The Impact of Different Planting Systems on the Bacterial Diversity of Rice Cultivated in Saline Soil Based on 16S rRNA Gene-Based Metagenomic Insights
Abstract
1. Introduction
2. Materials and Methods
2.1. Soil Sample Collection
2.2. Estimation of Physicochemical Properties of Soil Samples
2.3. Soil Enzyme Analysis
2.4. Genomic DNA Extraction
2.5. 16S rRNA Gene (V3–V4) Region Amplification and Data Processing
2.6. Data
2.7. Statistical Analysis
3. Results
3.1. Collection and Analyses of Soil Samples from Different Planting Systems
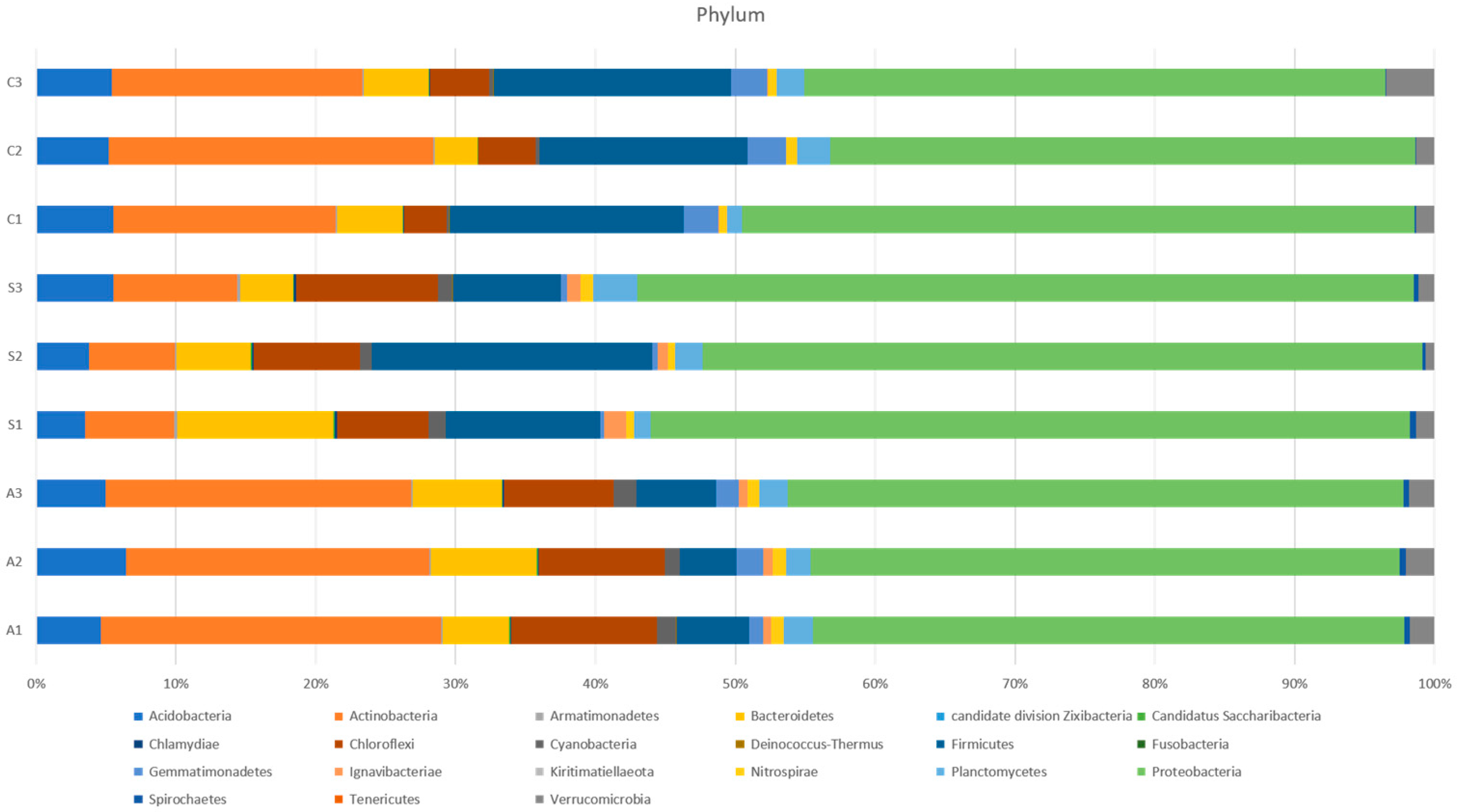
3.2. The 16S rRNA Gene-Based Metagenomic Analysis of Soil Samples
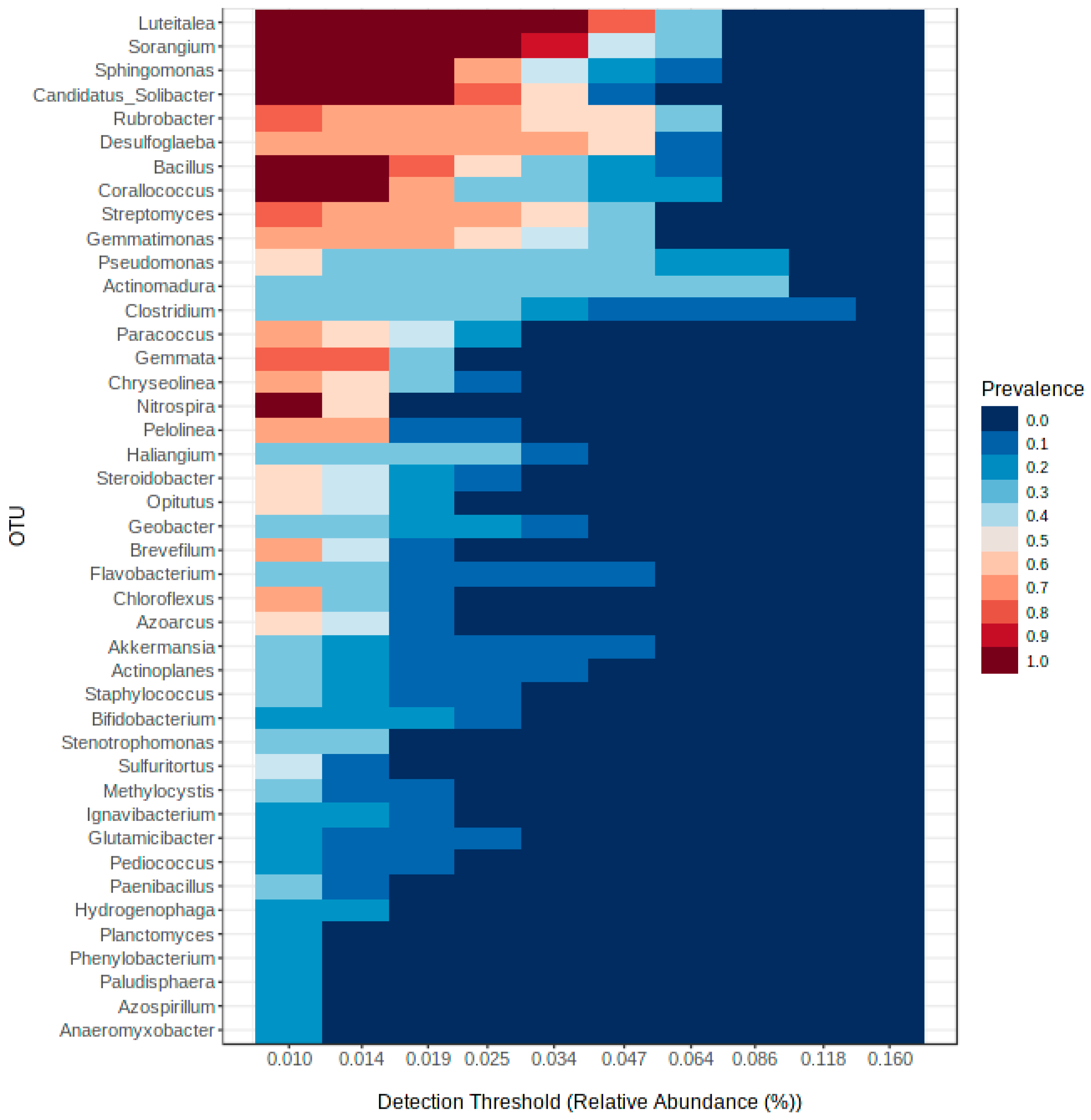
3.3. Soil Bacterial Community Differs in the Rhizosphere of Rice with Different Planting Systems
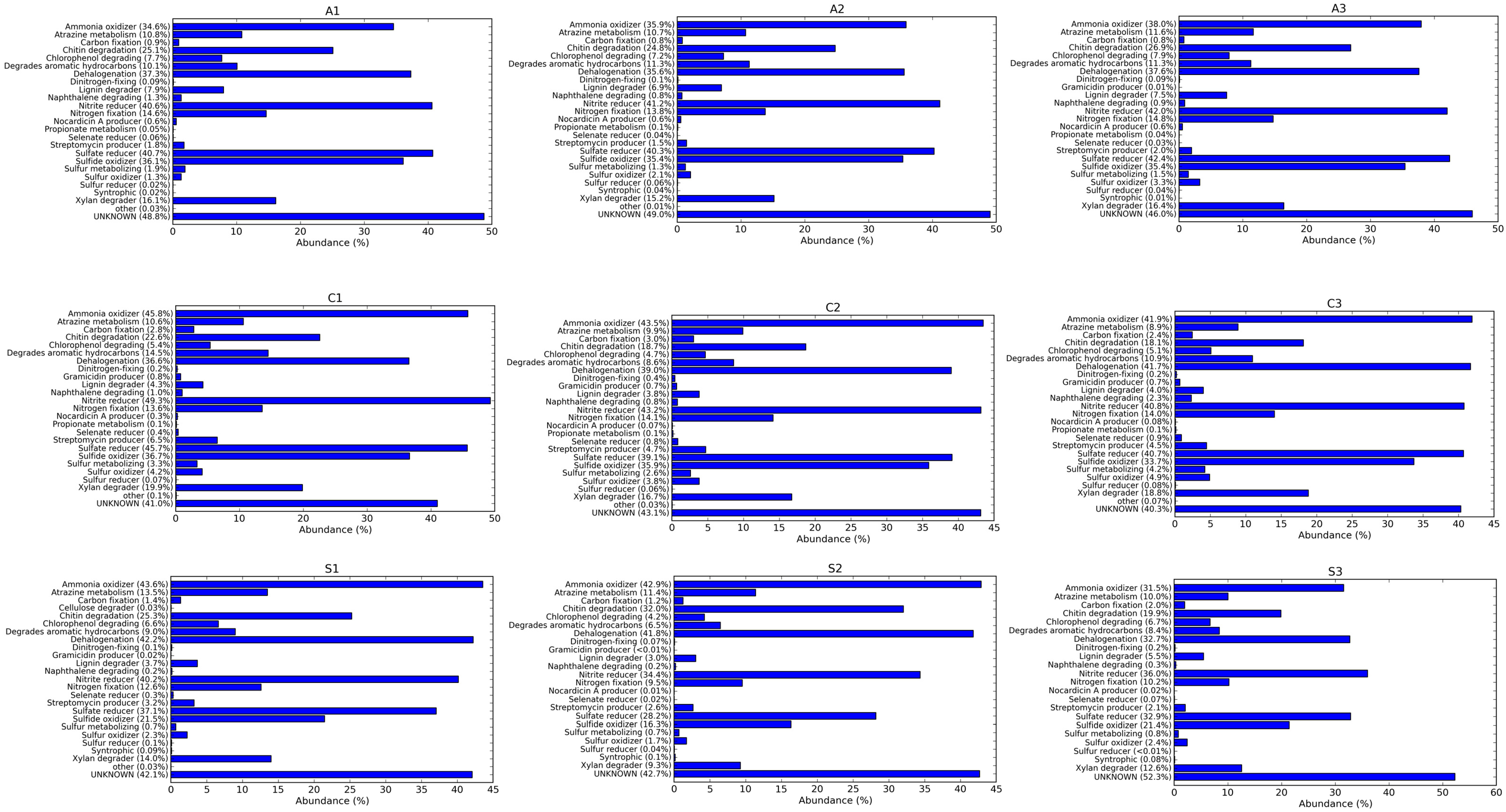
3.4. Comparative Analysis on the Bacterial Community Associated with Aerobic, Conventional and SRI Soil Samples
4. Discussion
4.1. Soil Salinity, Soil Characteristics and Microbial Community
4.2. Rhizosphere Bacterial Community Differs with Different Planting Systems
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Xu, Z.; Shao, T.; Lv, Z.; Yue, Y.; Liu, A.; Long, X.; Zhou, Z.; Gao, X.; Rengel, Z. The mechanisms of improving coastal saline soils by planting rice. Sci. Total Environ. 2020, 703, 135529. [Google Scholar] [CrossRef]
- Shrivastava, P.; Kumar, R. Soil salinity: A serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 2015, 22, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.R.; Mele, P.M. Subtle changes in rhizosphere microbial community structure in response to increased boron and sodium chloride concentrations. Soil Biol. Biochem. 2007, 39, 340–351. [Google Scholar] [CrossRef]
- Warrence, N.J.; Bauder, J.W.; Pearson, K.E. Basics of salinity and sodicity effects on soil physical properties. Dep. Land Resour. Environ. Sci. Mont. State Univ.-Bozeman MT 2002, 129, 1–29. [Google Scholar]
- Setia, R.; Smith, P.; Marschner, P.; Gottschalk, P.; Baldock, J.; Verma, V.; Setia, D.; Smith, J. Simulation of salinity effects on past, present, and future soil organic carbon stocks. Environ. Sci. Technol. 2012, 46, 1624–1631. [Google Scholar] [CrossRef] [PubMed]
- Saviozzi, A.; Cardelli, R.; Di Puccio, R. Impact of salinity on soil biological activities: A laboratory experiment. Commun. Soil Sci. Plant Anal. 2011, 42, 358–367. [Google Scholar] [CrossRef]
- Nannipieri, P.; Grego, S.; Ceccanti, B.; Bollag, J.; Stotzky, G. Ecological significance of the biological activity in soil. Soil Biochem. 1990, 293–356. [Google Scholar]
- Sharma, G.; Mishra, R. Soil microbial population numbers and enzyme activities in relation to altitude and forest degradation. Soil Biol. Biochem. 1992, 24, 761–767. [Google Scholar]
- Xiaobin, W.; Jingfeng, X.; Grant, C.; Bailey, L. Effects of placement of urea with a urease inhibitor on seedling emergence, N uptake and dry matter yield of wheat. Can. J. Plant Sci. 1995, 75, 449–452. [Google Scholar] [CrossRef][Green Version]
- Fitzgerald, J.W. Sulfate ester formation and hydrolysis: A potentially important yet often ignored aspect of the sulfur cycle of aerobic soils. Bacteriol. Rev. 1976, 40, 698–721. [Google Scholar] [CrossRef] [PubMed]
- Muthayya, S.; Sugimoto, J.D.; Montgomery, S.; Maberly, G.F. An overview of global rice production, supply, trade, and consumption. Ann. New York Acad. Sci. 2014, 1324, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, P.K.; Hazra, K.K.; Nath, C.P.; Das, A.; Acharya, C.L. Scope, constraints and challenges of intensifying rice (Oryza sativa) fallows through pulses. Indian J. Agron. 2016, 61, 122–128. [Google Scholar]
- Daliakopoulos, I.N.; Tsanis, I.K.; Koutroulis, A.; Kourgialas, N.N.; Varouchakis, A.E.; Karatzas, G.P.; Ritsema, C.J. The threat of soil salinity: A European scale review. Sci. Total Environ. 2016, 573, 727–739. [Google Scholar] [CrossRef] [PubMed]
- Ansari, F.A.; Ahmad, I. Isolation, functional characterization and efficacy of biofilm-forming rhizobacteria under abiotic stress conditions. Antonie Van Leeuwenhoek 2019, 112, 1827–1839. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Gong, J. A meta-analysis of the publicly available bacterial and archaeal sequence diversity in saline soils. World J. Microbiol. Biotechnol. 2013, 29, 2325–2334. [Google Scholar] [CrossRef] [PubMed]
- Nautiyal, C.S.; Dion, P. Molecular Mechanisms of Plant and Microbe Coexistence; Springer: Berlin/Heidelberg, Germany, 2008; Volume 15. [Google Scholar]
- De Datta, S.K. Principles and Practices of Rice Production; John Wiley & Sons, Inc.: Toronto, ON, Canada, 1981. [Google Scholar]
- Senthilkumar, K.; Bindraban, P.S.; Thiyagarajan, T.M.; De Ridder, N.; Giller, K.E. Modified rice cultivation in Tamil Nadu, India: Yield gains and farmers’(lack of) acceptance. Agric. Syst. 2008, 98, 82–94. [Google Scholar] [CrossRef]
- Zhao, L.; Wu, L.; Li, Y.; Lu, X.; Zhu, D.; Uphoff, N. Influence of the system of rice intensification on rice yield and nitrogen and water use efficiency with different N application rates. Exp. Agric. 2009, 45, 275–286. [Google Scholar] [CrossRef]
- Sinha, S.K.; Talati, J. Productivity impacts of the system of rice intensification (SRI): A case study in West Bengal, India. Agric. Water Manag. 2007, 87, 55–60. [Google Scholar] [CrossRef]
- Satyanarayana, A.; Thiyagarajan, T.M.; Uphoff, N. Opportunities for water saving with higher yield from the system of rice intensification. Irrig. Sci. 2007, 25, 99–115. [Google Scholar] [CrossRef]
- Stoop, W.A.; Uphoff, N.; Kassam, A. A review of agricultural research issues raised by the system of rice intensification (SRI) from Madagascar: Opportunities for improving farming systems for resource-poor farmers. Agric. Syst. 2002, 71, 249–274. [Google Scholar] [CrossRef]
- Uphoff, N. Higher yields with fewer external inputs? The system of rice intensification and potential contributions to agricultural sustainability. Int. J. Agric. Sustain. 2003, 1, 38–50. [Google Scholar] [CrossRef]
- Jana, K.; Karmakar, R.; Banerjee, S.; Sana, M.; Goswami, S.; Puste, A.M. Aerobic rice cultivation system: Eco-friendly and water saving technology under changed climate. Agr. Sci. Tech. 2018, 13, 1–5. [Google Scholar]
- Bouman, B.A.M.; Peng, S.; Castaneda, A.R.; Visperas, R.M. Yield and water use of irrigated tropical aerobic rice systems. Agric. Water Manag. 2005, 74, 87–105. [Google Scholar] [CrossRef]
- Souza, R.C.; Cantão, M.E.; Vasconcelos, A.T.R.; Nogueira, M.A.; Hungria, M. Soil metagenomics reveals differences under conventional and no-tillage with crop rotation or succession. Appl. Soil Ecol. 2013, 72, 49–61. [Google Scholar] [CrossRef]
- Chávez-Romero, Y.; Navarro-Noya, Y.E.; Reynoso-Martínez, S.C.; Sarria-Guzmán, Y.; Govaerts, B.; Verhulst, N.; Dendooven, L.; Luna-Guido, M. 16S metagenomics reveals changes in the soil bacterial community driven by soil organic C, N-fertilizer and tillage-crop residue management. Soil Tillage Res. 2016, 159, 1–8. [Google Scholar] [CrossRef]
- Imchen, M.; Kumavath, R.; Vaz, A.B.M.; Góes-Neto, A.; Barh, D.; Ghosh, P.; Kozyrovska, N.; Podolich, O.; Azevedo, V. 16S rRNA gene amplicon based metagenomic signatures of rhizobiome community in rice field during various growth stages. Front. Microbiol. 2019, 10, 2103. [Google Scholar] [CrossRef]
- Cycil, L.M.; DasSarma, S.; Pecher, W.; McDonald, R.; AbdulSalam, M.; Hasan, F. Metagenomic insights into the diversity of halophilic microorganisms indigenous to the Karak Salt Mine, Pakistan. Front. Microbiol. 2020, 11, 1567. [Google Scholar] [CrossRef]
- Jackson, M.L. Soil Chemical Analysis: Advanced Course; Parallel Press: Madison, WI, USA, 1973. [Google Scholar]
- Subbiah, B.V.; Bajaj, J.C. A soil test procedure for assessment of available nitrogen in rice soils. Curr. Sci. 1962, 31, 196. [Google Scholar]
- Olsen, S.R. Estimation of Available Phosphorus in Soils by Extraction with Sodium Bicarbonate; US Department of Agriculture: Washington, DC, USA, 1954. [Google Scholar]
- Stanford, G.; English, L. Use of the flame photometer in rapid soil tests for K and Ca. Agron. J. 1949, 41, 446–447. [Google Scholar] [CrossRef]
- Walkley, A.; Black, I.A. An examination of the Degtjareff method for determining soil organic matter, and a proposed modification of the chromic acid titration method. Soil Sci. 1934, 37, 29–38. [Google Scholar] [CrossRef]
- Lindsay, W.L.; Norvell, W. Development of a DTPA soil test for zinc, iron, manganese, and copper. Soil Sci. Soc. Am. J. 1978, 42, 421–428. [Google Scholar] [CrossRef]
- Casida Jr, L.; Klein, D.; Santoro, T. Soil dehydrogenase activity. Soil Sci. 1964, 98, 371–376. [Google Scholar] [CrossRef]
- Tabatabai, M.; Bremner, J. Use of p-nitrophenyl phosphate for assay of soil phosphatase activity. Soil Biol. Biochem. 1969, 1, 301–307. [Google Scholar] [CrossRef]
- Tabatabai, M.; Bremner, J. Assay of urease activity in soils. Soil Biol. Biochem. 1972, 4, 479–487. [Google Scholar] [CrossRef]
- Elsgaard, L.; Andersen, G.H.; Eriksen, J. Measurement of arylsulphatase activity in agricultural soils using a simplified assay. Soil Biol. Biochem. 2002, 34, 79–82. [Google Scholar] [CrossRef]
- Arndt, D.; Xia, J.; Liu, Y.; Zhou, Y.; Guo, A.C.; Cruz, J.A.; Sinelnikov, I.; Budwill, K.; Nesbø, C.L.; Wishart, D.S. METAGENassist: A comprehensive web server for comparative metagenomics. Nucleic Acids Res. 2012, 40, W88–W95. [Google Scholar] [CrossRef] [PubMed]
- Canfora, L.; Bacci, G.; Pinzari, F.; Lo Papa, G.; Dazzi, C.; Benedetti, A. Salinity and bacterial diversity: To what extent does the concentration of salt affect the bacterial community in a saline soil? PLoS ONE 2014, 9, e106662. [Google Scholar] [CrossRef]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef]
- Edwards, J.; Johnson, C.; Santos-Medellín, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 2015, 112, E911–E920. [Google Scholar] [CrossRef]
- Rath, K.M.; Fierer, N.; Murphy, D.V.; Rousk, J. Linking bacterial community composition to soil salinity along environmental gradients. ISME J. 2019, 13, 836–846. [Google Scholar] [CrossRef]
- Demisie, W.; Liu, Z.; Zhang, M. Effect of biochar on carbon fractions and enzyme activity of red soil. Catena 2014, 121, 214–221. [Google Scholar] [CrossRef]
- Schloter, M.; Dilly, O.; Munch, J.C. Indicators for evaluating soil quality. Agric. Ecosyst. Environ. 2003, 98, 255–262. [Google Scholar] [CrossRef]
- Sipahutar, M.K.; Piapukiew, J.; Vangnai, A.S. Efficiency of the formulated plant-growth promoting Pseudomonas fluorescens MC46 inoculant on triclocarban treatment in soil and its effect on Vigna radiata growth and soil enzyme activities. J. Hazard. Mater. 2018, 344, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Frankenberger Jr, W.; Bingham, F.T. Influence of salinity on soil enzyme activities. Soil Sci. Soc. Am. J. 1982, 46, 1173–1177. [Google Scholar] [CrossRef]
- Liu, S.H.; Kang, Y.H. Changes of soil microbial characteristics in saline-sodic soils under drip irrigation. J. Soil Sci. Plant Nutr. 2014, 14, 139–150. [Google Scholar] [CrossRef]
- Paul, K.I.; Polglase, P.J.; O’Connell, A.M.; Carlyle, J.C.; Smethurst, P.J.; Khanna, P.K. Defining the relation between soil water content and net nitrogen mineralization. Eur. J. Soil Sci. 2003, 54, 39–48. [Google Scholar] [CrossRef]
- Janssen, P.H. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 2006, 72, 1719–1728. [Google Scholar] [CrossRef]
- Ramette, A. Multivariate analyses in microbial ecology. FEMS Microbiol. Ecol. 2007, 62, 142–160. [Google Scholar] [CrossRef]
- Liszka, M.J.; Clark, M.E.; Schneider, E.; Clark, D.S. Nature versus nurture: Developing enzymes that function under extreme conditions. Annu. Rev. Chem. Biomol. Eng. 2012, 3, 77–102. [Google Scholar] [CrossRef]
- Fierer, N.; Leff, J.W.; Adams, B.J.; Nielsen, U.N.; Bates, S.T.; Lauber, C.L.; Owens, S.; Gilbert, J.A.; Wall, D.H.; Caporaso, J.G. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl. Acad. Sci. USA 2012, 109, 21390–21395. [Google Scholar] [CrossRef]
- Barns, S.M.; Takala, S.L.; Kuske, C.R. Wide distribution and diversity of members of the bacterial kingdom Acidobacterium in the environment. Appl. Environ. Microbiol. 1999, 65, 1731–1737. [Google Scholar] [CrossRef] [PubMed]
- Foesel, B.U.; Nägele, V.; Naether, A.; Wüst, P.K.; Weinert, J.; Bonkowski, M.; Lohaus, G.; Polle, A.; Alt, F.; Oelmann, Y. Determinants of Acidobacteria activity inferred from the relative abundances of 16S rRNA transcripts in German grassland and forest soils. Environ. Microbiol. 2014, 16, 658–675. [Google Scholar] [CrossRef] [PubMed]
- Björnsson, L.; Hugenholtz, P.; Tyson, G.W.; Blackall, L.L. Filamentous Chloroflexi (green non-sulfur bacteria) are abundant in wastewater treatment processes with biological nutrient removalccThe EMBL accession numbers for the sequences reported in this paper are X84472 (strain SBR1029 16S rDNA), X84474 (strain SBR1031 16S rDNA), X84498 (strain SBR1064 16S rDNA), X84565 (strain SBR2022 16S rDNA), X84576 (strain SBR2037 16S rDNA) and X84607 (strain SBR2076 16S rDNA). Microbiology 2002, 148, 2309–2318. [Google Scholar] [PubMed]
- Jackson, K.L.; Whitcraft, C.R.; Dillon, J.G. Diversity of Desulfobacteriaceae and overall activity of sulfate-reducing microorganisms in and around a salt pan in a southern California coastal wetland. Wetlands 2014, 34, 969–977. [Google Scholar] [CrossRef]
- Purkhold, U.; Pommerening-Röser, A.; Juretschko, S.; Schmid, M.C.; Koops, H.-P.; Wagner, M. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: Implications for molecular diversity surveys. Appl. Environ. Microbiol. 2000, 66, 5368–5382. [Google Scholar] [CrossRef]
- Liang, B.; Wang, L.-Y.; Mbadinga, S.M.; Liu, J.-F.; Yang, S.-Z.; Gu, J.-D.; Mu, B.-Z. Anaerolineaceae and Methanosaeta turned to be the dominant microorganisms in alkanes-dependent methanogenic culture after long-term of incubation. Amb Express 2015, 5, 37. [Google Scholar] [CrossRef]
- Belimov, A.A.; Safronova, V.I.; Sergeyeva, T.A.; Egorova, T.N.; Matveyeva, V.A.; Tsyganov, V.E.; Borisov, A.Y.; Tikhonovich, I.A.; Kluge, C.; Preisfeld, A. Characterization of plant growth promoting rhizobacteria isolated from polluted soils and containing 1-aminocyclopropane-1-carboxylate deaminase. Can. J. Microbiol. 2001, 47, 642–652. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Sun, Y.; Xie, X.; Kim, M.S.; Dowd, S.E.; Paré, P.W. A soil bacterium regulates plant acquisition of iron via deficiency-inducible mechanisms. Plant J. 2009, 58, 568–577. [Google Scholar] [CrossRef]
- Li, D.-M.; Alexander, M. Co-inoculation with antibiotic-producing bacteria to increase colonization and nodulation by rhizobia. Plant Soil 1988, 108, 211–219. [Google Scholar]
- Nimnoi, P.; Pongsilp, N.; Lumyong, S. Co-inoculation of soybean (Glycine max) with actinomycetes and Bradyrhizobium japonicum enhances plant growth, nitrogenase activity and plant nutrition. J. Plant Nutr. 2014, 37, 432–446. [Google Scholar] [CrossRef]
- Li, Z.; Ye, X.; Chen, P.; Ji, K.; Zhou, J.; Wang, F.; Dong, W.; Huang, Y.; Zhang, Z.; Cui, Z. Antifungal potential of Corallococcus sp. strain EGB against plant pathogenic fungi. Biol. Control 2017, 110, 10–17. [Google Scholar] [CrossRef]
- Hesse, C.; Schulz, F.; Bull, C.T.; Shaffer, B.T.; Yan, Q.; Shapiro, N.; Hassan, K.A.; Varghese, N.; Elbourne, L.D.H.; Paulsen, I.T. Genome-based evolutionary history of Pseudomonas spp. Environ. Microbiol. 2018, 20, 2142–2159. [Google Scholar] [CrossRef] [PubMed]
- Muleta, D.; Assefa, F.; Hjort, K.; Roos, S.; Granhall, U. Characterization of Rhizobacteria isolated from Wild Coffea arabica L. Eng. Life Sci. 2009, 9, 100–108. [Google Scholar] [CrossRef]
- Podile, A.R.; Kishore, G.K. Plant growth-promoting rhizobacteria. In Plant-Associated Bacteria; Springer: Dordrecht, The Netherlands, 2007; pp. 195–230. [Google Scholar]
- Teske, A.; Dhillon, A.; Sogin, M.L. Genomic markers of ancient anaerobic microbial pathways: Sulfate reduction, methanogenesis, and methane oxidation. Biol. Bull. 2003, 204, 186–191. [Google Scholar] [CrossRef] [PubMed]
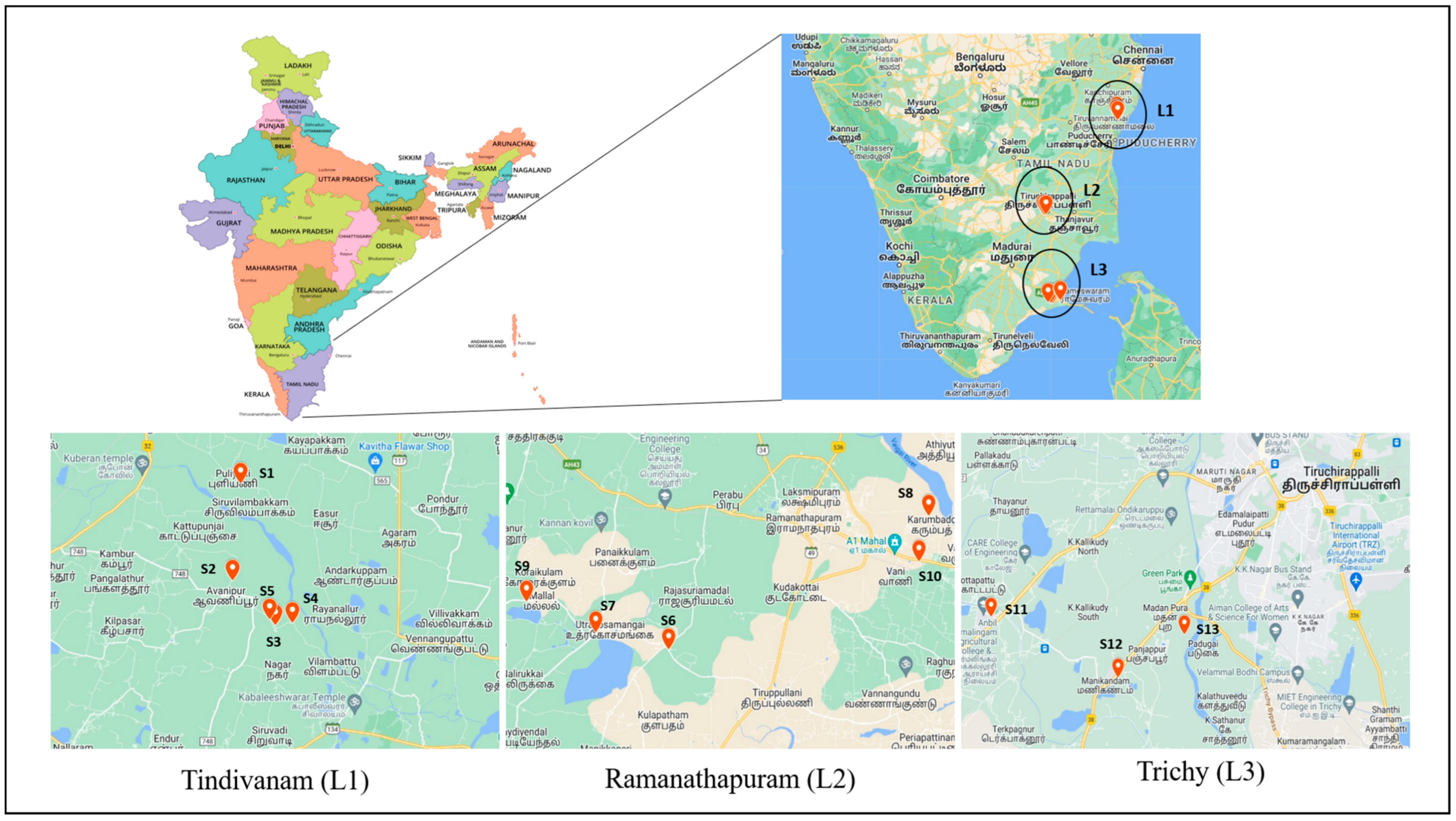


| Soil Sample No. | District | Rice Cultivar | Planting System | GPRS Co-Ordinates |
|---|---|---|---|---|
| S1 | Tindivanam (L1) | ADT 39 | Conventional | N 12.28324° E 079.821620° |
| S2 | Tindivanam (L1) | ADT 37 | SRI | N 12.28404° E 079.82057° |
| S3 | Tindivanam (L1) | ADT 39 | Conventional | N 12.26342° E 079.84108° |
| S4 | Tindivanam (L1) | BPT 5204 | Conventional | N 12.26443° E 079.84918° |
| S5 | Tindivanam (L1) | NLR 34449 | Conventional | N 12.26608° E 079.83827° |
| S6 | Ramanathapuram (L2) | Co51 | Conventional | N 09.30568° E 078.77067° |
| S7 | Ramanathapuram (L2) | Anna 4 | Aerobic | N 09.31370° E 078.73357° |
| S8 | Ramanathapuram (L2) | Co51 | Conventional | N 09.36709° E 078.90277° |
| S9 | Ramanathapuram (L2) | Co51 | Conventional | N 09.32794° E 078.69864° |
| S10 | Ramanathapuram (L2) | Co51 | Conventional | N 09.34616° E 078.89782° |
| S11 | Trichy (L3) | TRY 2 | SRI | N 10.75521° E 078.60307° |
| S12 | Trichy (L3) | Co51 | Conventional | N 10.73981° E 078.63986° |
| S13 | Trichy (L3) | ADT 36 | Conventional | N 10.75083° E 078.65900° |
| Soil Sample No. | pH | E.C (dSm−1) | Nitrogen (Kg ha−1) | Phosphorus (Kg ha−1) | Potassium (Kg ha−1) | Organic Carbon (%) | Soil Dehydrogenase (µg Triphenyl Formazan g−1 h−1) | Alkaline Phosphatase Activity (Phenol µg g−1 h−1) | Urease Activity (µg NH4-N g−1 h−1) | Arylsulphatase Activity (µg p Nitrophenol g−1 h−1) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 7.1 ± 0.21 | 6.25 ± 0.05 | 139.24 ± 6.53 | 16.9 ± 0.12 | 77.6 ± 1.16 | 0.64 ± 0.02 | 0.84 ± 0.044 f | 0.9 ± 0.038 g | 56 ± 1.283 e | 10.8 ± 0.469 f |
| 2 | 7.5 ± 0.21 | 7.84 ± 0.12 | 145.92 ± 0.59 | 17.7 ± 0.75 | 84.9 ± 4.06 | 0.72 ± 0.03 | 1.03 ± 0.002 e | 4.04 ± 0.204 e | 56 ± 2.351 e | 17.6 ± 0.247 d |
| 3 | 7.7 ± 0.39 | 6.96 ± 0.26 | 176.38 ± 1.07 | 21.2 ± 0.14 | 79.6 ± 3.5 | 0.68 ± 0.01 | 0.36 ± 0.016 h | 0.26 ± 0.012 h | 112 ± 4.104 c | 19.9 ± 0.371 c |
| 4 | 7.8 ± 0.11 | 7.59 ± 0.2 | 157.46 ± 3.49 | 19.4 ± 0.85 | 74.2 ± 2.74 | 0.87 ± 0.03 | 0.97 ± 0.008 e | 0.35 ± 0.09 h | 70 ± 0.204 d | 13.4 ± 0.093 e |
| 5 | 7.6 ± 0.08 | 8.06 ± 0.42 | 159.87 ± 5.64 | 17.6 ± 0.62 | 82.1 ± 2.38 | 0.84 ± 0.01 | 0.99 ± 0.028 e | 5.34 ± 0.012 d | 70 ± 0.353 d | 20.3 ± 0.118 bc |
| 6 | 7.2 ± 0.09 | 8.43 ± 0.38 | 164.25 ± 1.09 | 21.7 ± 0.23 | 102.5 ± 3.13 | 0.76 ± 0.01 | 1.44 ± 0.022 c | 7.69 ± 0.103 c | 224 ± 7.538 a | 21.5 ± 0.996 b |
| 7 | 6.7 ± 0.33 | 7.82 ± 0.36 | 168.64 ± 0.08 | 20.5 ± 0.04 | 85.6 ± 3.21 | 0.62 ± 0.03 | 1.53 ± 0.068 b | 10.81 ± 0.216 a | 140 ± 6.475 b | 14.5 ± 0.430 e |
| 8 | 6.8 ± 0.14 | 7.46 ± 0.04 | 140.78 ± 3.01 | 16.8 ± 0.1 | 83.5 ± 0.75 | 0.81 ± 0.01 | 0.67 ± 0.021 g | 0.31 ± 0.08 h | 112 ± 1.741 c | 7.5 ± 0.292 gh |
| 9 | 7 ± 0.14 | 8.22 ± 0.32 | 157.88 ± 1.48 | 15.2 ± 0.32 | 98.7 ± 4.5 | 0.85 ± 0.02 | 0.78 ± 0.040 f | 0.42 ± 0.014 gh | 112 ± 3.972 c | 8.8 ± 0.146 g |
| 10 | 6.6 ± 0.31 | 7.81 ± 0.25 | 143.28 ± 6.31 | 18.8 ± 0.08 | 92.4 ± 2.11 | 0.74 ± 0.01 | 0.38 ± 0.012 h | 0.54 ± 0.003 gh | 112 ± 1.179 c | 7.1 ± 0.047 h |
| 11 | 7.2 ± 0.08 | 5.67 ± 0.08 | 275.98 ± 3.41 | 28.7 ± 1.04 | 105.3 ± 2.67 | 0.69 ± 0.01 | 2.1 ± 0.084 a | 9.35 ± 0.253 b | 224 ± 8.137 a | 23.6 ± 0.745 a |
| 12 | 7 ± 0.06 | 4.59 ± 0.15 | 191.43 ± 5.37 | 26.1 ± 0.66 | 101.7 ± 1.47 | 0.76 ± 0.02 | 1.16 ± 0.037 d | 2.16 ± 0.005 f | 56 ± 0.644 e | 16.5 ± 0.339 d |
| 13 | 7.6 ± 0.34 | 4.94 ± 0.02 | 288.64 ± 11.12 | 25.3 ± 1.19 | 99.4 ± 3.81 | 0.84 ± 0.02 | 1.12 ± 0.001 d | 0.75 ± 0.024 gh | 56 ± 2.140 e | 17.4 ± 0.652 d |
| Soil Sample | Raw Reads | Quality Filtered Reads | No. of OTU’S | GC Content (%) |
|---|---|---|---|---|
| A1 | 289,490 | 288,410 | 29,846 | 57.5 |
| A2 | 275,378 | 274,158 | 27,537 | 57.5 |
| A3 | 406,084 | 304,290 | 33,972 | 57.5 |
| C1 | 278,482 | 277,174 | 41,482 | 56 |
| C2 | 262,030 | 260,678 | 31,512 | 56.5 |
| C3 | 247,480 | 244,152 | 30,186 | 57.5 |
| S1 | 254,354 | 252,488 | 29,072 | 58 |
| S2 | 244,802 | 243,724 | 26,713 | 59 |
| S3 | 258,600 | 256,730 | 23,127 | 58 |
| Total | 2,516,700 | 2,401,804 | 273,447 |
| Chao1 * | Shannon | Simpson | Fisher * | |
|---|---|---|---|---|
| Aerobic | 169.34 ± 3.2 ab | 3.8 ± 0.03 | 0.961 ± 0.01 | 25.5 ± 0.78 b |
| Conventional | 182 ± 2.1 ab | 4.01 ± 0.1 | 0.963 ± 0.01 | 28.17 ± 0.41 a |
| SRI | 189.67 ± 6.2 a | 4.05 ± 0.06 | 0.97 ± 0.01 | 28.5 ± 0.18 a |
| p value | 0.038 | 0.071 | 0.314 | 0.012 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Davidson Rokins, P.; Gopal, N.O.; Anandham, R.; Saraswathi, R. The Impact of Different Planting Systems on the Bacterial Diversity of Rice Cultivated in Saline Soil Based on 16S rRNA Gene-Based Metagenomic Insights. Agriculture 2022, 12, 1624. https://doi.org/10.3390/agriculture12101624
Davidson Rokins P, Gopal NO, Anandham R, Saraswathi R. The Impact of Different Planting Systems on the Bacterial Diversity of Rice Cultivated in Saline Soil Based on 16S rRNA Gene-Based Metagenomic Insights. Agriculture. 2022; 12(10):1624. https://doi.org/10.3390/agriculture12101624
Chicago/Turabian StyleDavidson Rokins, Pugazhenthi, Nellaiappan Olaganathan Gopal, Rangasamy Anandham, and Ramasamy Saraswathi. 2022. "The Impact of Different Planting Systems on the Bacterial Diversity of Rice Cultivated in Saline Soil Based on 16S rRNA Gene-Based Metagenomic Insights" Agriculture 12, no. 10: 1624. https://doi.org/10.3390/agriculture12101624
APA StyleDavidson Rokins, P., Gopal, N. O., Anandham, R., & Saraswathi, R. (2022). The Impact of Different Planting Systems on the Bacterial Diversity of Rice Cultivated in Saline Soil Based on 16S rRNA Gene-Based Metagenomic Insights. Agriculture, 12(10), 1624. https://doi.org/10.3390/agriculture12101624






