Genome-Wide Characterization of Glutamine Synthetase Family Genes in Cucurbitaceae and Their Potential Roles in Cold Response and Rootstock-Scion Signaling Communication
Abstract
:1. Introduction
2. Materials and Methods
2.1. Screening of GS Family Genes in Pumpkin and Cucumber
2.2. Phylogenetic Analysis of GS Proteins in Pumpkin and Cucumber
2.3. Structure and 5′-Upstream Regions Regulatory-Elements Analysis of GS Family Genes in Pumpkin and Cucumber
2.4. Evolution Analysis of GS Family Genes in Pumpkin and Cucumber
2.5. Gene Ontology Enrichment
2.6. Relative Gene Expression with mRNA Abundance of GS Family Genes
2.7. Semi-Quantitative RT-PCR Assays with Cold-Treated Pumpkin Seedlings
3. Results
3.1. Genome-Wide Characterization of GS Family Genes in Pumpkins and Cucumbers
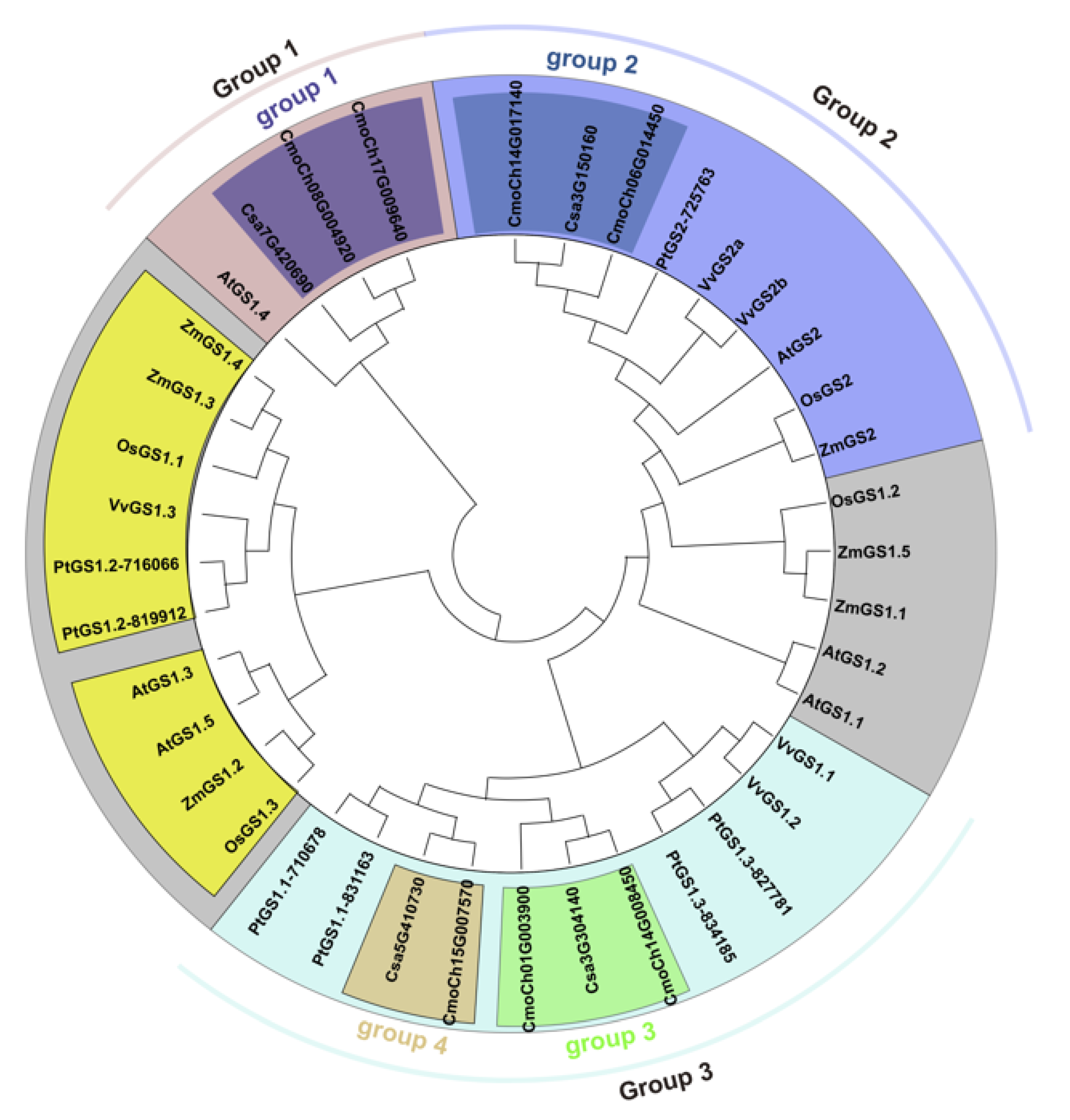
3.2. Evolutionary Relationships of GS Family Genes
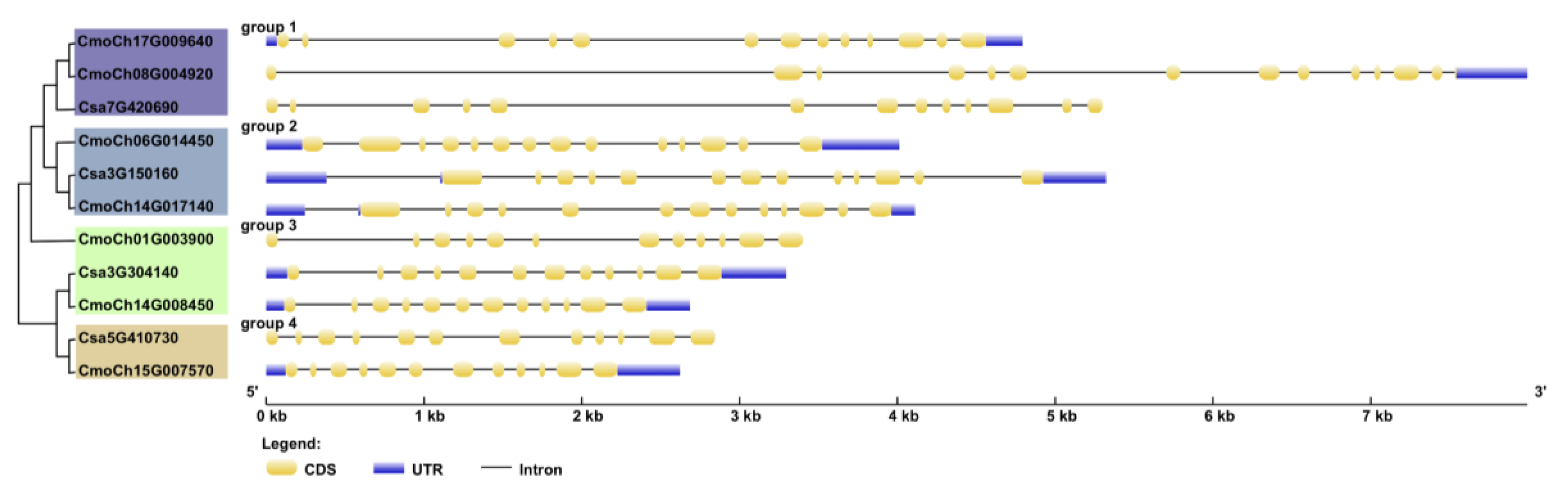
3.3. Structural Analysis of Cucurbitacea GS Family Genes
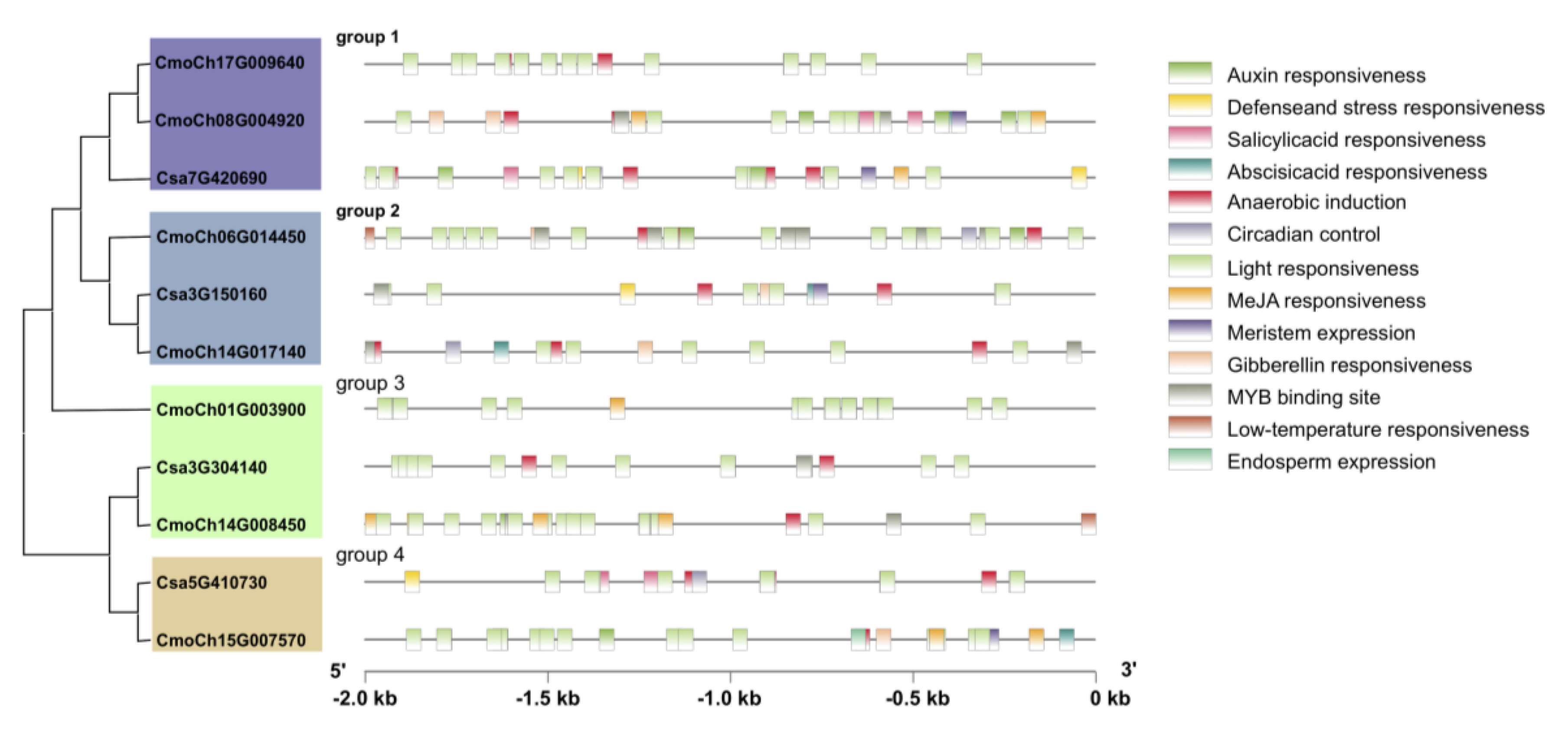
3.4. Regulatory Elements in Cucurbitacea GS Family Genes
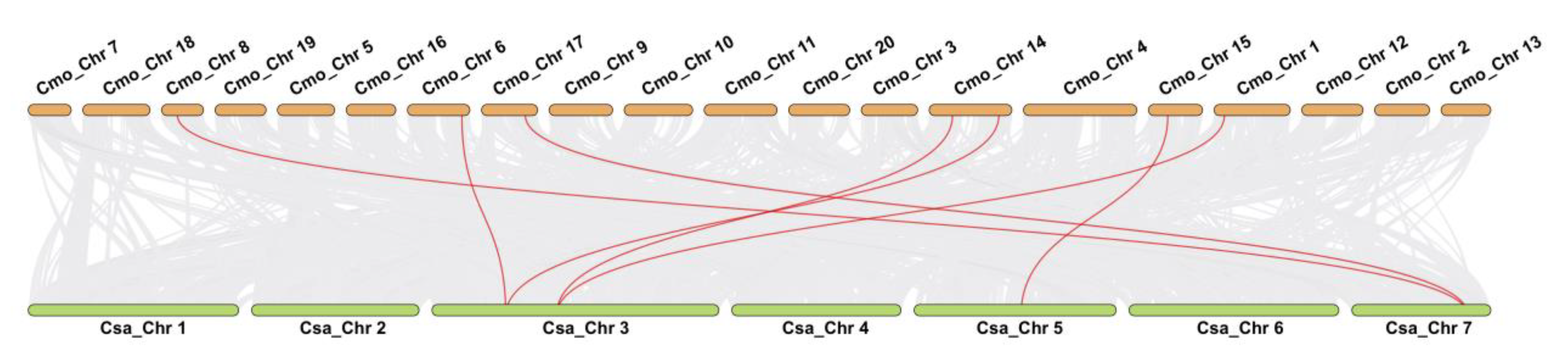
3.5. Distribution and Duplication of GS Family Genes in Pumpkin and Cucumber
3.6. Functional Annotations of GS Gene Members in Pumpkin and Cucumber
3.7. Analysis of the Expression Patterns of CmoGS Family Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bernard, S.M.; Habash, D. The importance of cytosolic glutamine synthetase in nitrogen assimilation and recycling. New Phytol. 2009, 182, 608–620. [Google Scholar] [CrossRef] [PubMed]
- Miflin, B.J.; Habash, D.Z. The role of glutamine synthetase and glutamate dehydrogenase in nitrogen assimilation and possibilities for improvement in the nitrogen utilization of crops. J. Exp. Bot. 2002, 53, 979–987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- James, D.; Borphukan, B.; Fartyal, D.; Achary, V.M.M.; Reddy, M.K. Transgenic manipulation of Glutamine Synthetase: A Target with Untapped Potential in Various Aspects of Crop Improvement. In Biotechnologies of Crop Improvement; Springer: Berlin/Heidelberg, Germany, 2018; Volume 2, pp. 367–416. [Google Scholar]
- Peterman, T.K.; Goodman, H.M. The glutamine synthetase gene family of Arabidopsis thaliana light-regulation and differential expression in leaves, roots and seeds. Mol. Genet. Genom. 1991, 230, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Li, M.G.; Villemur, R.; Hussey, P.J.; Silflow, C.D.; Gantt, J.S.; Snustad, D.P. Differential expression of six glutamine synthetase genes in Zea mays. Plant Mol. Biol. 1993, 23, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Sakakibara, H.; Kawabata, S.; Takahashi, H.; Hase, T.; Sugiyama, T. Molecular Cloning of the Family of Glutamine Synthetase Genes from Maize: Expression of Genes for Glutamine Synthetase and Ferredoxin-Dependent Glutamate Synthase in Photosynthetic and Non-Photosynthetic Tissues. Plant Cell Physiol. 1992, 33, 49–58. [Google Scholar] [CrossRef]
- Castro-Rodríguez, V.; García-Gutiérrez, A.; Canales, J.; Avila, C.; Kirby, E.G.; Cánovas, F.M. The glutamine synthetase gene family in Populus. BMC Plant Biol. 2011, 11, 119. [Google Scholar] [CrossRef] [Green Version]
- Lam, H.-M.; Coschigano, K.T.; Oliveira, I.C.; Melo-Oliveira, R.; Coruzzi, G.M. The Molecular-Genetics of Nitrogen Assimilation into Amino Acids in Higher Plants. Annu. Rev. Plant Biol. 1996, 47, 569–593. [Google Scholar] [CrossRef] [PubMed]
- Seabra, A.R.; Carvalho, H.; Pereira, P.J.B. Crystallization and preliminary crystallographic characterization of glutamine synthetase fromMedicago truncatula. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2009, 65, 1309–1312. [Google Scholar] [CrossRef] [Green Version]
- Unno, H.; Uchida, T.; Sugawara, H.; Kurisu, G.; Sugiyama, T.; Yamaya, T.; Sakakibara, H.; Hase, T.; Kusunoki, M. Atomic Structure of Plant Glutamine Synthetase-A key enzyme for plant productivity. J. Biol. Chem. 2006, 281, 29287–29296. [Google Scholar] [CrossRef] [Green Version]
- McNally, S.; Hirel, B.; Gadal, P.; Mann, A.; Stewart, G. Evidence for a specific isoform content related to their possible physiological role and their compartmentation within the leaf. Plant Physiol. 1983, 72, 22–25. [Google Scholar] [CrossRef] [Green Version]
- Andrews, M. The partitioning of nitrate assimilation between root and shoot of higher plants. Plant Cell Environ. 1986, 9, 511–519. [Google Scholar] [CrossRef]
- Hirel, B.; Lea, P.J.; Lea, P.J.; Morot-Gaudry, J.-F. Ammonia Assimilation. In Plant Nitrogen; Springer: Berlin/Heidelberg, Germany, 2001; pp. 79–99. [Google Scholar]
- Sun, H.; Huang, Q.-M.; Su, J. Highly effective expression of glutamine synthetase genes GS1 and GS2 in transgenic rice plants increases nitrogen-deficiency tolerance. J. Plant Physiol. Mol. Boil. 2005, 31, 492–498. [Google Scholar]
- Hachiya, T.; Inaba, J.; Wakazaki, M.; Sato, M.; Toyooka, K.; Miyagi, A.; Kawai-Yamada, M.; Sugiura, D.; Nakagawa, T.; Kiba, T.; et al. Excessive ammonium assimilation by plastidic glutamine synthetase causes ammonium toxicity in Arabidopsis thaliana. Nat. Commun. 2021, 12, 1–10. [Google Scholar] [CrossRef]
- Moison, M.; Marmagne, A.; Dinant, S.; Soulay, F.; Azzopardi, M.; Lothier, J.; Citerne, S.; Morin, H.; Legay, N.; Chardon, F.; et al. Three cytosolic glutamine synthetase isoforms localized in different-order veins act together for N remobilization and seed filling in Arabidopsis. J. Exp. Bot. 2018, 69, 4379–4393. [Google Scholar] [CrossRef]
- Wei, Y.; Xiong, S.; Zhang, Z.; Meng, X.; Wang, L.; Zhang, X.; Yu, M.; Yu, H.; Wang, X.; Ma, X. Localization, Gene Expression, and Functions of Glutamine Synthetase Isozymes in Wheat Grain (Triticum aestivum L.). Front. Plant Sci. 2021, 12. [Google Scholar] [CrossRef]
- Anwar, A.; Li, Y.; He, C.; Yu, X. 24-Epibrassinolide promotes NO3- and NH4+ ion flux rate and NRT1 gene expression in cucumber under suboptimal root zone temperature. BMC Plant Biol. 2019, 19, 1–5. [Google Scholar]
- Xin, M.; Wang, L.; Liu, Y.; Feng, Z.; Zhou, X.; Qin, Z. Transcriptome profiling of cucumber genome expression in response to long-term low nitrogen stress. Acta Physiol. Plant. 2017, 39, 130. [Google Scholar] [CrossRef]
- Liu, H.; Gao, Y.; Gao, C.; Liu, S.; Zhang, J.; Chen, G.; Zhang, S.; Wu, F. Study of the physiological mechanism of delaying cucumber senescence by wheat intercropping pattern. J. Plant Physiol. 2019, 234–235, 154–166. [Google Scholar] [CrossRef]
- Sun, H.; Wu, S.; Zhang, G.; Jiao, C.; Guo, S.; Ren, Y.; Zhang, J.; Zhang, H.; Gong, G.; Jia, Z.; et al. Karyotype Stability and Unbiased Fractionation in the Paleo-Allotetraploid Cucurbita Genomes. Mol. Plant 2017, 10, 1293–1306. [Google Scholar] [CrossRef] [Green Version]
- Condurso, C.; Verzera, A.; Dima, G.; Tripodi, G.; Crinò, P.; Paratore, A.; Romano, D. Effects of different rootstocks on aroma volatile compounds and carotenoid content of melon fruits. Sci. Hortic. 2012, 148, 9–16. [Google Scholar] [CrossRef]
- Pulgar, G.; Villora, G.; Moreno, D.; Romero, L. Improving the Mineral Nutrition in Grafted Watermelon Plants: Nitrogen Metabolism. Biol. Plant. 2000, 43, 607–609. [Google Scholar] [CrossRef]
- Xing, W.-W.; Li, L.; Gao, P.; Li, H.; Shao, Q.-S.; Shu, S.; Sun, J.; Guo, S.-R. Effects of grafting with pumpkin rootstock on carbohydrate metabolism in cucumber seedlings under Ca(NO3)2 stress. Plant Physiol. Biochem. 2015, 87, 124–132. [Google Scholar] [CrossRef] [PubMed]
- Britto, D.T.; Kronzucker, H. NH4+ toxicity in higher plants: A critical review. J. Plant Physiol. 2002, 159, 567–584. [Google Scholar] [CrossRef] [Green Version]
- Bateman, A.; Birney, E.; Cerruti, L.; Durbin, R.; Etwille, L.; Eddy, S.R.; Griffiths-Jones, S.; Howe, K.L.; Marshall, M.; Sonnhammer, E.L. The Pfam protein families database. Nucleic Acids Res. 2002, 30, 276–280. [Google Scholar] [CrossRef] [Green Version]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Bowers, J.E.; Wang, X.; Ming, R.; Alam, M.; Paterson, A.H. Synteny and Collinearity in Plant Genomes. Science 2008, 320, 486–488. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wei, G.; Tian, P.; Zhang, F.; Qin, H.; Miao, H.; Chen, Q.; Hu, Z.; Cao, L.; Wang, M.; Gu, X.; et al. Integrative Analyses of Nontargeted Volatile Profiling and Transcriptome Data Provide Molecular Insight into VOC Diversity in Cucumber Plants (Cucumis sativus). Plant Physiol. 2016, 172, 603–618. [Google Scholar] [CrossRef] [Green Version]
- Cheng, G.; You, X.; Wu, Y.; Zhang, J. Analysis on gene differential expression of cold-resistance cultivar ‘Ziyangyuanye’of Camellia sinensis after low temperature stress. J. Plant Res. Environ. 2013, 22, 38–43. [Google Scholar]
- Miao, G.H.; Hirel, B.; Marsolier, M.C.; Ridge, R.W.; Verma, D.P.S. Ammonia-regulated expression of a soybean gene encoding cytosolic Glutamine-Synthetase in transgenic lotus-corniculatus. Plant Cell 1991, 3, 11–22. [Google Scholar] [PubMed] [Green Version]
- Teixeira, J.; Pereira, S. High salinity and drought act on an organ-dependent manner on potato glutamine synthetase expression and accumulation. Environ. Exp. Bot. 2007, 60, 121–126. [Google Scholar] [CrossRef]
- Scarpeci, T.E.; Marro, M.L.; Bortolotti, S.; Boggio, S.B.; Valle, E.M. Plant nutritional status modulates glutamine synthetase levels in ripe tomatoes (Solanum lycopersicum cv. Micro-Tom). J. Plant Physiol. 2007, 164, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.; Landherr, L.L.; Frohlich, M.W.; Leebens-Mack, J.; Ma, H.; De Pamphilis, C.W. Patterns of gene duplication in the plant SKP1 gene family in angiosperms: Evidence for multiple mechanisms of rapid gene birth. Plant J. 2007, 50, 873–885. [Google Scholar] [CrossRef]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-Wide Analysis of the ERF Gene Family in Arabidopsis and Rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Wu, N.; Song, W.; Yin, G.; Qin, Y.; Yan, Y.; Hu, Y. Soybean (Glycine max) expansin gene superfamily origins: Segmental and tandem duplication events followed by divergent selection among subfamilies. BMC Plant Biol. 2014, 14, 93. [Google Scholar] [CrossRef] [Green Version]
- Kumada, Y.; Benson, D.; Hillemann, D.; Hosted, T.; Rochefort, D.; Thompson, C.; Wohlleben, W.; Tateno, Y. Evolution of the glutamine synthetase gene, one of the oldest existing and functioning genes. Proc. Natl. Acad. Sci. USA 1993, 90, 3009–3013. [Google Scholar] [CrossRef] [Green Version]
- Pesole, G.; Bozzetti, M.P.; Lanave, C.; Preparata, G.; Saccone, C. Glutamine synthetase gene evolution: A good molecular clock. Proc. Natl. Acad. Sci. USA 1991, 88, 522–526. [Google Scholar] [CrossRef] [Green Version]
- Tingey, S.V.; Tsai, F.Y.; Edwards, J.W.; Walker, E.; Coruzzi, G.M. Chloroplast and cytosolic glutamine synthetase are encoded by homologous nuclear genes which are differentially expressed in vivo. J. Biol. Chem. 1988, 263, 9651–9657. [Google Scholar] [CrossRef]
- Seabra, A.R.; Vieira, C.P.; Cullimore, J.V.; Carvalho, H.G. Medicago truncatula contains a second gene encoding a plastid located glutamine synthetase exclusively expressed in developing seeds. BMC Plant Biol. 2010, 10, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Wei, Y.; Shi, L.; Ma, X.; Theg, S.M. New isoforms and assembly of glutamine synthetase in the leaf of wheat (Triticum aestivum L.). J. Exp. Bot. 2015, 66, 6827–6834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomsen, H.C.; Eriksson, D.; Møller, I.S.; Schjoerring, J.K. Cytosolic glutamine synthetase: A target for improvement of crop nitrogen use efficiency? Trends Plant Sci. 2014, 19, 656–663. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Lu, X.; Liu, M.; Xiang, C.; Liu, W.; Wang, C.; Zhang, X.; Wang, T.; Liu, Z.; Gao, L.; et al. Genome-Wide Characterization of Glutamine Synthetase Family Genes in Cucurbitaceae and Their Potential Roles in Cold Response and Rootstock-Scion Signaling Communication. Agriculture 2021, 11, 1156. https://doi.org/10.3390/agriculture11111156
Li X, Lu X, Liu M, Xiang C, Liu W, Wang C, Zhang X, Wang T, Liu Z, Gao L, et al. Genome-Wide Characterization of Glutamine Synthetase Family Genes in Cucurbitaceae and Their Potential Roles in Cold Response and Rootstock-Scion Signaling Communication. Agriculture. 2021; 11(11):1156. https://doi.org/10.3390/agriculture11111156
Chicago/Turabian StyleLi, Xiaojun, Xiaohong Lu, Mengshuang Liu, Chenggang Xiang, Wenqian Liu, Cuicui Wang, Xiaojing Zhang, Tao Wang, Zixi Liu, Lihong Gao, and et al. 2021. "Genome-Wide Characterization of Glutamine Synthetase Family Genes in Cucurbitaceae and Their Potential Roles in Cold Response and Rootstock-Scion Signaling Communication" Agriculture 11, no. 11: 1156. https://doi.org/10.3390/agriculture11111156
APA StyleLi, X., Lu, X., Liu, M., Xiang, C., Liu, W., Wang, C., Zhang, X., Wang, T., Liu, Z., Gao, L., & Zhang, W. (2021). Genome-Wide Characterization of Glutamine Synthetase Family Genes in Cucurbitaceae and Their Potential Roles in Cold Response and Rootstock-Scion Signaling Communication. Agriculture, 11(11), 1156. https://doi.org/10.3390/agriculture11111156





