Research on pH Value Detection Method during Maize Silage Secondary Fermentation Based on Computer Vision
Abstract
1. Introduction
2. Materials and Methods
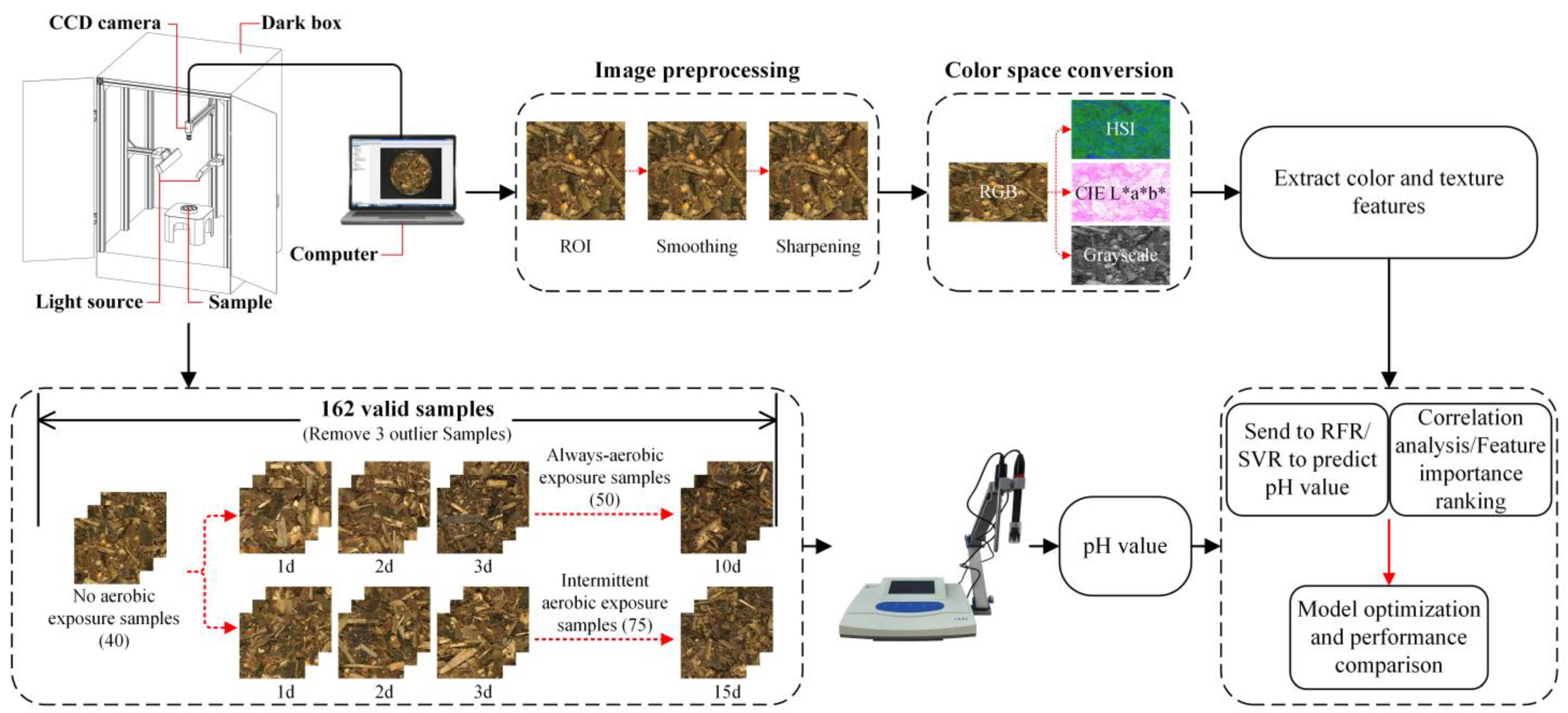
2.1. Sample Preparation
- (1)
- No aerobic exposure treatment: 40 samples were randomly selected from the collected samples for direct analysis.
- (2)
- Intermittent aerobic exposure treatment: 75 samples were randomly selected from the collected samples, opened in 1–15 days, opened for 1 h every day, and placed in polystyrene boxes with several holes (10 mm in diameter) around them. Then, the lid was covered to reduce moisture evaporation. This process was carried out at room temperature (18–22 °C), and five samples were randomly taken daily for analysis.
- (3)
- Always-aerobic exposure treatment: 50 samples were randomly selected from the collected samples, kept unsealed for 1–10 days, placed in polystyrene boxes with several holes, and covered with lids. This process was carried out at room temperature (18–22 °C), and five samples were randomly taken daily for analysis.
2.2. Construction of Computer Vision System
2.3. Chemical Analysis and Image Acquisition
2.4. Image Preprocessing and Feature Extraction
2.5. Data Analysis
2.5.1. Random Forest Regression (RFR)
2.5.2. Support Vector Regression (SVR)
2.6. Model Evaluation Indices
3. Results and Discussions
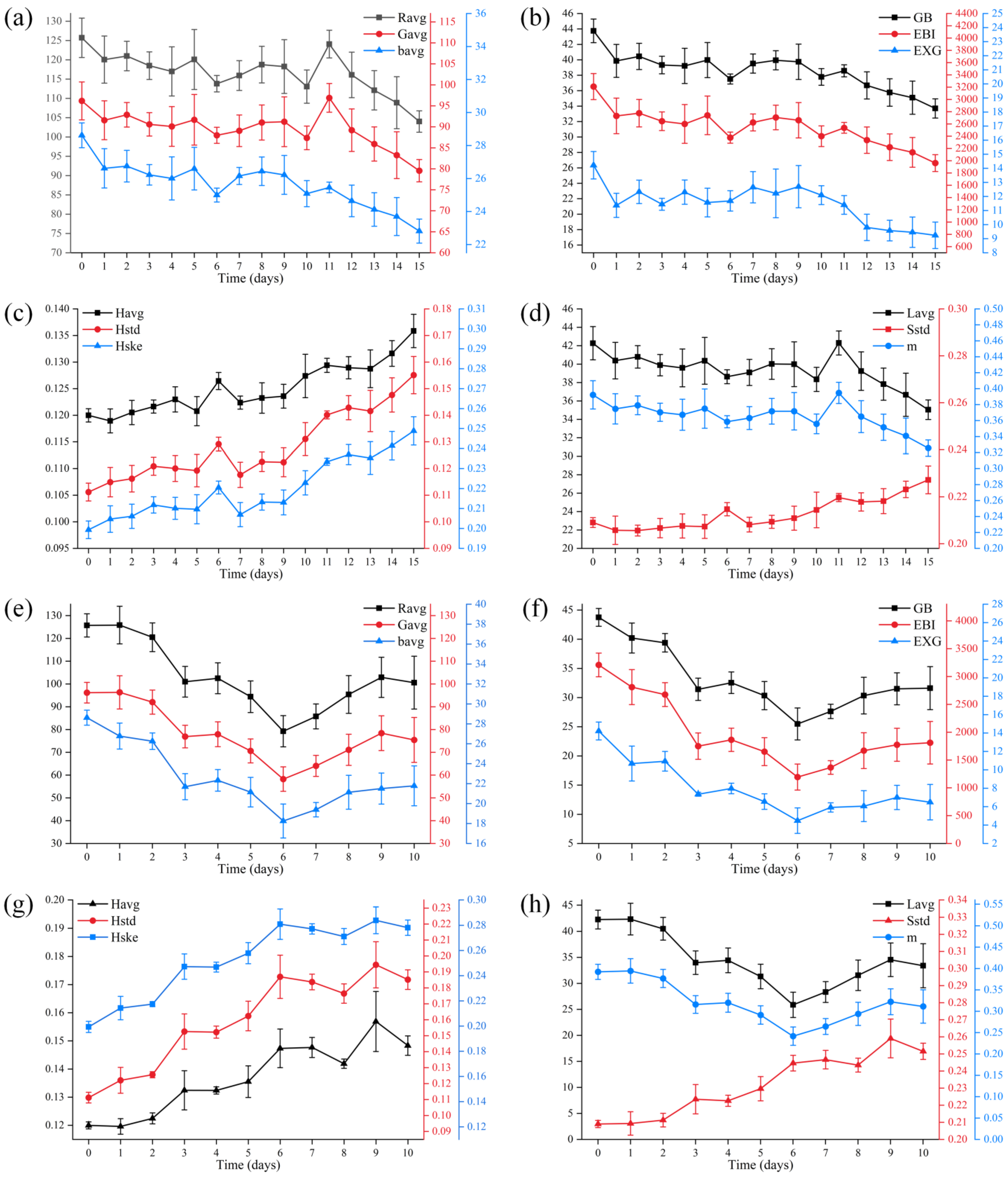
3.1. Correlation between Image Features and pH Value
3.2. Changes in Image Features during Secondary Fermentation
3.3. Changes in pH Value during Secondary Fermentation
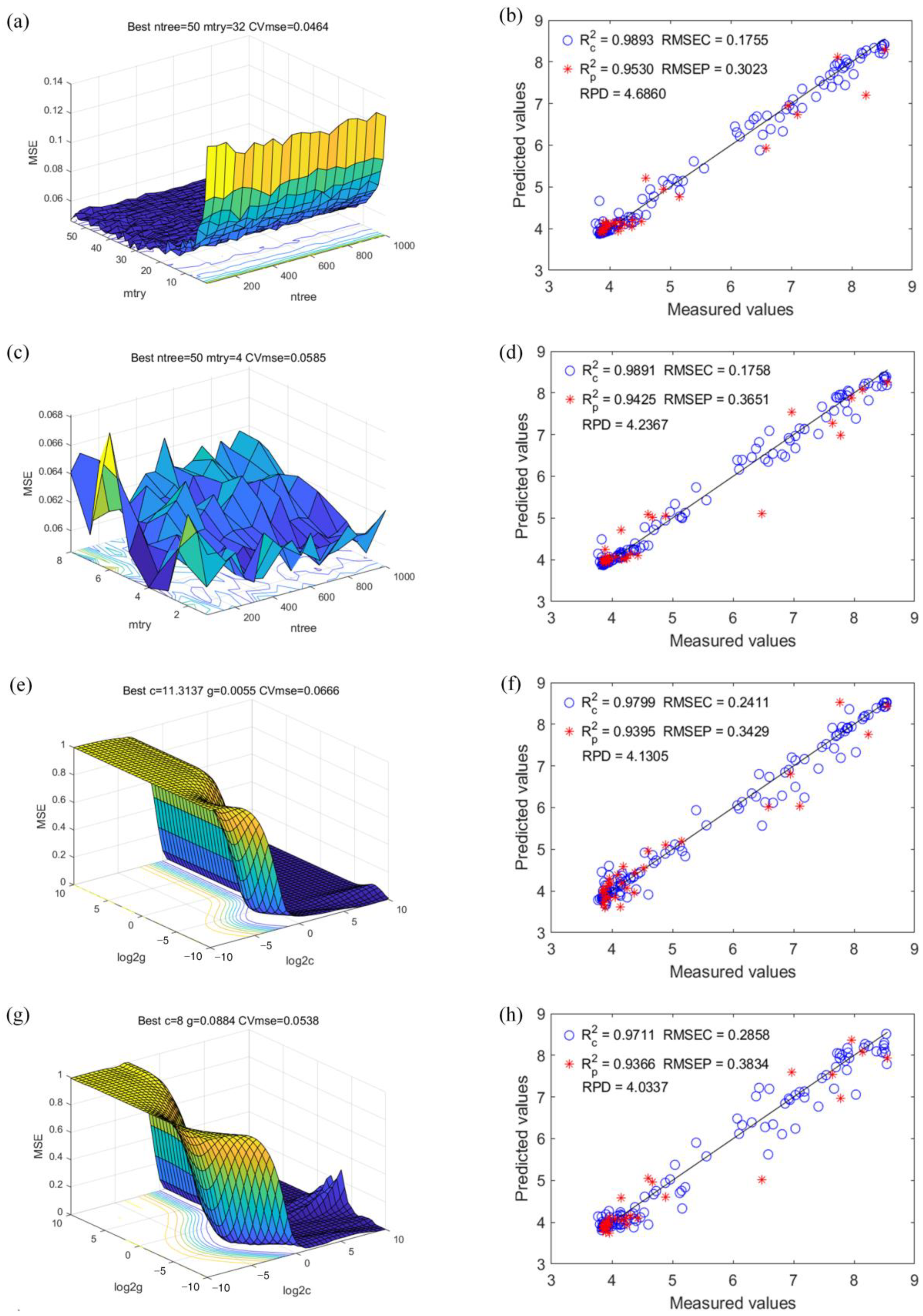
3.4. RFR
3.5. SVR
3.6. Model Comparison and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Bai, J.; Franco, M.; Ding, Z.; Hao, L.; Ke, W.; Wang, M.; Xie, D.; Li, Z.; Zhang, Y.; Ai, L.; et al. Effect of Bacillus Amyloliquefaciens and Bacillus Subtilis on Fermentation, Dynamics of Bacterial Community and Their Functional Shifts of Whole-Plant Corn Silage. J. Anim. Sci. Biotechnol. 2022, 13, 7. [Google Scholar] [CrossRef]
- Konieczna, A.; Roman, K.; Roman, M.; Śliwiński, D.; Roman, M. Energy Efficiency of Maize Production Technology: Evidence from Polish Farms. Energies 2021, 14, 170. [Google Scholar] [CrossRef]
- Weinberg, Z.G.; Ashbell, G. Engineering Aspects of Ensiling. Biochem. Eng. J. 2003, 13, 181–188. [Google Scholar] [CrossRef]
- Sun, Y.; Li, M.; Zhou, H.; Shan, G.; Cheng, Q.; Jungbluth, K.H.; Buescher, W.; Maack, C.; Lipski, A.; Wang, Z.; et al. In Situ Measurements and Simulation of Oxygen Diffusion and Heat Transfer in Maize Silage Relative to the Silo Surface. Comput. Electron. Agric. 2017, 137, 1–8. [Google Scholar] [CrossRef]
- Driehuis, F.; Elferink, S.J.W.H.O. The Impact of the Quality of Silage on Animal Health and Food Safety: A Review. Vet. Q. 2000, 22, 212–216. [Google Scholar] [CrossRef] [PubMed]
- Basso, F.C.; Bernardes, T.F.; Roth, A.P.d.T.P.; Lodo, B.N.; Berchielli, T.T.; Reis, R.A. Fermentation and Aerobic Stability of Corn Silage Inoculated with Lactobacillus Buchneri. Rev. Bras. Zootec. 2012, 41, 1789–1794. [Google Scholar] [CrossRef]
- Kung, L.; Shaver, R.D.; Grant, R.J.; Schmidt, R.J. Silage Review: Interpretation of Chemical, Microbial, and Organoleptic Components of Silages. J. Dairy Sci. 2018, 101, 4020–4033. [Google Scholar] [CrossRef] [PubMed]
- Borreani, G.; Tabacco, E. Improving Corn Silage Quality in the Top Layer of Farm Bunker Silos through the Use of a Next-Generation Barrier Film with High Impermeability to Oxygen. J. Dairy Sci. 2014, 97, 2415–2426. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.H.; Shao, T.; Zhang, J.G. Determination of Aerobic Deterioration of Corn Stalk Silage Caused by Aerobic Bacteria. Anim. Feed Sci. Technol. 2013, 183, 124–131. [Google Scholar] [CrossRef]
- Pereira-Crespo, S.; Sainz-Ramirez, A.; Andrea Plata-Reyes, D.; Gomez-Miranda, A.; Gonzalez-Alcantara, F.; Botana, A.; Gonzalez, L.; Veiga, M.; Resch, C.; Lorenzana, R.; et al. Prediction of the Fermentative Quality of Sunflower Silage by Nearinfrared Reflectance Spectroscopy (NIRS) on Oven-Dried Samples. Rev. Mex. Cienc. Pecu. 2021, 12, 609–620. [Google Scholar] [CrossRef]
- Harris, P.A.; Nelson, S.; Carslake, H.B.; Argo, C.M.; Wolf, R.; Fabri, F.B.; Brolsma, K.M.; van Oostrum, M.J.; Ellis, A.D. Comparison of NIRS and Wet Chemistry Methods for the Nutritional Analysis of Haylages for Horses. J. Equine Vet. Sci. 2018, 71, 13–20. [Google Scholar] [CrossRef]
- Sørensen, L.K. Prediction of Fermentation Parameters in Grass and Corn Silage by Near Infrared Spectroscopy. J. Dairy Sci. 2004, 87, 3826–3835. [Google Scholar] [CrossRef]
- Xian, L.; Lu-Jia, H.; Zeng-Ling, Y.; Qiong-Fei, L. Rapid prediction of pH value and fermentation products in silage by near infrared spectroscopy. Chin. J. Anal. Chem. 2007, 35, 1285–1289. [Google Scholar]
- Park, H.-S.; Kim, J.-H.; Choi, K.-C.; Kim, H.-S. Mathematical Transformation Influencing Accuracy of Near Infrared Spectroscopy (NIRS) Calibrations for the Prediction of Chemical Composition and Fermentation Parameters in Corn Silage. J. Korean Soc. Grassl. Forage Sci. 2016, 36, 50–57. [Google Scholar] [CrossRef]
- Hetta, M.; Mussadiq, Z.; Wallsten, J.; Halling, M.; Swensson, C.; Geladi, P. Prediction of Nutritive Values, Morphology and Agronomic Characteristics in Forage Maize Using Two Applications of NIRS Spectrometry. Acta Agric. Scand. Sect. B—Soil Plant Sci. 2017, 67, 326–333. [Google Scholar] [CrossRef]
- Wang, Y.; Li, L.; Liu, Y.; Cui, Q.; Ning, J.; Zhang, Z. Enhanced Quality Monitoring during Black Tea Processing by the Fusion of NIRS and Computer Vision. J. Food Eng. 2021, 304, 110599. [Google Scholar] [CrossRef]
- Bhargava, A.; Bansal, A. Fruits and Vegetables Quality Evaluation Using Computer Vision: A Review. J. King Saud Univ.-Comput. Inf. Sci. 2021, 33, 243–257. [Google Scholar] [CrossRef]
- Islam, N.; Rashid, M.M.; Wibowo, S.; Xu, C.-Y.; Morshed, A.; Wasimi, S.A.; Moore, S.; Rahman, S.M. Early Weed Detection Using Image Processing and Machine Learning Techniques in an Australian Chilli Farm. Agriculture 2021, 11, 387. [Google Scholar] [CrossRef]
- Bhujel, A.; Kim, N.-E.; Arulmozhi, E.; Basak, J.K.; Kim, H.-T. A Lightweight Attention-Based Convolutional Neural Networks for Tomato Leaf Disease Classification. Agriculture 2022, 12, 228. [Google Scholar] [CrossRef]
- Santos Pereira, L.F.; Barbon, S.; Valous, N.A.; Barbin, D.F. Predicting the Ripening of Papaya Fruit with Digital Imaging and Random Forests. Comput. Electron. Agric. 2018, 145, 76–82. [Google Scholar] [CrossRef]
- Jiang, Y.; Bian, B.; Wang, X.; Chen, S.; Li, Y.; Sun, Y. Identification of Tomato Maturity Based on Multinomial Logistic Regression with Kernel Clustering by Integrating Color Moments and Physicochemical Indices. J. Food Process Eng. 2020, 43, e13504. [Google Scholar] [CrossRef]
- Zulkifli, N.; Hashim, N.; Harith, H.H.; Mohamad Shukery, M.F.; Onwude, D.I. Prediction of the Ripening Stages of Papayas Using Discriminant Analysis and Support Vector Machine Algorithms. J. Sci. Food Agric. 2021, 102, 3266–3276. [Google Scholar] [CrossRef] [PubMed]
- Lopes, J.F.; Ludwig, L.; Barbin, D.F.; Grossmann, M.V.E.; Barbon, S. Computer Vision Classification of Barley Flour Based on Spatial Pyramid Partition Ensemble. Sensors 2019, 19, 2953. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Tan, Q.; Zhang, Y.; Zha, X.; Yang, S.; Yang, R. Research on Maize Seed Classification and Recognition Based on Machine Vision and Deep Learning. Agriculture 2022, 12, 232. [Google Scholar] [CrossRef]
- Piedad, E.J.; Larada, J.I.; Pojas, G.J.; Ferrer, L.V.V. Postharvest Classification of Banana (Musa Acuminata) Using Tier-Based Machine Learning. Postharvest Biol. Technol. 2018, 145, 93–100. [Google Scholar] [CrossRef]
- Qian, L.; Daren, L.; Qingliang, N.; Danfeng, H.; Liying, C. Non-Destructive Monitoring of Netted Muskmelon Quality Based on Its External Phenotype Using Random Forest. PLoS ONE 2019, 14, e0221259. [Google Scholar] [CrossRef]
- Ivorra, E.; Camilo Sarria-Gonzalez, J.; Giron-Hernandez, J. Computer Vision Techniques for Modelling the Roasting Process of Coffee (Coffea Arabica L.) Var. Castillo. Czech J. Food Sci. 2020, 38, 388–396. [Google Scholar] [CrossRef]
- Sanaeifar, A.; Bakhshipour, A.; de la Guardia, M. Prediction of Banana Quality Indices from Color Features Using Support Vector Regression. Talanta 2016, 148, 54–61. [Google Scholar] [CrossRef]
- Dong, C.; Liang, G.; Hu, B.; Yuan, H.; Jiang, Y.; Zhu, H.; Qi, J. Prediction of Congou Black Tea Fermentation Quality Indices from Color Features Using Non-Linear Regression Methods. Sci. Rep. 2018, 8, 10535. [Google Scholar] [CrossRef]
- Sabzi, S.; Javadikia, H.; Ignacio Arribas, J. A Three-Variety Automatic and Non-Intrusive Computer Vision System for the Estimation of Orange Fruit PH Value. Measurement 2020, 152, 107298. [Google Scholar] [CrossRef]
- Keramat-Jahromi, M.; Mohtasebi, S.S.; Mousazadeh, H.; Ghasemi-Varnamkhasti, M.; Rahimi-Movassagh, M. Real-Time Moisture Ratio Study of Drying Date Fruit Chips Based on on-Line Image Attributes Using KNN and Random Forest Regression Methods. Measurement 2021, 172, 108899. [Google Scholar] [CrossRef]
- Teixeira, C.; Fontaneli, R.S. Sensory Evaluation of Winter Cereal Silage. J. Chem. Chem. Eng. 2018, 11, 102–106. [Google Scholar]
- Tharangani, R.M.H.; Yakun, C.; Zhao, L.S.; Ma, L.; Liu, H.L.; Su, S.L.; Shan, L.; Yang, Z.N.; Kononoff, P.J.; Weiss, W.P.; et al. Corn Silage Quality Index: An Index Combining Milk Yield, Silage Nutritional and Fermentation Parameters. Anim. Feed Sci. Technol. 2021, 273, 114817. [Google Scholar] [CrossRef]
- Bakhshipour, A.; Zareiforoush, H.; Bagheri, I. Application of Decision Trees and Fuzzy Inference System for Quality Classification and Modeling of Black and Green Tea Based on Visual Features. J. Food Meas. Charact. 2020, 14, 1402–1416. [Google Scholar] [CrossRef]
- Peng, L.; Rui-mei, W.; Pu-xiang, Y.; Wen-jin, L.; Jian-ping, W.; Yang, T.; Xiao, H.; Shi-rong, A. Study of Sensory Quality Evaluation of Tea Using Computer Vision Technology and Forest Random Method. Spectrosc. Spectr. Anal. 2019, 39, 193–198. [Google Scholar] [CrossRef]
- Gonzalez, R.C.; Woods, R.E. Digital Image Processing; Pearson: New York, NY, USA, 2018; ISBN 978-0-13-335672-4. [Google Scholar]
- Garcia-Lamont, F.; Cervantes, J.; López, A.; Rodriguez, L. Segmentation of Images by Color Features: A Survey. Neurocomputing 2018, 292, 1–27. [Google Scholar] [CrossRef]
- Stricker, M.A.; Orengo, M. Similarity of Color Images. In Proceedings of the Storage and Retrieval for Image and Video Databases III, San Diego/La Jolla, CA, USA, 23 March 1995; Volume 2420, pp. 381–392. [Google Scholar]
- McGuire, R.G. Reporting of Objective Color Measurements. HortScience 1992, 27, 1254–1255. [Google Scholar] [CrossRef]
- Meyer, G.E.; Mehta, T.; Kocher, M.F.; Mortensen, D.A.; Samal, A. Textural Imaging and Discriminant Analysis For Distinguishingweeds For Spot Spraying. Trans. ASAE 1998, 41, 1189–1197. [Google Scholar] [CrossRef]
- Woebbecke, D.M.; Meyer, G.E.; Von Bargen, K.; Mortensen, D.A. Color Indices for Weed Identification under Various Soil, Residue, and Lighting Conditions. Trans. ASAE USA 1995, 38, 259–269. [Google Scholar] [CrossRef]
- Golzarian, M.R.; Frick, R.A. Classification of Images of Wheat, Ryegrass and Brome Grass Species at Early Growth Stages Using Principal Component Analysis. Plant Methods 2011, 7, 28. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef]
- Kennard, R.W.; Stone, L.A. Computer Aided Design of Experiments. Technometrics 1969, 11, 137–148. [Google Scholar] [CrossRef]
- Munera, S.; Hernández, F.; Aleixos, N.; Cubero, S.; Blasco, J. Maturity Monitoring of Intact Fruit and Arils of Pomegranate Cv. ‘Mollar de Elche’ Using Machine Vision and Chemometrics. Postharvest Biol. Technol. 2019, 156, 110936. [Google Scholar] [CrossRef]
- Jain, A.; Nandakumar, K.; Ross, A. Score Normalization in Multimodal Biometric Systems. Pattern Recognit. 2005, 38, 2270–2285. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, L. Classification of Fruits Using Computer Vision and a Multiclass Support Vector Machine. Sensors 2012, 12, 12489–12505. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- An, G.; Xing, M.; He, B.; Liao, C.; Huang, X.; Shang, J.; Kang, H. Using Machine Learning for Estimating Rice Chlorophyll Content from In Situ Hyperspectral Data. Remote Sens. 2020, 12, 3104. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and Regression by RandomForest. R News 2002, 2, 6. [Google Scholar]
- Goldstein, B.A.; Polley, E.C.; Briggs, F.B.S. Random Forests for Genetic Association Studies. Stat. Appl. Genet. Mol. Biol. 2011, 10, 32. [Google Scholar] [CrossRef]
- Oliveira, S.; Oehler, F.; San-Miguel-Ayanz, J.; Camia, A.; Pereira, J.M.C. Modeling Spatial Patterns of Fire Occurrence in Mediterranean Europe Using Multiple Regression and Random Forest. For. Ecol. Manag. 2012, 275, 117–129. [Google Scholar] [CrossRef]
- Leroux, L.; Castets, M.; Baron, C.; Escorihuela, M.-J.; Bégué, A.; Lo Seen, D. Maize Yield Estimation in West Africa from Crop Process-Induced Combinations of Multi-Domain Remote Sensing Indices. Eur. J. Agron. 2019, 108, 11–26. [Google Scholar] [CrossRef]
- Li, J.; Mao, X. Comparison of Canopy Closure Estimation of Plantations Using Parametric, Semi-Parametric, and Non-Parametric Models Based on GF-1 Remote Sensing Images. Forests 2020, 11, 597. [Google Scholar] [CrossRef]
- Ghasemi-Varnamkhasti, M.; Mohtasebi, S.S.; Siadat, M.; Ahmadi, H.; Razavi, S.H. From Simple Classification Methods to Machine Learning for the Binary Discrimination of Beers Using Electronic Nose Data. Eng. Agric. Environ. Food 2015, 8, 44–51. [Google Scholar] [CrossRef]
- Yao, K.; Sun, J.; Zhang, L.; Zhou, X.; Tian, Y.; Tang, N.; Wu, X. Nondestructive Detection for Egg Freshness Based on Hyperspectral Imaging Technology Combined with Harris Hawks Optimization Support Vector Regression. J. Food Saf. 2021, 41, e12888. [Google Scholar] [CrossRef]
- Jin, G.; Wang, Y.; Li, L.; Shen, S.; Deng, W.-W.; Zhang, Z.; Ning, J. Intelligent Evaluation of Black Tea Fermentation Degree by FT-NIR and Computer Vision Based on Data Fusion Strategy. LWT 2020, 125, 109216. [Google Scholar] [CrossRef]
- Porep, J.U.; Kammerer, D.R.; Carle, R. On-Line Application of near Infrared (NIR) Spectroscopy in Food Production. Trends Food Sci. Technol. 2015, 46, 211–230. [Google Scholar] [CrossRef]
- Olivares Díaz, E.; Kawamura, S.; Matsuo, M.; Kato, M.; Koseki, S. Combined Analysis of Near-Infrared Spectra, Colour, and Physicochemical Information of Brown Rice to Develop Accurate Calibration Models for Determining Amylose Content. Food Chem. 2019, 286, 297–306. [Google Scholar] [CrossRef]
- Mouazen, A.M.; Dridi, S.; Rouissi, H.; De Baerdemaeker, J.; Ramon, H. Prediction of Selected Ewe’s Milk Properties and Differentiating between Pasture and Box Feeding Using Visible and near Infrared Spectroscopy. Biosyst. Eng. 2009, 104, 353–361. [Google Scholar] [CrossRef]
- Prieto, N.; Pawluczyk, O.; Dugan, M.E.R.; Aalhus, J.L. A Review of the Principles and Applications of Near-Infrared Spectroscopy to Characterize Meat, Fat, and Meat Products. Appl. Spectrosc. 2017, 71, 1403–1426. [Google Scholar] [CrossRef]
- Wilkinson, J.M.; Davies, D.R. The Aerobic Stability of Silage: Key Findings and Recent Developments. Grass Forage Sci. 2013, 68, 1–19. [Google Scholar] [CrossRef]
- Merry, R.J.; Davies, D.R. Propionibacteria and Their Role in the Biological Control of Aerobic Spoilage in Silage. Le Lait 1999, 79, 149–164. [Google Scholar] [CrossRef]
- Brüning, D.; Gerlach, K.; Weiß, K.; Südekum, K.-H. Effect of Compaction, Delayed Sealing and Aerobic Exposure on Maize Silage Quality and on Formation of Volatile Organic Compounds. Grass Forage Sci. 2018, 73, 53–66. [Google Scholar] [CrossRef]




| NO. | Name | Description | Type |
|---|---|---|---|
| 1–3 | Ravg/Rstd/Rske | Mean value/Standard deviation/Skewness of the R channel | Color |
| 4–6 | Gavg/Gstd/Gske | Mean value/Standard deviation/Skewness of the G channel | |
| 7–9 | Bavg/Bstd/Bske | Mean value/Standard deviation/Skewness of the B channel | |
| 10–12 | Havg/Hstd/Hske | Mean value/Standard deviation/Skewness of the H channel | |
| 13–15 | Savg/Sstd/Sske | Mean value/Standard deviation/Skewness of the S channel | |
| 16–18 | Iavg/Istd/Iske | Mean value/Standard deviation/Skewness of the I channel | |
| 19–21 | Lavg/Lstd/Lske | Mean value/Standard deviation/Skewness of the L* channel | |
| 22–24 | aavg/astd/aske | Mean value/Standard deviation/Skewness of the a* channel | |
| 25–26 | bavg/bstd | Mean value/Standard deviation of the b* channel | |
| 27 | hab | Hue angle | |
| 28 | EXG | Excess green (2 × Gavg − Ravg − Bavg) | |
| 29 | GB | Green minus blue (Gavg − Bavg) | |
| 30 | EBI | Additional blue index ((Bavg − Gavg) × (Bavg − Ravg)) | |
| 31 | m | Mean | Texture |
| 32 | δ | Deviation | |
| 33 | R | Smoothness | |
| 34 | μ3 | Third moment | |
| 35 | U | Consistency | |
| 36 | e | Entropy | |
| 37–40 | E0°/E45°/E90°/E135° | Energy in four orientations (0°, 45°, 90°, and 135°) | |
| 41–44 | I0°/I45°/I 90°/I 135° | Contrast in four orientations (0°, 45°, 90°, and 135°) | |
| 45–48 | H0°/H 45°/H 90°/H 135° | Entropy in four orientations (0°, 45°, 90°, and 135°) | |
| 49–52 | U0°/U 45°/U 90°/U 135° | Uniformity in four orientations (0°, 45°, 90°, and 135°) | |
| 53–56 | C0°/C 45°/C 90°/C 135° | Correlation in four orientations (0°, 45°, 90°, and 135°) |
| Row | Model Parameters | Values and Specifications |
|---|---|---|
| 1 | ntree | 50–1000 (value every 50) |
| 2 | mtry | 1–number of feature inputs (value every 1) |
| Row | Model Parameters | Values and Specifications |
|---|---|---|
| 1 | c | 2−10–210 (value every 20.5) |
| 2 | g | 2−10–210 (value every 20.5) |
| Models | Description | Calibration Set | Prediction Set | RPD | ||
|---|---|---|---|---|---|---|
| RMSEC | RMSEP | |||||
| RFR model 1 | After optimization | 0.9893 | 0.1755 | 0.9530 | 0.3023 | 4.6860 |
| RFR model 2 | After resizing and optimization | 0.9891 | 0.1758 | 0.9425 | 0.3651 | 4.2367 |
| SVR model 1 | After optimization | 0.9799 | 0.2411 | 0.9395 | 0.3429 | 4.1305 |
| SVR model 2 | After resizing and optimization | 0.9711 | 0.2858 | 0.9366 | 0.3834 | 4.0337 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, X.; Tian, H.; Zhao, K.; Li, D.; Xiao, Z.; Yu, Y.; Liu, F. Research on pH Value Detection Method during Maize Silage Secondary Fermentation Based on Computer Vision. Agriculture 2022, 12, 1623. https://doi.org/10.3390/agriculture12101623
Ren X, Tian H, Zhao K, Li D, Xiao Z, Yu Y, Liu F. Research on pH Value Detection Method during Maize Silage Secondary Fermentation Based on Computer Vision. Agriculture. 2022; 12(10):1623. https://doi.org/10.3390/agriculture12101623
Chicago/Turabian StyleRen, Xianguo, Haiqing Tian, Kai Zhao, Dapeng Li, Ziqing Xiao, Yang Yu, and Fei Liu. 2022. "Research on pH Value Detection Method during Maize Silage Secondary Fermentation Based on Computer Vision" Agriculture 12, no. 10: 1623. https://doi.org/10.3390/agriculture12101623
APA StyleRen, X., Tian, H., Zhao, K., Li, D., Xiao, Z., Yu, Y., & Liu, F. (2022). Research on pH Value Detection Method during Maize Silage Secondary Fermentation Based on Computer Vision. Agriculture, 12(10), 1623. https://doi.org/10.3390/agriculture12101623






