Approach for Image-Based Semantic Segmentation of Canopy Cover in Pea–Oat Intercropping
Abstract
1. Introduction
2. Materials and Methods
2.1. Field Experiment and Image Acquisition
2.2. Image Processing and CNN Architecture
2.3. CNN Training
2.4. Evaluation
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Vandermeer, J. The Ecology of Intercropping, 1st ed.; Cambridge University Press: Cambridge, UK, 1989; ISBN 0521345928. [Google Scholar]
- Jensen, E.S.; Bedoussac, L.; Carlsson, G.; Journet, E.; Justes, E.; Hauggaard-Nielsen, H. Enhancing Yields in Organic Crop Production by Eco-Functional Intensification. Sustain. Agric. Res. 2015, 4, 42–50. [Google Scholar] [CrossRef]
- Verret, V.; Pelzer, E.; Bedoussac, L.; Jeu, M. Tracking on-farm innovative practices to support crop mixture design: The case of annual mixtures including a legume crop. Eur. J. Agron. 2020, 115. [Google Scholar] [CrossRef]
- Hauggaard-Nielsen, H.; Gooding, M.; Ambus, P.; Corre-Hellou, G.; Crozat, Y.; Dahlmann, C.; Dibet, A. Pea-Barley intercropping for efficient symbiotic N2-fixation, soil N acquisition and use of other nutrients in European organic cropping systems. Field Crop. Res. 2009, 113, 64–71. [Google Scholar] [CrossRef]
- Jensen, E.S. Intercropping of grain legumes and cereals improves the use of soil N resources and reduces the requirement for synthetic fertilizer N: A global-scale analysis. Agron. Sustain. Dev. 2020, 40, 1–9. [Google Scholar] [CrossRef]
- Bedoussac, L.; Journet, E.; Hauggaard-Nielsen, H.; Naudin, C.; Corre-Hellou, G.; Jensen, E.S. Ecological principles underlying the increase of productivity achieved by cereal-grain legume intercrops in organic farming. A review. Agron. Sustain. Dev. 2015, 35, 911–935. [Google Scholar] [CrossRef]
- Gaudio, N.; Escobar-Gutiérrez, A.J.; Casadebaig, P.; Evers, J.B.; Gérard, F.; Louarn, G.; Colbach, N.; Munz, S.; Launay, M.; Marrou, H.; et al. Current knowledge and future research opportunities for modeling annual crop mixtures. A review. Agron. Sustain. Dev. 2019, 39, 1–20. [Google Scholar] [CrossRef]
- Munz, S.; Graeff-Hönninger, S.; Lizaso, J.I.; Chen, Q.; Claupein, W. Modeling light availability for a subordinate crop within a strip-intercropping system. Field Crop. Res. 2014, 155, 77–89. [Google Scholar] [CrossRef]
- Shepherd, M.J.; Lindsey, L.E.; Lindsey, A.J. Soybean Canopy Cover Measured with Canopeo Compared with Light Interception. Agric. Environ. Lett. 2018, 3, 1–3. [Google Scholar] [CrossRef]
- Kusumam, K.; Kranjík, T.; Pearson, S.; Cielniak, G.; Duckett, T. Can You Pick a Broccoli? 3D-Vision Based Detection and Localisation of Broccoli Heads in the Field. In Proceedings of the 2016 IEEE International Conference on Intelligent Robots and Systems (IROS), Daejeon, Korea, 9–14 October 2016; pp. 646–651. [Google Scholar]
- Riehle, D.; Reiser, D.; Griepentrog, H.W. Robust index-based semantic plant/background segmentation for RGB-images. Comput. Electron. Agric. 2020, 169, 105201. [Google Scholar] [CrossRef]
- Åstrand, B.; Baerveldt, A.J. An agricultural mobile robot with vision-based perception for mechanical weed control. Auton. Robots 2002, 13, 21–35. [Google Scholar] [CrossRef]
- Kamilaris, A.; Prenafeta-Boldú, F.X. Deep learning in agriculture: A survey. Comput. Electron. Agric. 2018, 147, 70–90. [Google Scholar] [CrossRef]
- Lottes, P.; Hoeferlin, M.; Sander, S.; Stachniss, C. An Effective Classification System for Separating Sugar Beets and Weeds for Precision Farming Applications. In Proceedings of the IEEE International Conference on Robotics and Automation (ICRA), Stockholm, Sweden, 16–21 May 2016; pp. 5157–5163. [Google Scholar]
- Milioto, A.; Lottes, P.; Stachniss, C. Real-time Semantic Segmentation of Crop and Weed for Precision Agriculture Robots Leveraging Background Knowledge in CNNs. In Proceedings of the IEEE International Conference on Robotics and Automation (ICRA), Brisbane, QLD, Australia, 21–25 May 2018; pp. 2229–2235. [Google Scholar]
- Bargoti, S.; Underwood, J. Image Segmentation for Fruit Detection and Yield Estimation in Apple Orchards. J. Field. Robot. 2016, 34, 1–22. [Google Scholar] [CrossRef]
- Dias, P.A.; Tabb, A.; Medeiros, H. Multispecies fruit flower detection using a refined semantic segmentation network. IEEE Robot. Autom. Lett. 2018, 3, 3003–3010. [Google Scholar] [CrossRef]
- Sankaran, S.; Mishra, A.; Ehsani, R.; Davis, C. A review of advanced techniques for detecting plant diseases. Comput. Electron. Agric. 2010. [Google Scholar] [CrossRef]
- Reiser, D.; Vázquez-Arellano, M.; Paraforos, D.S.; Garrido-Izard, M.; Griepentrog, H.W. Iterative individual plant clustering in maize with assembled 2D LiDAR data. Comput. Ind. 2018, 99. [Google Scholar] [CrossRef]
- Krogh Mortensen, A.; Dyrmann, M.; Karstoft, H.; Nyholm Jorgensen, R.; Gislum, R. Semantic Segmentation of Mixed Crops using Deep Convolutional Neural Network. In Proceedings of the CIGR-AgEng Conference, Aarhus, Denmark, 26–29 June 2016; pp. 1–6. [Google Scholar]
- Lottes, P.; Behley, J.; Milioto, A.; Stachniss, C. Robust joint stem detection and crop—Weed classification using image sequences for plant—Specific treatment in precision farming. J. Field Robot. 2020, 37, 20–34. [Google Scholar] [CrossRef]
- Dutta, A.; Gitahi, J.M.; Ghimire, P.; Mink, R.; Peteinatos, G.; Engels, J.; Hahn, M.; Gerhards, R. Weed Detection in Close-range Imagery of Agricultural Fields using Neural Networks. Publ. DGPF 2018, 27, 633–645. [Google Scholar]
- Bosilj, P.; Duckett, T. Transfer learning between crop types for semantic segmentation of crops versus weeds in precision agriculture. J. Field Robot. 2020, 37, 7–19. [Google Scholar] [CrossRef]
- Reiser, D.; Vázquez-Arellano, M.; Izard, M.G.; Paraforos, D.S.; Sharipov, G.; Griepentrog, H.W. Clustering of Laser Scanner Perception Points of Maize Plants. Adv. Anim. Biosci. 2017, 8, 204–209. [Google Scholar] [CrossRef]
- Taylor, L.; Nitschke, G. Improving Deep Learning with Generic Data Augmentation. In Proceedings of the 2018 IEEE Symposium Series on Computational Intelligence (SSCI), Bangalore, India, 18–21 November 2018; pp. 1542–1547. [Google Scholar]
- Bechar, A.; Vigneault, C. Agricultural robots for field operations: Concepts and components. Biosyst. Eng. 2016, 149, 94–111. [Google Scholar] [CrossRef]
- Shorten, C.; Khoshgoftaar, T.M. A survey on Image Data Augmentation for Deep Learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Meier, U. Growth Stages of Mono- and Dicotyledonous Plants; BBCH Monograph, Open Agrar Repositorium; Blackwell Wissenschafts-Verlag: Berlin, Germany, 2018; ISBN 9783955470715. [Google Scholar]
- Software, I. Open Source Software ImagJ. 2019. Available online: https://imagej.net/Welcome (accessed on 1 May 2020).
- Mohammad, H.; Axel, D.; David, W.; Antoine, B.; Aaron, C.; Yoshua, B.; Chris, P.; Pierre-Marc, J.; Hugo, L. Brain tumor segmentation with Deep Neural Networks. Med. Image Anal. 2017, 35, 18–31. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]


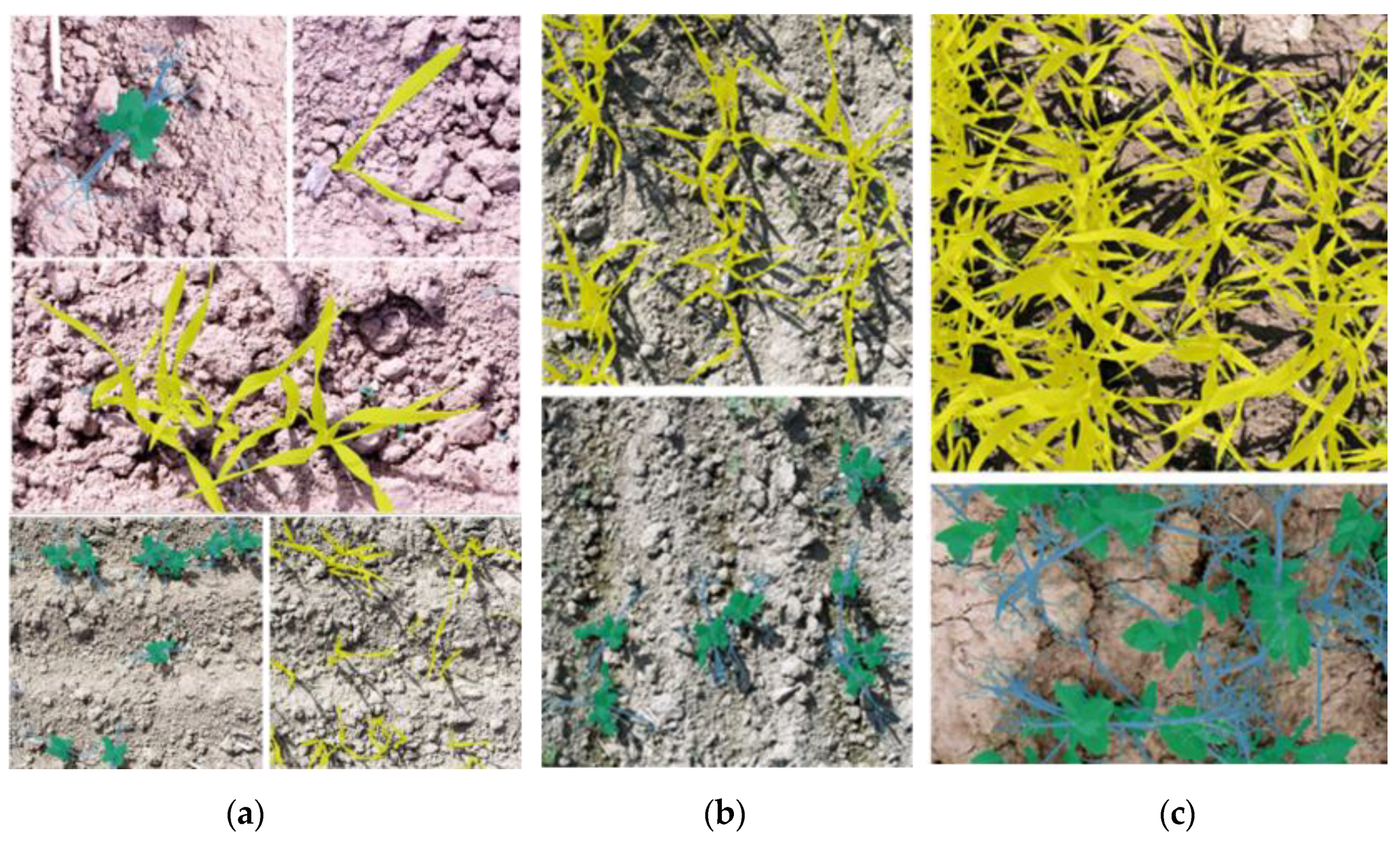
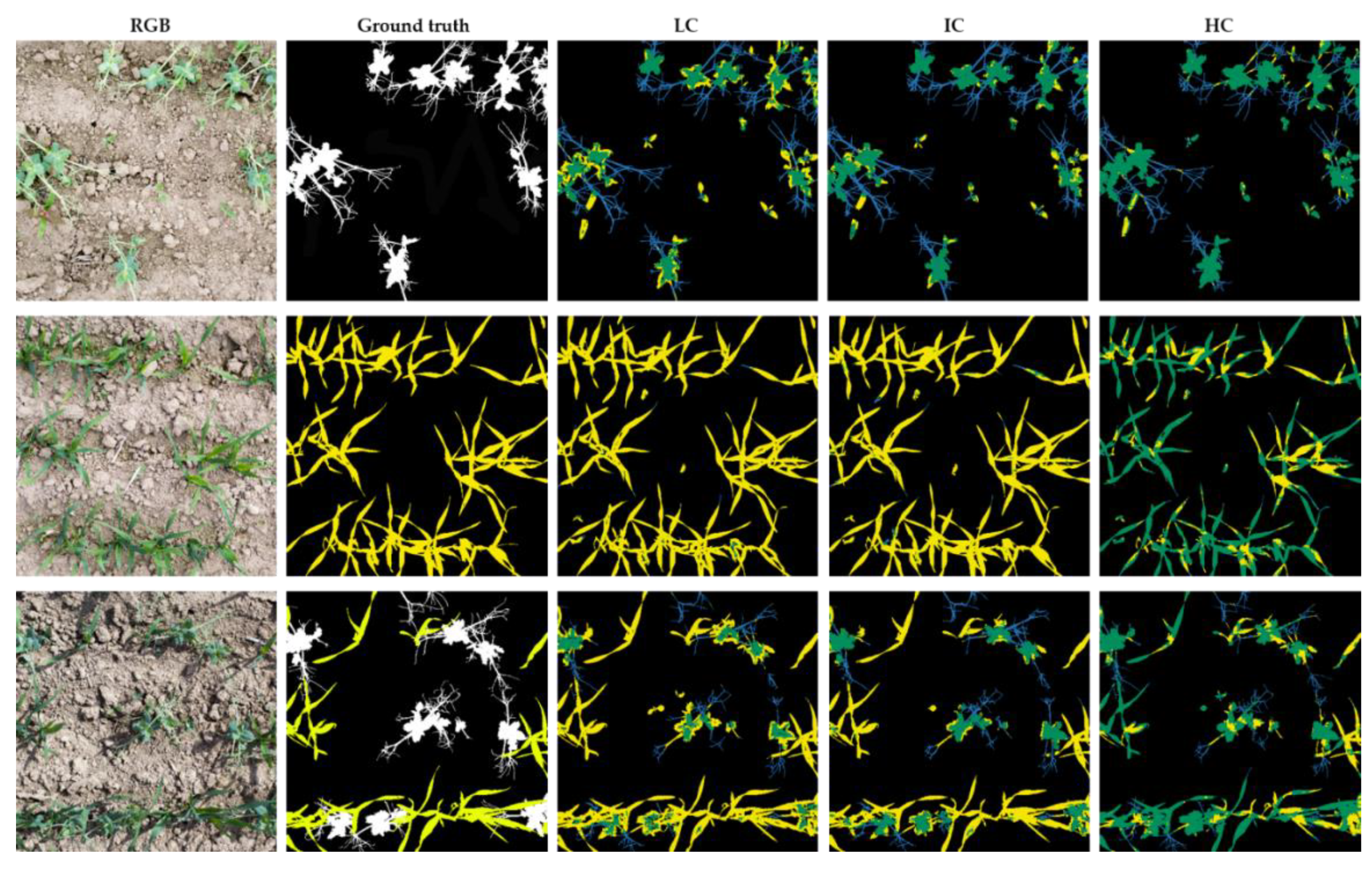
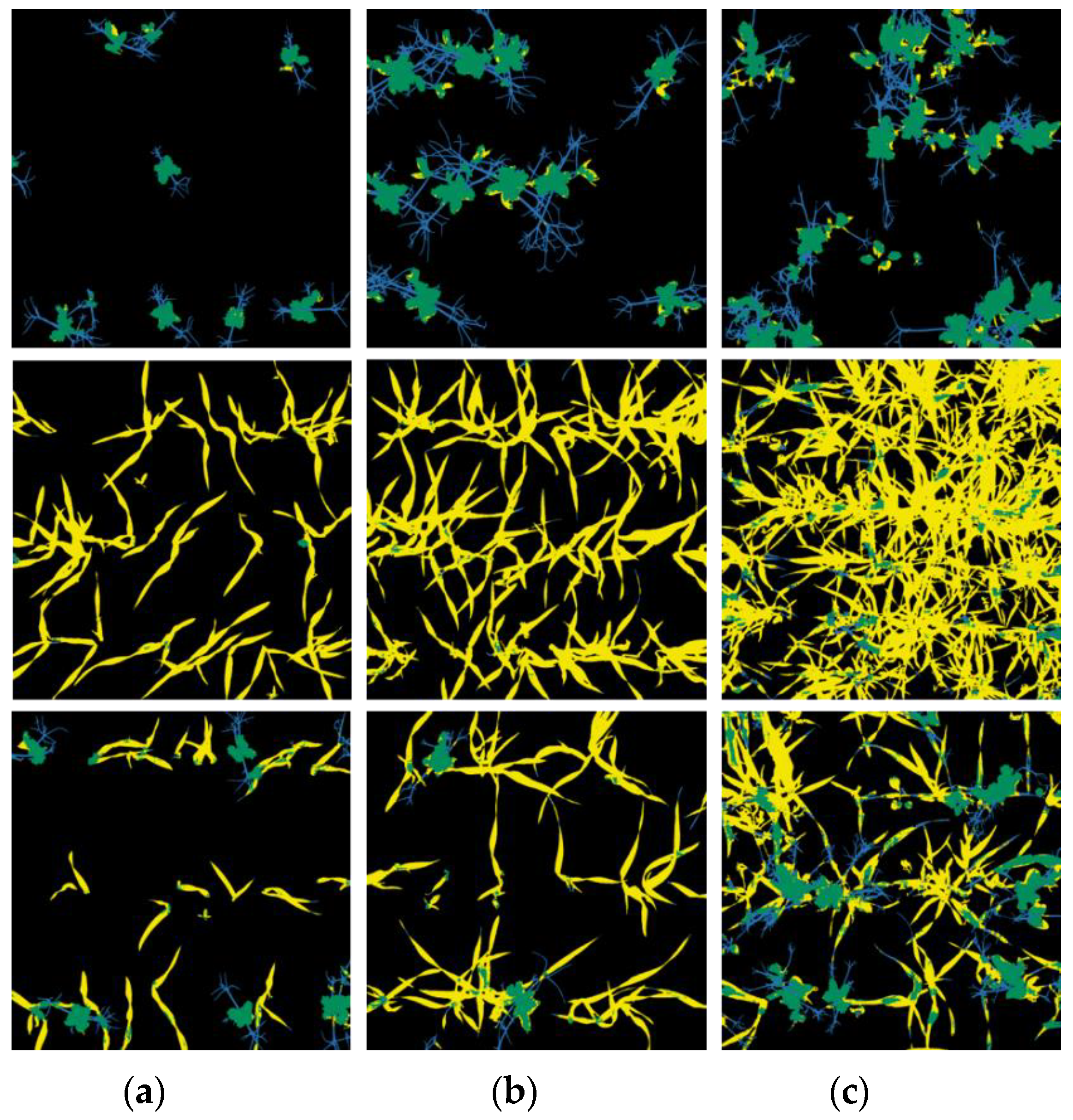
| Date | Name of Dataset | Cover in Sole Crops [%] | Cover in Intercrops [%] | ||||
|---|---|---|---|---|---|---|---|
| Pea | Oat | Weed | Pea | Oat | Weed | ||
| 25 April | Low cover | 7.0 | 14.1 | 0.4 | 3.8 | 7.9 | 0.6 |
| 2 May | Intermediate cover | 10.1 | 20.7 | 1.2 | 4.8 | 13.0 | 1.4 |
| 16 May | High cover | 17.6 | 51.8 | 1.9 | 11.4 | 30.3 | 2.0 |
| Low Cover (LC) | Intermediate Cover (IC) | High Cover (HC) | LC + IC | LC + IC + HC | |
|---|---|---|---|---|---|
| No. of images | 42 | 7 | 7 | 49 | 56 |
| No. of pea plants | 102 | 40 | 39 | 142 | 181 |
| No. of oat plants | 158 | 98 | 64 | 256 | 320 |
| Pea pixels | 3.628.537 | 2.518.875 | 2.129.794 | 6.147.412 | 8.277.206 |
| Oat pixels | 3.473.450 | 3.456.233 | 4.020.441 | 6.929.683 | 10.950.124 |
| Soil pixels | 87.489.759 | 41.344.890 | 5.614.212 | 128.834.649 | 134.448.861 |
| Dataset | Network | IoU [%] | Precision [%] | Recall [%] | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| mIoU | Soil | Pea | Oat | Soil | Pea | Oat | Soil | Pea | Oat | ||
| Low Cover | LC | 81.1 | 96.7 | 67.7 | 79.1 | 97.9 | 78.7 | 95.1 | 98.7 | 82.9 | 82.5 |
| IC | 69.1 | 94.1 | 55.8 | 57.5 | 95.8 | 65.1 | 97.9 | 98.2 | 79.6 | 58.2 | |
| HC | 41.4 | 85.6 | 30.2 | 8.4 | 91.9 | 31.9 | 98.0 | 92.6 | 85.3 | 8.4 | |
| LC + IC | 77.7 | 96.1 | 60.7 | 76.4 | 97.3 | 78.3 | 93.9 | 98.8 | 73.0 | 80.3 | |
| LC + IC + HC | 81.1 | 96.6 | 67.9 | 78.7 | 97.9 | 77.4 | 95.5 | 98.7 | 84.7 | 81.8 | |
| Int. Cover | LC | 75.5 | 93.5 | 55.6 | 77.3 | 96.8 | 81.8 | 82.5 | 96.5 | 63.5 | 92.4 |
| IC | 80.3 | 94.9 | 64.5 | 81.4 | 97.1 | 82.5 | 90.2 | 97.7 | 74.7 | 89.3 | |
| HC | 46.0 | 80.7 | 31.8 | 25.5 | 89.3 | 33.6 | 88.6 | 89.4 | 85.2 | 26.3 | |
| LC + IC | 75.8 | 93.6 | 55.9 | 77.8 | 96.6 | 77.7 | 85.8 | 96.9 | 66.6 | 89.3 | |
| LC + IC + HC | 81.4 | 95.1 | 66.5 | 82.5 | 97.7 | 81.6 | 88.3 | 97.3 | 78.3 | 92.6 | |
| High Cover | LC | 50.3 | 64.2 | 35.8 | 50.7 | 76.8 | 40.7 | 87.9 | 79.7 | 75.2 | 54.5 |
| IC | 40.1 | 59.5 | 33.0 | 27.9 | 69.1 | 37.8 | 95.1 | 81.1 | 71.9 | 28.3 | |
| HC | 66.3 | 78.6 | 47.5 | 72.9 | 86.5 | 64.3 | 88.1 | 89.6 | 64.5 | 80.9 | |
| LC + IC | 41.3 | 58.1 | 30.1 | 35.8 | 70.4 | 34.1 | 90.9 | 76.9 | 71.7 | 37.1 | |
| LC + IC + HC | 67.5 | 78.1 | 51.1 | 73.3 | 88.3 | 62.3 | 86.2 | 87.1 | 74.1 | 83.0 | |
| Dataset | Network | Sole Crops | Intercrops | ||
|---|---|---|---|---|---|
| Pea IoU [%] | Oat IoU [%] | Pea IoU [%] | Oat IoU [%] | ||
| Low Cover | LC | 81.7 | 83.1 | 60.2 | 73.7 |
| LC + IC + HC | 82.2 | 84.3 | 58.3 | 70.4 | |
| Int. Cover | IC | 76.5 | 86.6 | 52.6 | 79.6 |
| LC + IC + HC | 76.6 | 89.7 | 56.8 | 79.9 | |
| High Cover | HC | 74.4 | 82.1 | 25.6 | 63.3 |
| LC + IC + HC | 72.3 | 82.7 | 38.5 | 64.2 | |
| Dataset | Network | Sole Crops | Intercrops | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pea Cover [%] | Oat Cover [%] | Pea Cover [%] | Oat Cover [%] | ||||||||||
| Δa | Δr | Δa | Δr | Δa | Δr | Δa | Δr | ||||||
| Low Cover | LC + IC + HC | 6.3 | −0.7 | −10.0 | 12.6 | −1.5 | −10.6 | 4.7 | 0.9 | 23.7 | 6.2 | −1.7 | −21.5 |
| LC | 6.2 | −0.8 | −11.4 | 12.4 | −1.7 | −12.1 | 4.3 | 0.5 | 13.2 | 6.5 | −1.4 | −17.7 | |
| Int. Cover | LC + IC + HC | 9.5 | −0.6 | −5.9 | 20.6 | −0.1 | −0.5 | 4.3 | −0.5 | −10.4 | 13.2 | 0.2 | 1.5 |
| IC | 9.3 | −0.8 | −7.9 | 19.2 | −1.5 | −7.2 | 3.6 | −1.2 | −25.0 | 13.0 | 0.0 | 0.0 | |
| High Cover | LC + IC + HC | 17.0 | −0.6 | −3.4 | 50.5 | −1.3 | −2.5 | 13.8 | 2.4 | 21.1 | 28.1 | −2.2 | −7.3 |
| HC | 16.5 | −1.1 | −6.3 | 46.5 | −5.3 | −10.2 | 9.3 | −2.1 | −18.4 | 29.1 | −1.2 | −4.0 | |
| Mean ( | LC + IC + HC | −0.6 | −6.4 | −1.0 | −4.5 | 0.9 | 11.5 | −1.2 | −9.1 | ||||
| single net | −0.9 | −8.5 | −2.8 | −9.8 | −0.9 | −10.1 | −0.9 | −7.2 | |||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Munz, S.; Reiser, D. Approach for Image-Based Semantic Segmentation of Canopy Cover in Pea–Oat Intercropping. Agriculture 2020, 10, 354. https://doi.org/10.3390/agriculture10080354
Munz S, Reiser D. Approach for Image-Based Semantic Segmentation of Canopy Cover in Pea–Oat Intercropping. Agriculture. 2020; 10(8):354. https://doi.org/10.3390/agriculture10080354
Chicago/Turabian StyleMunz, Sebastian, and David Reiser. 2020. "Approach for Image-Based Semantic Segmentation of Canopy Cover in Pea–Oat Intercropping" Agriculture 10, no. 8: 354. https://doi.org/10.3390/agriculture10080354
APA StyleMunz, S., & Reiser, D. (2020). Approach for Image-Based Semantic Segmentation of Canopy Cover in Pea–Oat Intercropping. Agriculture, 10(8), 354. https://doi.org/10.3390/agriculture10080354






