas, H.; Naeem, M.K.; Rubab, M.; Widemann, E.; Uzair, M.; Zahra, N.; Shafiq, S. Role of Wheat Phosphorus Starvation Tolerance 1 Genes in Phosphorus Acquisition and Root Architecture. Genes 2022, 13, 487. [Google Scholar] [CrossRef]
- Dharshini, S.; Hoang, N.V.; Mahadevaiah, C.; Padmanabhan, T.S.; Alagarasan, G.; Suresha, G.S.; Appunu, C. Root transcriptome analysis of Saccharum spontaneum uncovers key genes and pathways in response to low-temperature stress. Env. Expt. Bot. 2020, 171, 103935. [Google Scholar] [CrossRef]
- Yoshida, M. Fructan structure and metabolism in overwintering plants. Plants 2021, 10, 933. [Google Scholar] [CrossRef]
- Halder, T.; Choudhary, M.; Liu, H.; Chen, Y.; Yan, G.; Siddique, K.H. Wheat Proteomics for Abiotic Stress Tolerance and Root System Architecture: Current Status and Future Prospects. Proteomes 2022, 10, 17. [Google Scholar] [CrossRef]
- Komatsu, S.; Nakamura, T.; Sugimoto, Y.; Sakamoto, K. Proteomic and metabolomic analyses of soybean root tips under flooding stress. Protein Pept. Lett. 2014, 21, 865–884. [Google Scholar] [CrossRef]
- Ma, J.; Luo, W.; Zhang, H. Identification of quantitative trait loci for seedling root traits from Tibetan semi-wild wheat (Triticum aestivum ssp. tibetanum). Genome 2017, 25, 1068–1075. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, R.; Chang, X.; Jing, R. Mapping QTLs for seedling root traits in a doubled haploid wheat population under different water regimes. Euphytica 2013, 189, 51–66. [Google Scholar] [CrossRef]
- Mathew, I.; Shimelis, H.; Shayanowako, A.I.T.; Laing, M.; Chaplot, V. Genome-wide association study of drought tolerance and biomass allocation in wheat. PLoS ONE 2019, 14, e0225383. [Google Scholar] [CrossRef] [PubMed]
- Oyiga, B.C.; Palczak, J.; Wojciechowski, T.; Lynch, J.P.; Naz, A.A.; Léon, J.; Ballvora, A. Genetic components of root architecture and anatomy adjustments to water-deficit stress in spring barley. Plant Cell Env. 2019, 43, 692–711. [Google Scholar] [CrossRef]
- Zaidi, P.H.; Seetharam, K.; Krishna, G.; Krishnamurthy, L.; Gajanan, S.; Babu, R.; Zerka, M.; Vinayan, M.T.; Vivek, B.S. Genomic regions associated with root traits under drought stress in tropical maize (Zea mays L.). PLoS ONE 2016, 11, e0164340. [Google Scholar] [CrossRef]
- Grondin, A.; Dixit, S.; Torres, R.; Venkateshwarlu, C.; Rogers, E.; Mitchell-Olds, T.; Benfey, P.N.; Kumar, A.; Henry, A. Physiological mechanisms contributing to the QTL qDTY3.2 effects on improved performance of rice Moroberekan×Swarna BC2F3:4 lines under drought. Rice 2018, 11, 43. [Google Scholar] [CrossRef]
- Colombi, T.; Herrmann, A.M.; Vallenback, P.; Keller, T. Cortical cell diameter is key to energy costs of root growth in wheat. Plant Physiol. 2019, 180, 2049–2060. [Google Scholar] [CrossRef]
- Zhang, X.; Mi, Y.; Mao, H.; Liu, S.; Chen, L.; Qin, F. Genetic variation in ZmTIP1 contributes to root hair elongation and drought tolerance in maize. Plant Biotechnol. J. 2020, 18, 1271–1283. [Google Scholar] [CrossRef]
- Peng, Z.; Wang, M.; Li, F.; Lv, H.; Li, C.; Xia, G. A proteomic study of the response to salinity and drought stress in an lntrogression strain of bread wheat. Mol. Cell. Proteom. 2009, 8, 2676–2686. [Google Scholar] [CrossRef]
- Van den Berg, T.; Korver, R.A.; Testerink, C.; Tusscher, K.H. Modeling halotropism: A key role for root tip architecture and reflux loop remodeling in redistributing auxin. Development 2016, 143, 3350–3362. [Google Scholar] [CrossRef]
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salt tolerance mechanisms of plants. Ann. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Robin, A.H.; Matthew, C.; Uddin, M.J.; Bayazid, K.N. Salinity-induced reduction in root surface area and changes in major root and shoot traits at the phytomer level in wheat. J. Exp. Bot. 2016, 67, 3719–3729. [Google Scholar] [CrossRef]
- Karlova, R.; Boer, D.; Hayes, S.; Testerink, C. Root plasticity under abiotic stress. Plant Physiol. 2021, 187, 1057–1070. [Google Scholar] [CrossRef]
- Yang, X.; Dong, G.; Palaniappan, K.; Mi, G.; Baskin, T.I. Temperature-compensated cell production rate and elongation zone length in the root of Arabidopsis thaliana. Plant Cell Environ. 2017, 40, 264–276. [Google Scholar] [CrossRef]
- Eysholdt-Derzso’, E.; Sauter, M. Hypoxia and the group VII ethylene response transcription factor HRE2 promote adventitious root elongation in Arabidopsis. Plant Biol. 2019, 21, 103–108. [Google Scholar] [CrossRef]
- Yamauchi, T.; Colmer, T.D.; Pedersen, O.; Nakazono, M. Regulation of root traits for internal aeration and tolerance to soil waterlogging-flooding stress. Plant Physiol. 2018, 176, 1118–1130. [Google Scholar] [CrossRef]
- Schneider, H.M.; Lynch, J.P. Should root plasticity be a crop breeding target? Front. Plant Sci. 2020, 11, 546. [Google Scholar] [CrossRef]
- Sojikul, P.; Saithong, T.; Kalapanulak, S.; Pisuttinusart, N.; Limsirichaikul, S.; Tanaka, M.; Utsumi, Y.; Sakurai, T.; Seki, M.; Narangajavana, J. Genome-wide analysis reveals phytohormone action during cassava storage root initiation. Plant Mol. Biol. 2015, 88, 531–554. [Google Scholar] [CrossRef]
- Patil, S.; Srividhya, A.; Veeraghattapu, R.; Deborah, D.A.K.; Kadambari, G.M.; Nagireddy, R.; Siddiq, E.A.; Vemireddy, L.R. Molecular dissection of a genomic region governing root traits associated with drought tolerance employing a combinatorial approach of QTL mapping and RNA-seq in rice. Plant Mol. Biol. Rep. 2017, 35, 457–468. [Google Scholar] [CrossRef]
- Muthurajan, R.; Rahman, H.; Manoharan, M.; Ramanathan, V.; Nallathambi, J. Drought responsive transcriptome profiling in roots of contrasting rice genotypes. Indian J. Plant Physiol. 2018, 23, 393–407. [Google Scholar] [CrossRef]
- Müller, M.; Munné-Bosch, S. Ethylene response factors: A key regulatory hub in hormone and stress signaling. Plant Physiol. 2015, 169, 32–41. [Google Scholar] [CrossRef]
- Bhaskarla, V.; Zinta, G.; Ford, R.; Jain, M.; Varshney, R.K.; Mantri, N. Comparative root transcriptomics provide insights into drought adaptation strategies in chickpea (Cicer arietinum L.). Int. J. Mol. Sci. 2020, 21, 1781. [Google Scholar] [CrossRef]
- Torres, G.A.; Lelandais-Brière, C.; Besin, E.; Jubier, M.F.; Roche, O.; Mazubert, C.; Hartmann, C. Characterization of the expression of Phaseolus vulgaris OCT1, a dehydration-regulated gene that encodes a new type of phloem transporter. Plant Mol. Biol. 2003, 51, 341–349. [Google Scholar] [CrossRef]
- Moon, S.; Chandran, A.K.N.; Gho, Y.S.; Park, S.A.; Kim, S.R.; Yoo, Y.H.; Jung, K.H. Integrated omics analysis of root-preferred genes across diverse rice varieties including Japonica and indica cultivars. J. PlantPhysiol. 2018, 220, 11–23. [Google Scholar] [CrossRef]
- Kong, W.; Zhong, H.; Gong, Z.; Fang, X.; Sun, T.; Deng, X.; Li, Y. Meta-analysis of salt stress transcriptome responses in different rice genotypes at the seedling stage. Plants 2019, 8, 64. [Google Scholar] [CrossRef]
- Joshi, S.; Nath, J.; Singh, A.K.; Pareek, A.; Joshi, R. Ion transporters and their regulatory signal transduction mechanisms for salinity tolerance in plants. Physiol. Plant. 2022, 174, e13702. [Google Scholar] [CrossRef]
- Huang, G.T.; Ma, S.L.; Bai, L.P.; Zhang, L.; Ma, H.; Jia, P.; Liu, J.; Zhong, M.; Guo, Z.F. Signal transduction during cold, salt, and drought stresses in plants. Mol. Biol. Rep. 2012, 39, 969–987. [Google Scholar] [CrossRef]
- Aung, B.; Gao, R.; Gruber, M.Y.; Yuan, Z.C.; Sumarah, M.; Hannoufa, A. MsmiR156 affects global gene expression and promotes root regenerative capacity and nitrogen fixation activity in alfalfa. Tran. Res. 2017, 26, 541–557. [Google Scholar] [CrossRef]
- Mangrauthia, S.K.; Bhogireddy, S.; Agarwal, S.; Prasanth, V.V.; Voleti, S.R.; Neelamraju, S.; Subrahmanyam, D. Genome-wide changes in microRNA expression during short and prolonged heat stress and recovery in contrasting rice cultivars. J. Expt. Bot. 2017, 68, 2399–2412. [Google Scholar] [CrossRef]
- Mo, C.; Wan, S.; Xia, Y.; Ren, N.; Zhou, Y.; Jiang, X. Expression patterns and identified protein-protein interactions suggest that cassava CBL-CIPK signal networks function in responses to abiotic stresses. Front. Plant Sci. 2018, 9, 269. [Google Scholar] [CrossRef]
- Carrasco-Gil, S.; Ortega-Villasante, C.; Sobrino-Plata, J.; Barón-Sola, Á.; Millán, R.; Hernandez, L.E. Attenuation of Mercury Phytotoxicity with a High Nutritional Level of Nitrate in Alfalfa Plants Grown Hydroponically. Res. Sqr. 2021. In print. [Google Scholar] [CrossRef]
- Krouk, G.; Lacombe, B.; Bielach, A.; Perrine-Walker, F.; Malinska, K.; Mounier, E. Nitrate-regulated auxin transport by NRT1. 1 defines a mechanism for nutrient sensing in plants. Dev. Cell 2010, 18, 927–937. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Jiang, H.; Wang, H.; Cui, J.; Wang, J.; Hu, J.; Guo, L.; Qian, Q.; Xue, D. Transcriptome analysis of rice seedling roots in response to potassium deficiency. Sci. Rep. 2017, 7, 5523. [Google Scholar] [CrossRef] [PubMed]
- Balzergue, C.; Dartevelle, T.; Godon, C.; Laugier, E.; Meisrimler, C. Low phosphate activates STOP1-ALMT1 to rapidly inhibit root cell elongation. Nat. Commum. 2017, 8, 15300. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, H.; Ai, Q.; Liang, G.; Yu, D. Two bHLH Transcription Factors, bHLH34 and bHLH104, Regulate Iron Homeostasis in Arabidopsis thaliana. Plant Physiol. 2016, 170, 2478–2493. [Google Scholar] [CrossRef] [PubMed]
- Joshi, R.; Pareek, A.; Singla-Pareek, S.L. Plant metallothioneins: Classification, distribution, function, and regulation. In Plant Metal Interaction; Ahmad, P., Ed.; Elsevier: London, UK, 2016; pp. 239–261. [Google Scholar]
- Ruzicka, K.; Ljung, K.; Vanneste, S.; Podhorska, R.; Beeckman, T.; Friml, J. Ethylene regulates root growth through effects on auxin biosynthesis and transportdependent auxin distribution. Plant Cell 2007, 19, 2197–2212. [Google Scholar] [CrossRef]
- Brinke, A.; Reifferscheid, G.; Klein, R.; Feiler, U.; Buchinger, S. Transcriptional changes measured in rice roots after exposure to arsenite-contaminated sediments. Environ. Sci. Pollut. Res. 2018, 25, 2707–2717. [Google Scholar] [CrossRef]
- Kumar, G.; Kushwaha, H.R.; Panjabi-Sabharwal, V.; Kumari, S.; Joshi, R.; Karan, R.; Mittal, S.; Pareek, S.L.S.; Pareek, A. Clustered metallothionein genes are co-regulated in rice and ectopic expression of OsMT1e-Pconfers multiple abiotic stress tolerance in tobacco via ROS scavenging. BMC Plant Biol. 2012, 12, 107. [Google Scholar] [CrossRef]
- Komatsu, S.; Wada, T.; Abaléa, Y.; Nouri, M.Z.; Nanjo, Y.; Nakayama, N.; Shimamura, S.; Yamamoto, R.; Nakamura, T.; Furukawa, K. Analysis of plasma membrane proteome in soybean and application to flooding stress response. J. Proteome Res. 2009, 8, 4487–4499. [Google Scholar] [CrossRef]
- Nanjo, Y.; Maruyama, K.; Yasue, H.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Komatsu, S. Transcriptional responses to flooding stress in roots including hypocotyl of soybean seedlings. Plant Mol. Biol. 2011, 77, 129–144. [Google Scholar] [CrossRef]
- Nanjo, Y.; Nouri, M.Z.; Komatsu, S. Quantitative proteomic analyses of crop seedlings subjected to stress conditions; a commentary. Phytochemistry 2011, 2, 1263–1272. [Google Scholar] [CrossRef]
- O’Malley, R.C.; Huang, S.C.; Song, L.; Lewsey, M.G.; Bartlett, A.; Nery, J.R.; Galli, M.; Gallavotti, A.; Ecker, J.R. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 2016, 165, 1280–1292. [Google Scholar] [CrossRef]
- Hsieh, P.H.; Kan, C.C.; Wu, H.Y.; Yang, H.C.; Hsieh, M.H. Early molecular events associated with nitrogen deficiency in rice seedling roots. Sci. Rep. 2018, 8, 12207. [Google Scholar] [CrossRef]
- Shi, X.; Wang, X.; Cheng, F.; Cao, H.; Liang, H.; Lu, J.; Kong, Q.; Bie, Z. iTRAQ-based quantitative proteomics analysis of cold stress-induced mechanisms in grafted watermelon seedlings. J. Proteom. 2019, 192, 311–320. [Google Scholar] [CrossRef]
- Wu, G.; Tian, N.; She, F.; Cao, A.; Wu, W.; Zheng, S.; Yang, N. Characteristics analysis of Early Responsive to Dehydration genes in Arabidopsis thaliana (AtERD). Plant Sign. Behav. 2022, 31, 2105021. [Google Scholar] [CrossRef]
- Lee, D.G.; Ahsan, N.; Lee, S.H.; Lee, J.J.; Bahk, J.D.; Kang, K.Y. Chilling stress-induced proteomic changes in rice roots. J. Plant Physiol. 2009, 166, 1–11. [Google Scholar] [CrossRef]
- Renaut, J.; Lutts, S.; Hoffmann, L.; Hausman, J.F. Responses of poplar to chilling temperatures: Proteomic and physiological aspects. Plant Biol. 2004, 6, 81–90. [Google Scholar] [CrossRef]
- Mohammadi, P.P.; Moieni, A.; Komatsu, S. Comparative proteome analysis of drought-sensitive and drought-tolerant rapeseed roots and their hybrid F1 line under drought stress. Amino Acids 2012, 43, 2137–2152. [Google Scholar] [CrossRef]
- Yoshimura, K.; Masuda, A.; Kuwano, M.; Yokota, A.; Akashi, K. Programmed proteome response for drought avoidance/tolerance in the root of a C3 xerophyte (wild watermelon) under water deficits. Plant Cell Physiol. 2008, 49, 226–241. [Google Scholar] [CrossRef]
- Du, C.X.; Fan, H.F.; Guo, S.R.; Tezuka, T.; Li, J. Proteomic analysis of cucumber seedling roots subjected to salt stress. Phytochemistry 2010, 71, 1450–1459. [Google Scholar] [CrossRef]
- Zhou, S.; Sauvé, R.J.; Liu, Z.; Reddy, S.; Bhatti, S.; Hucko, S.D. Identification of salt-induced changes in leaf and root proteomes of the wild tomato, Solanum chilense. J. Am. Soc. Hort. Sci. 2011, 136, 288–302. [Google Scholar] [CrossRef]
- Jiang, Y.; Yang, B.; Harris, N.S.; Deyholos, M.K. Comparative proteomic analysis of NaCl stress-responsive proteins in Arabidopsis roots. J. Expt. Bot. 2007, 58, 3591–3607. [Google Scholar] [CrossRef]
- Komatsu, S.; Hiraga, S.; Yanagawa, Y. Proteomics techniques for the development of flood tolerant crops. J. Proteom. Res. 2012, 11, 68–78. [Google Scholar] [CrossRef]
- Yin, X.; Sakata, K.; Komatsu, S. Phosphoproteomics reveals the effect of ethylene in soybean root under flooding stress. J. Proteom. Res. 2014, 13, 5618–5634. [Google Scholar] [CrossRef]
- Oh, M.W.; Nanjo, Y.; Komatsu, S. Identification of nuclear proteins in soybean under flooding stresses using proteomic technique. Protein Pept. Lett. 2014, 21, 458–467. [Google Scholar] [CrossRef]
- Utsumi, Y.; Tanaka, M.; Utsumi, C.; Takahashi, S.; Matsui, A.; Fukushima, A.; Seki, M. Integrative omics approaches revealed a crosstalk among phytohormones during tuberous root development in cassava. Plant Mol. Biol. 2020, 109, 249–262. [Google Scholar] [CrossRef]
- Kang, B.; Zhang, Z.; Wang, L.; Zheng, L.; Mao, W.; Li, M. OsCYP2, a chaperone involved in degradation of auxin-responsive proteins, plays crucial roles in rice lateral root initiation. Plant J. 2013, 74, 86–97. [Google Scholar] [CrossRef]
- Zheng, H.; Li, S.; Ren, B.; Zhang, J.; Ichii, M.; Taketa, S. LATERAL ROOTLESS2, a cyclophilin protein, regulates lateral root initiation and auxin signalling pathway in rice. Mol. Plants 2013, 6, 1719–1721. [Google Scholar] [CrossRef]
- Hassan, S.; Mathesius, U. The role of flavonoids in root-rhizosphere signalling: Opportunities and challenges for improving plant-microbe interactions. J. Expt. Bot. 2012, 63, 3429–3444. [Google Scholar] [CrossRef]
- Jain, M.; Ghanashyam, C.; Bhattacharjee, A. Comprehensive expression analysis suggests overlapping and specific roles of rice glutathione S-transferase genes during development and stress responses. BMC Genom. 2010, 11, 73. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, J.P.; Behera, L.; Sharma, S.S.; Praveena, J.; Nayak, S.K.; Samal, K.C. Omics Studies and Systems Biology Perspective towards Abiotic Stress Response in Plants. Am. J. Plant Sci. 2020, 11, 2172. [Google Scholar] [CrossRef]
- Komatsu, S.; Oh, M.W.; Jang, H.Y.; Kwon, S.J.; Kim, H.R.; Ko, J.H.; Woo, S.H.; Nanjo, Y. Proteomic analyses of soybean root tips during germination. Protein Peptide Lett. 2014, 21, 1308–1319. [Google Scholar] [CrossRef]
- Khan, N.; Bano, A.; Rahman, M.A.; Guo, J.; Kang, Z.; Babar, M.A. Comparative physiological and metabolic analysis reveals a complex mechanism involved in drought tolerance in chickpea (Cicer arietinum L.) induced by PGPR and PGRs. Sci. Rep. 2019, 9, 2097. [Google Scholar] [CrossRef] [PubMed]
- Ristova, D.; Giovannetti, M.; Metesch, K.; Busch, W. Natural genetic variation shapes root system responses to phytohormonaes in Arabidopsis. Plant J. 2018, 96, 468–481. [Google Scholar] [CrossRef]
- Laffont, C.; Blanchet, S.; Lapierre, C.; Brocard, L.; Ratet, P.; Crespi, M.; Mathesius, U.; Frugier, F. The compact root architecture1 gene regulates lignification, flavonoid production, and polar auxin transport in Medicago truncatula. Plant Physiol. 2010, 153, 1597–1607. [Google Scholar] [CrossRef]
- Rabara, R.C.; Tripathi, P.; Rushton, P.J. Comparative metabolome profile between tobacco and soybean grown under water-stressed conditions. BioMed Res. Int. 2017, 2017, 3065251. [Google Scholar] [CrossRef]
- Ho, W.W.H.; Hill, C.B.; Doblin, M.S.; Shelden, M.C.; van de Meene, A.; Rupasinghe, T.; Roessner, U. Integrative multi-omics analyses of barley rootzones under salinity stress reveal two distinctive salt tolerance mechanisms. Plant Commum. 2020, 1, 100031. [Google Scholar] [CrossRef]

| Stress Responsive Genes/QTLs | Gene Names | Function | Reference | |
|---|---|---|---|---|
| Genomics | AtBZR1 | Brassinazole-Resistant 1 | RGR regulation under Zn deficiency | [40] |
| AtPHR1 | Phosphare Response 1 | Responsible for crosstalk among Fe and P signalling to control root growth | ||
| AtVIM1 | Variant in Methylation 1 | RGR regulation under P–Zn deficiency | ||
| AtVDAC3 | Voltage-Dependent Anion-Selective Channel Protein 3 | Primary root growth control under P–Fe deficiency | ||
| OsDRO1 | Deep Rooting 1 | Deep rooting under drought stress | [41] | |
| OsDRO2 | Deep Rooting 2 | Root angle maintenance under drought stress | [42] | |
| CaLG04 | QTLs | Root length maintenance under drought stress | [43] | |
| CaLG06 | Root surface area maintenance under drought stress | |||
| OsqDTY3.2 | Maintain whole plant water level under drought stress | [44] | ||
| Transcriptomics | SlPIP1;7 | Plasma membrane Intrinsic Protein 1;7 | Promotes root growth along with tolerance to drought | [45] |
| CaTIP2/3 | Tonoplast Intrinsic Protein 2/3 | Drought tolerance | [46] | |
| CaNIP6;3 | NOD26 Like Protein 6;3 | Drought tolerance | ||
| TaPSTOL1 | Phosphorus Starvation Tolerance | Early root growth under phosphorus deficiency | [47] | |
| AtNTR1.1 | Nitrate Transporter 1.1 | Gets slightly reduced in roots under high nitrate condition | [48] | |
| Ci1-FFT | fructan 1-fructosyltransferase | Freezing tolerance in roots | [49] | |
| Proteomics | TaRab24 | RAS oncogene family protein 24 | Drought-stress tolerance and root growth improvement | [50] |
| TaPPDK, TaLEA1 and TaLEA2 | Pyruvate phosphate dikinase, Late Embryogenesis abundant 1&2 | Increase root depth, root biomass as well as tolerance to salinity stress | [50] | |
| GmRACK1 | Receptor for Activated C Kinase 1 | Regulation of flooding stress | [51] |
| Abiotic Stress | Morphological and Root Traits | References |
|---|---|---|
| Drought stress | Increased crown root angle and length | [56] |
| Lateral root development and aerenchyma formation | [44] | |
| Shallow or deep root | [22] | |
| Fewer but large size cortical cells | [58] | |
| Increased root hair length and density | [59] | |
| Salinity stress | Cellular dehydration and ionic imbalance | [60] |
| Enhanced lignin and suberin deposition in cell wall | [20] | |
| Root bending away from saline environment | [61] | |
| Growth of both primary and lateral root get arrested | [62] | |
| Decreased root hair length and density | [63] | |
| Temperature stress | Compact roots during cold stress and elongated roots during heat stress | [64] |
| Seminal root elongation in warm climates | [65] | |
| Cell elongation in root elongation zone and reduced meristem size under warm conditions | ||
| Flooding stress | Adventitious root formation | [66] |
| Formation of aerenchyma and oxygen loss barriers | [67] | |
| Nutrient stress | Reduced primary root length in Fe and Zn deficiency | [38] |
| P deficiency promotes early primary root growth and root hair proliferation | [40] | |
| Lateral root growth inhibition in nitrate deficiency | [48] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Joshi, S.; Chinnusamy, V.; Joshi, R. Root System Architecture and Omics Approaches for Belowground Abiotic Stress Tolerance in Plants. Agriculture 2022, 12, 1677. https://doi.org/10.3390/agriculture12101677
Joshi S, Chinnusamy V, Joshi R. Root System Architecture and Omics Approaches for Belowground Abiotic Stress Tolerance in Plants. Agriculture. 2022; 12(10):1677. https://doi.org/10.3390/agriculture12101677
Chicago/Turabian StyleJoshi, Shubham, Viswanathan Chinnusamy, and Rohit Joshi. 2022. "Root System Architecture and Omics Approaches for Belowground Abiotic Stress Tolerance in Plants" Agriculture 12, no. 10: 1677. https://doi.org/10.3390/agriculture12101677
APA StyleJoshi, S., Chinnusamy, V., & Joshi, R. (2022). Root System Architecture and Omics Approaches for Belowground Abiotic Stress Tolerance in Plants. Agriculture, 12(10), 1677. https://doi.org/10.3390/agriculture12101677






