Improving the Yield of Xenocoumacin 1 by PBAD Promoter Replacement in Xenorhabdus nematophila CB6
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Media, and Growth Conditions
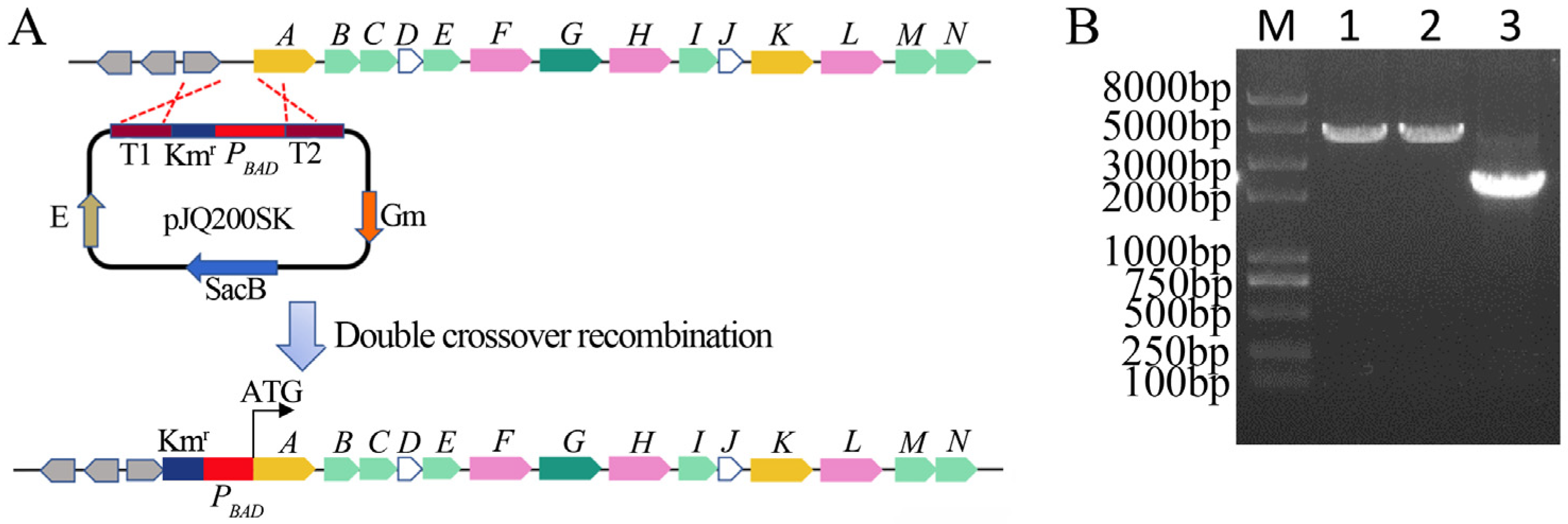
2.2. Construction of ΔPBAD-xcnA Mutant Strain
2.3. Monitoring the Growth of ΔPBAD-xcnA Mutant Strain
2.4. Analysis of the Yield of Xcns
2.5. Quantitative Real-Time PCR (qRT-PCR)
2.6. Antimicrobial Activity Assay
3. Results
3.1. Construction of ΔPBAD-xcnA Strain
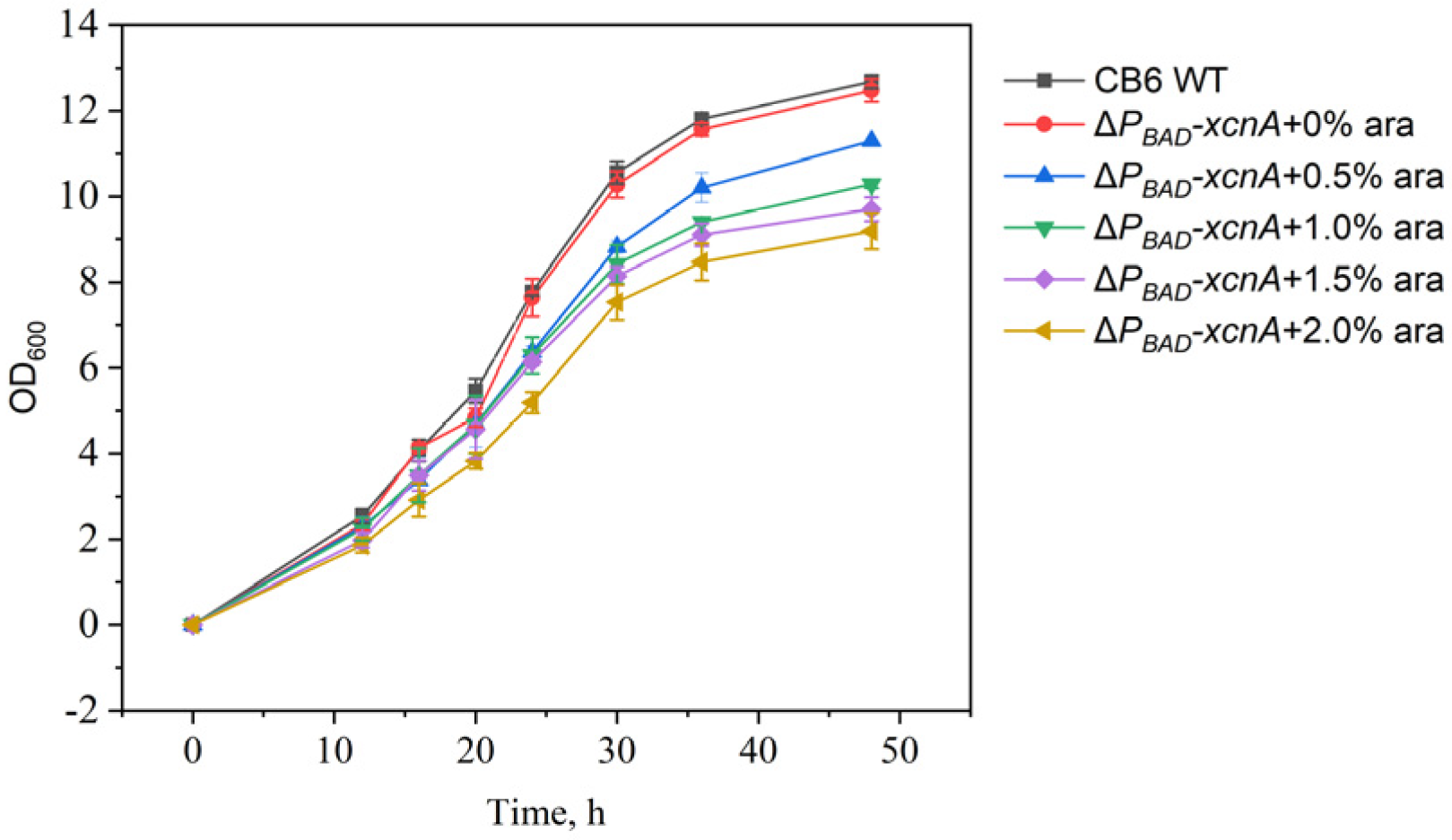
3.2. Growth Profile of ΔPBAD-xcnA Strain
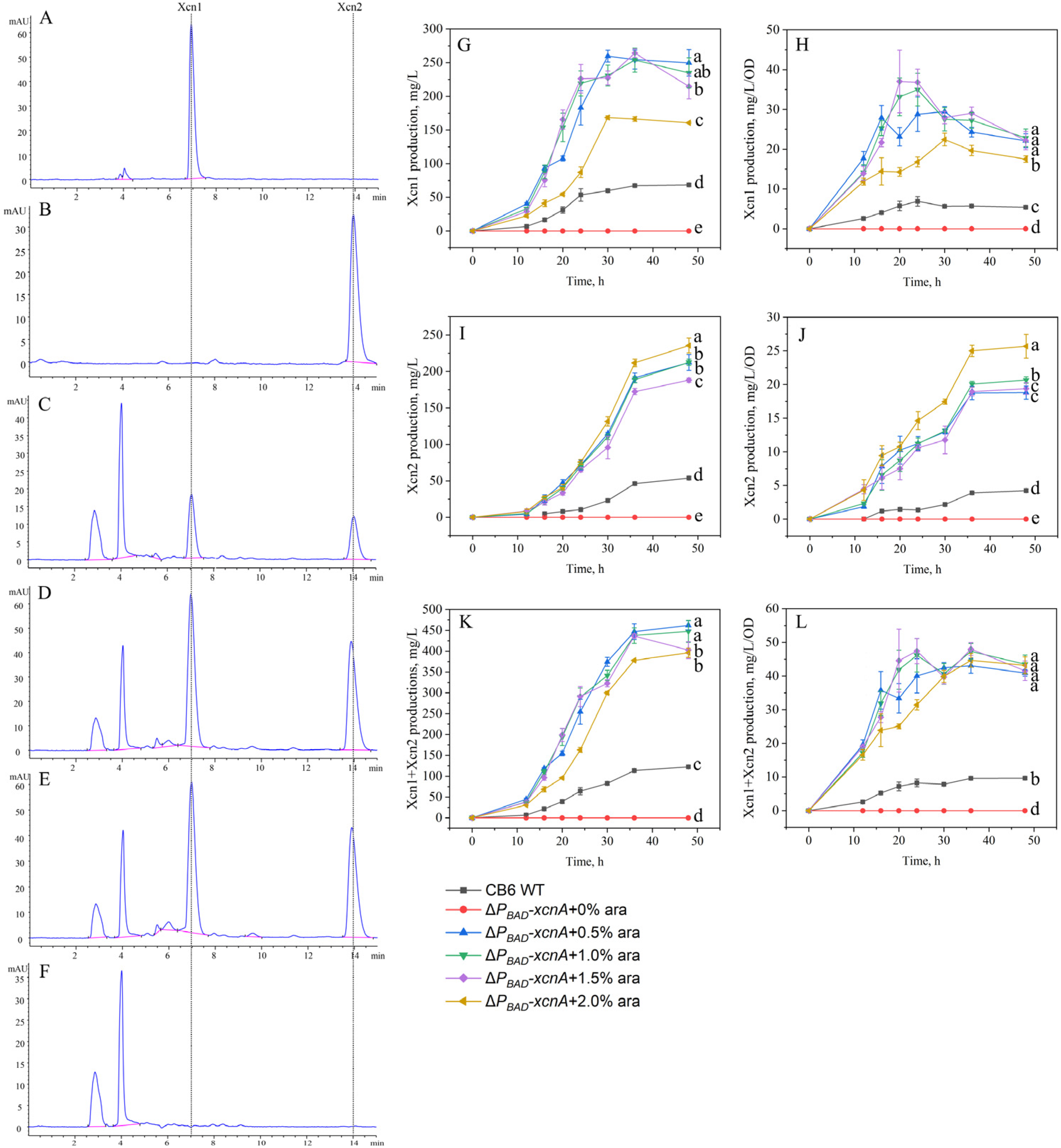
3.3. Effects of PBAD Promoter Replacement on the Yield of Xcn1
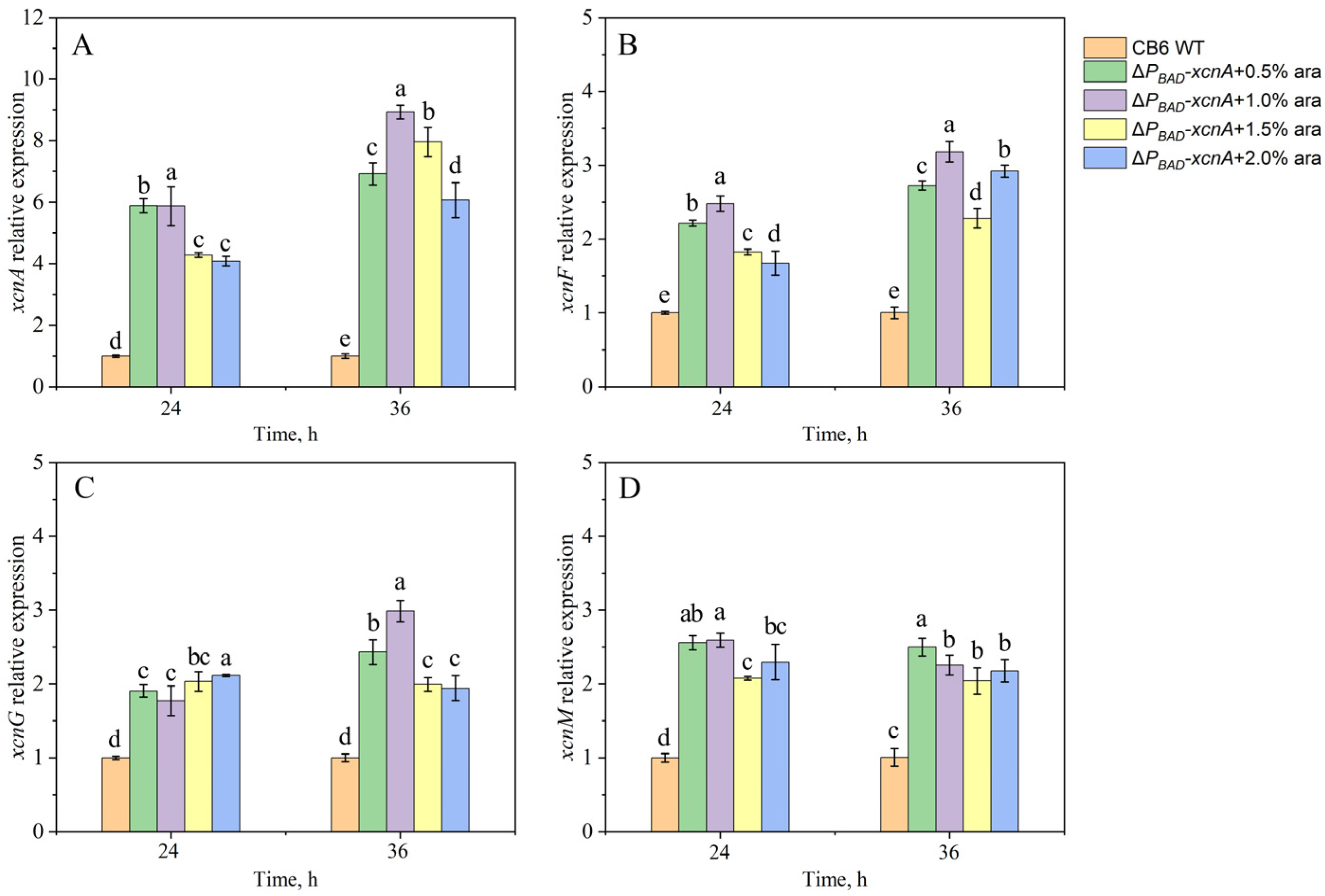
3.4. Transcriptional Analysis of the Xcn Genes
3.5. Antimicrobial Activity of ΔPBAD-xcnA Mutant Strain
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shi, Y.M.; Bode, H.B. Chemical language and warfare of bacterial natural products in bacteria-nematode-insect interactions. Nat. Prod. Rep. 2018, 35, 309–335. [Google Scholar] [CrossRef]
- Herbert, E.E.; Goodrich-Blair, H. Friend and foe: The two faces of Xenorhabdus nematophila. Nat. Rev. Microbiol. 2007, 5, 634–646. [Google Scholar] [CrossRef] [PubMed]
- Forst, S.; Nealson, K. Molecular biology of the symbiotic-pathogenic bacteria Xenorhabdus spp. and Photorhabdus spp. Microbiol. Res. 1996, 60, 21–43. [Google Scholar] [CrossRef]
- Park, D.; Ciezki, K.; van der Hoeven, R.; Singh, S.; Reimer, D.; Bode, H.B.; Forst, S. Genetic analysis of xenocoumacin antibiotic production in the mutualistic bacterium Xenorhabdus nematophila. Mol. Microbiol. 2009, 73, 938–949. [Google Scholar] [CrossRef]
- Adeolu, M.; Alnajar, S.; Naushad, S.; Gupta, R.S. Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: Proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 5575–5599. [Google Scholar]
- Goodrich-Blair, H.; Clarke, D.J. Mutualism and pathogenesis in Xenorhabdus and Photorhabdus: Two roads to the same destination. Mol. Microbiol. 2007, 64, 260–268. [Google Scholar] [CrossRef] [PubMed]
- Goodrich-Blair, H. They′ve got a ticket to ride: Xenorhabdus nematophila—Steinernema carpocapsae symbiosis. Curr. Opin. Microbiol. 2007, 10, 225–230. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Patel, T.; Rickman, T.; Goodrich-Blair, H.; Hussa, E.A. High levels of the Xenorhabdus nematophila transcription factor Lrp promote mutualism with the Steinernema carpocapsae nematode host. Appl. Environ. Microbiol. 2017, 83, e00276-17. [Google Scholar] [CrossRef] [Green Version]
- Hillman, K.; Goodrich-Blair, H. Are you my symbiont? Microbial polymorphic toxins and antimicrobial compounds as honest signals of beneficial symbiotic defensive traits. Curr. Opin. Microbiol. 2016, 31, 184–190. [Google Scholar] [CrossRef] [Green Version]
- Shi, H.; Zeng, H.; Yang, X.; Zhao, J.; Chen, M.; Qiu, D. An insecticidal protein from Xenorhabdus ehlersii triggers prophenoloxidase activation and hemocyte decrease in Galleria mellonella. Curr. Microbiol. 2012, 64, 604–610. [Google Scholar] [CrossRef]
- Liu, H.; Zeng, H.; Yao, Q.; Yuan, J.; Zhang, Y.; Qiu, D.; Yang, X.; Yang, H.; Liu, Z. Steinernema glaseri surface enolase: Molecular cloning, biological characterization, and role in host immune suppression. Mol. Biochem. Parasit. 2012, 185, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Jian, H.; Zhang, J.; Liu, Z.; Chen, Z.; Yang, X.; Yang, H.; Huang, D. An intracellular toxic protein (Xin) isolated from Xenorhabdus nematophilus strain BJ. Prog. Nat. Sci. 2002, 12, 310–312. [Google Scholar]
- Snyder, H.; Stock, S.P.; Kim, S.-K.; Flores-Lara, Y.; Forst, S. New insights into the colonization and release processes of Xenorhabdus nematophila and the morphology and ultrastructure of the bacterial receptacle of its nematode host, Steinernema carpocapsae. Appl. Environ. Microbiol. 2007, 73, 5338–5346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Li, L.; Ziyan, N.; Song, P.; Wang, Q. Relationship between insecticidal activity of Tc toxin from Xenorhabdus nematophila and the insect midgut environments. Toxicon 2019, 158, S85. [Google Scholar] [CrossRef]
- Richards, G.R.; Goodrich-Blair, H. Examination of Xenorhabdus nematophila lipases in pathogenic and mutualistic host interactions reveals a role for xlpA in nematode progeny production. Appl. Environ. Microbiol. 2010, 76, 221–229. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.; Orr, D.; Divinagracia, E.; McGraw, J.; Dorff, K.; Forst, S. Role of secondary metabolites in establishment of the mutualistic partnership between Xenorhabdus nematophila and the entomopathogenic nematode Steinernema carpocapsae. Appl. Environ. Microbiol. 2015, 81, 754–764. [Google Scholar] [CrossRef] [Green Version]
- Tobias, N.J.; Heinrich, A.K.; Eresmann, H.; Wright, P.R.; Neubacher, N.; Backofen, R.; Bode, H.B. Photorhabdus-nematode symbiosis is dependent on hfq-mediated regulation of secondary metabolites. Environ. Microbiol. 2017, 19, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Challinor, V.L.; Zhao, L.; Reimer, D.; Adihou, H.; Grün, P.; Kaiser, M.; Bode, H.B. Biosynthesis of the antibiotic nematophin and its elongated derivatives in entomopathogenic bacteria. Org. Lett. 2017, 19, 806–809. [Google Scholar] [CrossRef] [PubMed]
- Nollmann, F.I.; Heinrich, A.K.; Brachmann, A.O.; Morisseau, C.; Mukherjee, K.; Casanova-Torres, Á.M.; Strobl, F.; Kleinhans, D.; Kinski, S.; Schultz, K. A Photorhabdus natural product inhibits insect juvenile hormone epoxide hydrolase. ChemBioChem 2015, 16, 766. [Google Scholar] [CrossRef]
- Li, J.; Chen, G.; Webster, J.M.; Czyzewska, E. Antimicrobial metabolites from a bacterial symbiont. J. Nat. Prod. 1995, 58, 1081–1086. [Google Scholar] [CrossRef]
- Muangpat, P.; Yooyangket, T.; Fukruksa, C.; Suwannaroj, M.; Yimthin, T.; Sitthisak, S.; Chantratita, N.; Vitta, A.; Tobias, N.J.; Bode, H.B. Screening of the antimicrobial activity against drug resistant bacteria of Photorhabdus and Xenorhabdus associated with entomopathogenic nematodes from Mae Wong National Park, Thailand. Front. Microbiol. 2017, 8, 1142. [Google Scholar] [CrossRef] [PubMed]
- Martens, E.C.; Heungens, K.; Goodrich-Blair, H. Early colonization events in the mutualistic association between Steinernema carpocapsae nematodes and Xenorhabdus nematophila bacteria. J. Bacteriol. 2003, 185, 3147–3154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Chen, G.; Webster, J.M. Nematophin, a novel antimicrobial substance produced by Xenorhabdus nematophilus (Enterobactereaceae). Can. J. Microbiol. 1997, 43, 770–773. [Google Scholar] [CrossRef]
- Sundar, L.; Chang, F. Antimicrobial activity and biosynthesis of indole antibiotics produced by Xenorhabdus nematophilus. J. Gen. Microbiol. 1993, 139, 3139–3148. [Google Scholar] [CrossRef] [Green Version]
- Huang, W.; Zhu, C.; Yang, X.; Yang, H.; Xu, H.; Xie, Y.; Jian, H. Isolation and structural identification of main component CB6–1 produced by Xenorhabdus nematophilus var. pekingensis. Chinese J. Antibiot. 2005, 30, 513–515. [Google Scholar]
- Yang, X.; Qiu, D.; Yang, H.; Liu, Z.; Zeng, H.; Yuan, J. Antifungal activity of xenocoumacin 1 from Xenorhabdus nematophilus var. pekingensis against Phytophthora infestans. World J. Microb. Biotechnol. 2011, 27, 523–528. [Google Scholar] [CrossRef]
- Lang, G.; Kalvelage, T.; Peters, A.; Wiese, J.; Imhoff, J.F. Linear and cyclic peptides from the entomopathogenic bacterium Xenorhabdus nematophilus. J. Nat. Prod. 2008, 71, 1074–1077. [Google Scholar] [CrossRef]
- Zhang, S.; Fang, X.; Tang, Q.; Ge, J.; Wang, Y.; Zhang, X. CpxR negatively regulates the production of xenocoumacin 1, a dihydroisocoumarin derivative produced by Xenorhabdus nematophila. Microbiologyopen 2019, 8, e00674. [Google Scholar] [CrossRef] [PubMed]
- Bozhüyük, K.A.; Zhou, Q.; Engel, Y.; Heinrich, A.; Pérez, A.; Bode, H.B. Natural products from Photorhabdus and other entomopathogenic bacteria. Curr. Top. Microbiol. Immunol. 2017, 402, 55–79. [Google Scholar]
- Bode, H.B. Entomopathogenic bacteria as a source of secondary metabolites. Curr. Opin. Chem. Biol. 2009, 13, 224–230. [Google Scholar] [CrossRef]
- Böszörményi, E.; Érsek, T.; Fodor, A.; Fodor, A.; Földes, L.S.; Hevesi, M.; Hogan, J.; Katona, Z.; Klein, M.; Kormány, A. Isolation and activity of Xenorhabdus antimicrobial compounds against the plant pathogens Erwinia amylovora and Phytophthora nicotianae. J. Appl. Microbiol. 2009, 107, 746–759. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, S.W.; Proschak, A.; Jaskolla, T.W.; Karas, M.; Bode, H.B. Structure elucidation and biosynthesis of lysine-rich cyclic peptides in Xenorhabdus nematophila. Org. Biomol. Chem. 2011, 9, 3130–3132. [Google Scholar] [CrossRef]
- Gualtieri, M.; Aumelas, A.; Thaler, J.-O. Identification of a new antimicrobial lysine-rich cyclolipopeptide family from Xenorhabdus nematophila. J. Antibiot. 2009, 62, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Ji, D.; Yi, Y.; Kang, G.-H.; Choi, Y.-H.; Kim, P.; Baek, N.-I.; Kim, Y. Identification of an antibacterial compound, benzylideneacetone, from Xenorhabdus nematophila against major plant-pathogenic bacteria. FEMS Microbiol. Lett. 2004, 239, 241–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, S.; Zhang, S.; Fang, X.; Liu, Q.; Gao, J.; Bilal, M.; Wang, Y.; Zhang, X. Regulation of antimicrobial activity and xenocoumacins biosynthesis by pH in Xenorhabdus nematophila. Microb. Cell Fact. 2017, 16, 203. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Liu, Q.; Han, Y.; Han, J.; Yan, Z.; Wang, Y.; Zhang, X. Nematophin, an antimicrobial dipeptide compound from Xenorhabdus nematophila YL001 as a potent biopesticide for Rhizoctonia solani control. Front. Microbiol. 2019, 10, 1765. [Google Scholar] [CrossRef] [Green Version]
- McInerney, B.V.; Taylor, W.C.; Lacey, M.J.; Akhurst, R.J.; Gregson, R.P. Biologically active metabolites from Xenorhabdus spp., Part 2. Benzopyran-1-one derivatives with gastroprotective activity. J. Nat. Prod. 1991, 54, 785–795. [Google Scholar] [CrossRef]
- Maxwell, P.W.; Chen, G.; Webster, J.M.; Dunphy, G.B. Stability and activities of antibiotics produced during infection of the insect Galleria mellonella by two isolates of Xenorhabdus nematophilus. Appl. Environ. Microbiol. 1994, 60, 715–721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, W.; Yang, X.; Yang, H. Identification and activity of antibacterial substance from Xenorhabdus nematophila var. Pekingense. Nat. Prod. Res. Devel. 2006, 18, 25–28. [Google Scholar]
- Zhou, T.; Yang, X.; Qiu, D.; Zeng, H. Inhibitory effects of xenocoumacin 1 on the different stages of Phytophthora capsici and its control effect on Phytophthora blight of pepper. BioControl 2017, 62, 151–160. [Google Scholar] [CrossRef]
- Yuan, J.; Yang, X.; Jian, H.; Yang, H.; Pang, Z. Selection of High Antibiotics Production Strain from Xenorhabdus nematophila. Chinese J. Biol. Control 2004, 20, 45–48. [Google Scholar]
- Zhang, Y.; Li, J.; Zhong, J.; Qiu, D.; Yang, X. Screening of High Yield Pekingmycin-producing Strains of Xenorhabdus nematophila var. pekinense by Cultivation in 96-well Microtiter Plates. Chinese J. Biol. Control 2013, 29, 437–442. [Google Scholar]
- Yang, X.; Yang, H.; Jian, H.; Liu, Z. Effect of fermentation conditions on antibiotic production of Xenorhabdus nematophila. Microbiology 2001, 12, 12–16. [Google Scholar]
- Wang, Y.; Li, Y.; Zhang, Q.; Zhang, X. Enhanced antibiotic activity of Xenorhabdus nematophila by medium optimization. Bioresour. Technol. 2008, 99, 1708–1715. [Google Scholar] [CrossRef]
- Han, Y.; Gao, J.; Zhang, S.; Han, J.; Yan, Z.; Ta, Y.; Wang, Y. Increasing the Production of Xenocoumacin 1 by Optimizing the Fermentation Process of Xenorhabdus nematophila. 2021. [Google Scholar] [CrossRef]
- Dong, Y.; Li, X.; Duan, J.; Qin, Y.; Yang, X.; Ren, J.; Li, G. Improving the Yield of Xenocoumacin 1 Enabled by In Situ Product Removal. ACS Omega 2020, 5, 20391–20398. [Google Scholar] [CrossRef] [PubMed]
- Jubelin, G.; Lanois, A.; Severac, D.; Rialle, S.; Longin, C.; Gaudriault, S.; Givaudan, A. FliZ is a global regulatory protein affecting the expression of flagellar and virulence genes in individual Xenorhabdus nematophila bacterial cells. PLoS Genet. 2013, 9, e1003915. [Google Scholar] [CrossRef]
- Brachmann, A.O.; Kirchner, F.; Kegler, C.; Kinski, S.C.; Schmitt, I.; Bode, H.B. Triggering the production of the cryptic blue pigment indigoidine from Photorhabdus luminescens. J. Biotechnol. 2012, 157, 96–99. [Google Scholar] [CrossRef]
- Bode, E.; Brachmann, A.O.; Kegler, C.; Simsek, R.; Dauth, C.; Zhou, Q.; Kaiser, M.; Klemmt, P.; Bode, H.B. Simple “on-demand” production of bioactive natural products. ChemBioChem 2015, 16, 1115–1119. [Google Scholar] [CrossRef] [PubMed]
- Van, D.; Forst, S. OpnS, an Outer Membrane Porin of Xenorhabdus nematophila, Confers a Competitive Advantage for Growth in the Insect Host. J. Bacteriol. 2009, 191, 5471–5479. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zhou, T.; Zeng, H.; Qiu, D.; Yang, X.; Wang, B.; Chen, M.; Guo, L.; Wang, S. Global transcriptional responses of Bacillus subtilis to xenocoumacin 1. J. Appl. Microbiol. 2011, 111, 652–662. [Google Scholar] [CrossRef]
- Qin, Y.; Fu, Y.; Kang, W.; Li, H.; Gao, H.; Vitalievitch, K.S.; Liu, H. Isolation and identification of a cold-adapted bacterium and its characterization for biocontrol and plant growth-promoting activity. Ecol. Eng. 2017, 105, 362–369. [Google Scholar] [CrossRef]
- Park, S.Y.; Yang, D.; Ha, S.H.; Lee, S.Y. Metabolic engineering of microorganisms for the production of natural compounds. Adv. Biosyst. 2018, 2, 1700190. [Google Scholar] [CrossRef] [Green Version]
- Choi, K.R.; Jang, W.D.; Yang, D.; Cho, J.S.; Park, D.; Lee, S.Y. Systems metabolic engineering strategies: Integrating systems and synthetic biology with metabolic engineering. Trends Biotechnol. 2019, 37, 817–837. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Park, S.Y.; Park, Y.S.; Eun, H.; Lee, S.Y. Metabolic engineering of Escherichia coli for natural product biosynthesis. Trends Biotechnol. 2020, 38, 745–765. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Hwang, S.; Kim, W.; Lee, Y.; Kim, J.H.; Cho, S.; Kim, H.U.; Yoon, Y.J.; Oh, M.-K.; Palsson, B.O. Systems and synthetic biology to elucidate secondary metabolite biosynthetic gene clusters encoded in Streptomyces genomes. Nat. Prod. Rep. 2021, 38, 1330–1361. [Google Scholar] [CrossRef]
- Robertsen, H.L.; Weber, T.; Kim, H.U.; Lee, S.Y. Toward systems metabolic engineering of Streptomycetes for secondary metabolites production. Biotechnol. J. 2018, 13, 1700465. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.-P.; Bu, Q.-T.; Li, J.-F.; Xie, H.; Su, Y.-T.; Du, Y.-L.; Li, Y.-Q. Genome-based rational engineering of Actinoplanes deccanensis for improving fidaxomicin production and genetic stability. Bioresour. Technol. 2021, 330, 124982. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Wang, M.; Ni, X.; Xia, H. Optimization of tetramycin production in Streptomyces ahygroscopicus S91. J. Biol. Eng. 2021, 15, 16. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, H.; Li, S.; Zhang, Y.; Cheng, X.; Xiang, W.; Wang, X. Engineering of primary metabolic pathways for titer improvement of milbemycins in Streptomyces bingchenggensis. Appl. Microbiol. Biotechnol. 2021, 105, 1875–1887. [Google Scholar] [CrossRef]
- Li, L.; Jiang, W.; Lu, Y. New strategies and approaches for engineering biosynthetic gene clusters of microbial natural products. Biotechnol. Adv. 2017, 35, 936–949. [Google Scholar] [CrossRef]
- Dhakal, D.; Sohng, J.K.; Pandey, R.P. Engineering Actinomycetes for biosynthesis of macrolactone polyketides. Microb. Cell Fact. 2019, 18, 137. [Google Scholar] [CrossRef] [PubMed]
- Hug, J.J.; Krug, D.; Müller, R. Bacteria as genetically programmable producers of bioactive natural products. Nat. Rev. Chem. 2020, 4, 172–193. [Google Scholar] [CrossRef]
- Brachmann, A.O.; Brameyer, S.; Kresovic, D.; Hitkova, I.; Kopp, Y.; Manske, C.; Schubert, K.; Bode, H.B.; Heermann, R. Pyrones as bacterial signaling molecules. Nat. Chem. Biol. 2013, 9, 573–578. [Google Scholar] [CrossRef] [PubMed]
- Guzman, L.-M.; Belin, D.; Carson, M.J.; Beckwith, J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995, 177, 4121–4130. [Google Scholar] [CrossRef] [Green Version]
- Hu, M.; Xiong, B.; Li, Z.; Liu, L.; Li, S.; Zhang, C.; Zhang, X.; Bi, C. A novel gene expression system for Ralstonia eutropha based on the T7 promoter. BMC Microbiol. 2020, 20, 121. [Google Scholar] [CrossRef] [PubMed]

| Strain or Plasmid | Relevant Genotype, Phenotype, or Characteristic(s) | Reference or Source |
|---|---|---|
| Strains | ||
| X. nematophila | ||
| CB6 | Wildtype, phase I variant; Ampr | Laboratory stock |
| PBAD-xcnA | CB6ΔPBAD-xcnA::Kmr | This study |
| E. coli | ||
| DH5α | F-φ80d lacZΔM15 Δ(lacZYA-argF) U169 end A1 recA1 hsdR17(rk−, mk+) supE44λ- thi-1 gyrA96 relA1 phoA | TransGen Biotech |
| S17-l λpir | recA, thi, pro, hsdR-M+. RP4-2Tc::Mu Km::Tn7 in the chromosome | Laboratory stock |
| Plasmids | ||
| pBAD30 | Expression vector; Ampr, ori p15A, ori F1 | Laboratory stock |
| pYBA-1132 | Expression vector; Kmr, ori pBR322 | Laboratory stock |
| pJQ200SK | Suicide vector; Gmr, Sucs, SacB, ori p15A, oriT | Laboratory stock |
| pJQPBAD-xcnA | Plasmid pJQ200SK carrying 859 bp upstream homology arm, 1075 bp kanamycin resistant cassette, 1245 bp PBAD promoter fragment and 853 bp downstream homology arm | This study |
| Primer | Sequence (5′→3′) | Use |
|---|---|---|
| XcnA-5DT | CGGTATCGATGTTATTGCTGTTTGTATTTGTG | Mutant construction |
| XcnA-P1 | CATAGGCTCCTTAAGTTGGTGCCATAATTAATAG | Mutant construction |
| Km-F | ACCAACTTAAGGAGCCTATGGAAACTGGGA | Mutant construction |
| Km-R | CAGACAATTGTCAGAAGAACTCGTCAAGAAG | Mutant construction |
| PBAD-F | GTTCTTCTGACAATTGTCTGATTCGTTACC | Mutant construction |
| PBAD-R | TCTTCTTCATATGCTAGCCTCCTGTTAGCCCA | Mutant construction |
| XcnA-P2 | AGGCTAGCATATGAAGAAGACGATTTTTGAGTTG | Mutant construction |
| XcnA-3DT | ATTCGATATCAAGCCCCATTACCATCTTCAA | Mutant construction |
| XcnA-Pro-TF | CAGGTATAGTAAATTAATAGGGG | Mutant construction |
| XcnA-Pro-TR | GTTTCTCTTCACTACTCCAACG | Mutant construction |
| infusion-F | CAGCAATAACATCGATACCGTCGACCTCG | Mutant construction |
| infusion-R | AATGGGGCTTGATATCGAATTCCTGCAGCC | Mutant construction |
| xcnA-Fwd | GCATAGAGCCTCGTGAATTAGAG | qPCR analysis |
| xcnA-Rev | GATATGTTCTCTCACCCGGGG | qPCR analysis |
| xcnF-Fwd | GCTGGCGTGATCAAATCTCTTC | qPCR analysis |
| xcnF-Rev | CAGGCAATTGACTTTATTCCCATC | qPCR analysis |
| xcnG-Fwd | CTGTTGTGGTGATGGCGAATG | qPCR analysis |
| xcnG-Rev | GAAAACAGGGAAAGGAAGGCAC | qPCR analysis |
| xcnM-Fwd | CGTATTGATGTTCATGGAGTTGATG | qPCR analysis |
| xcnM-Rev | ATGATCTTGAATATGCTCAGCCAAC | qPCR analysis |
| recA-Fwd | GCTGAAATTCTATGCGTCTGTCC | qPCR analysis |
| recA-Rev | CTGTTTGAATGGTGCTGCAACTTTG | qPCR analysis |
| Samples | Inhibition Zone Against B. subtilis (mm) | Inhibition Rate of P. capsici (%) * |
|---|---|---|
| 50 μg/mL Xcn1 | 11.30 ± 0.26 c | ND |
| CB6 wildtype | 11.83 ± 0.29 b | 78.80 ± 0.92 a |
| ΔPBAD-xcnA + 0% ara | 9.67 ± 0.28 d | 24.80 ± 1.39 b |
| ΔPBAD-xcnA + 0.5% ara | 16.34 ± 0.58 a | 77.80 ± 1.59 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, Y.; Jia, F.; Li, X.; Li, B.; Ren, J.; Yang, X.; Li, G. Improving the Yield of Xenocoumacin 1 by PBAD Promoter Replacement in Xenorhabdus nematophila CB6. Agriculture 2021, 11, 1251. https://doi.org/10.3390/agriculture11121251
Qin Y, Jia F, Li X, Li B, Ren J, Yang X, Li G. Improving the Yield of Xenocoumacin 1 by PBAD Promoter Replacement in Xenorhabdus nematophila CB6. Agriculture. 2021; 11(12):1251. https://doi.org/10.3390/agriculture11121251
Chicago/Turabian StyleQin, Youcai, Fenglian Jia, Xiaohui Li, Beibei Li, Jie Ren, Xiufen Yang, and Guangyue Li. 2021. "Improving the Yield of Xenocoumacin 1 by PBAD Promoter Replacement in Xenorhabdus nematophila CB6" Agriculture 11, no. 12: 1251. https://doi.org/10.3390/agriculture11121251
APA StyleQin, Y., Jia, F., Li, X., Li, B., Ren, J., Yang, X., & Li, G. (2021). Improving the Yield of Xenocoumacin 1 by PBAD Promoter Replacement in Xenorhabdus nematophila CB6. Agriculture, 11(12), 1251. https://doi.org/10.3390/agriculture11121251






