Microbial Analysis of Saliva to Identify Oral Diseases Using a Point-of-Care Compatible qPCR Assay
Abstract
1. Introduction
2. Experimental Section
2.1. Study Population
2.2. Study Design
2.3. Sampling of Saliva
2.4. DNA Extraction for qPCR
2.5. Oligonucleotide Design
2.6. Primer and Probe Specificity
2.7. Duplex qPCR
2.8. Conversion into Genome Equivalent
2.9. Reference Microbial Testing
2.10. Statistics
3. Results
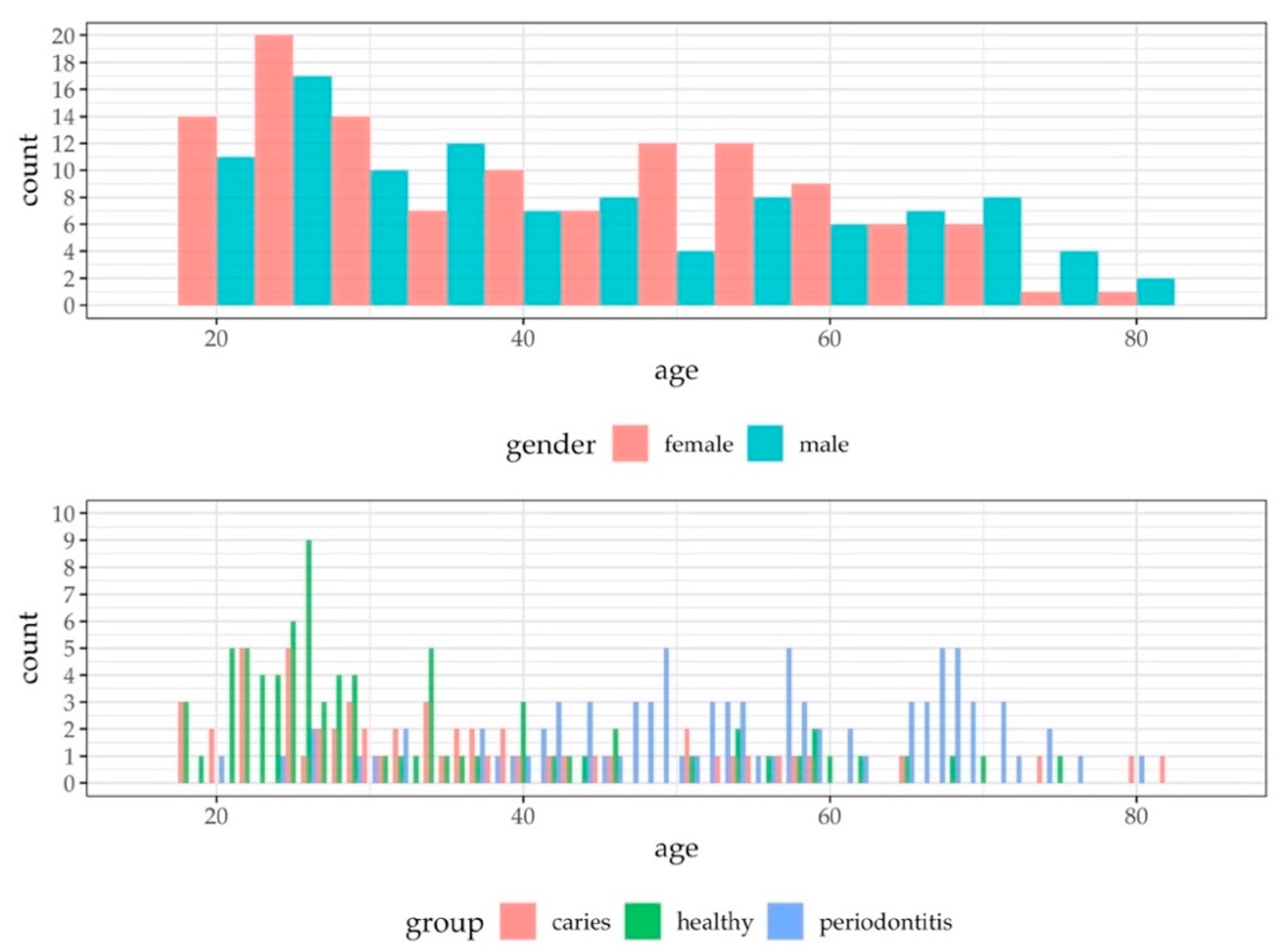
3.1. Study Population
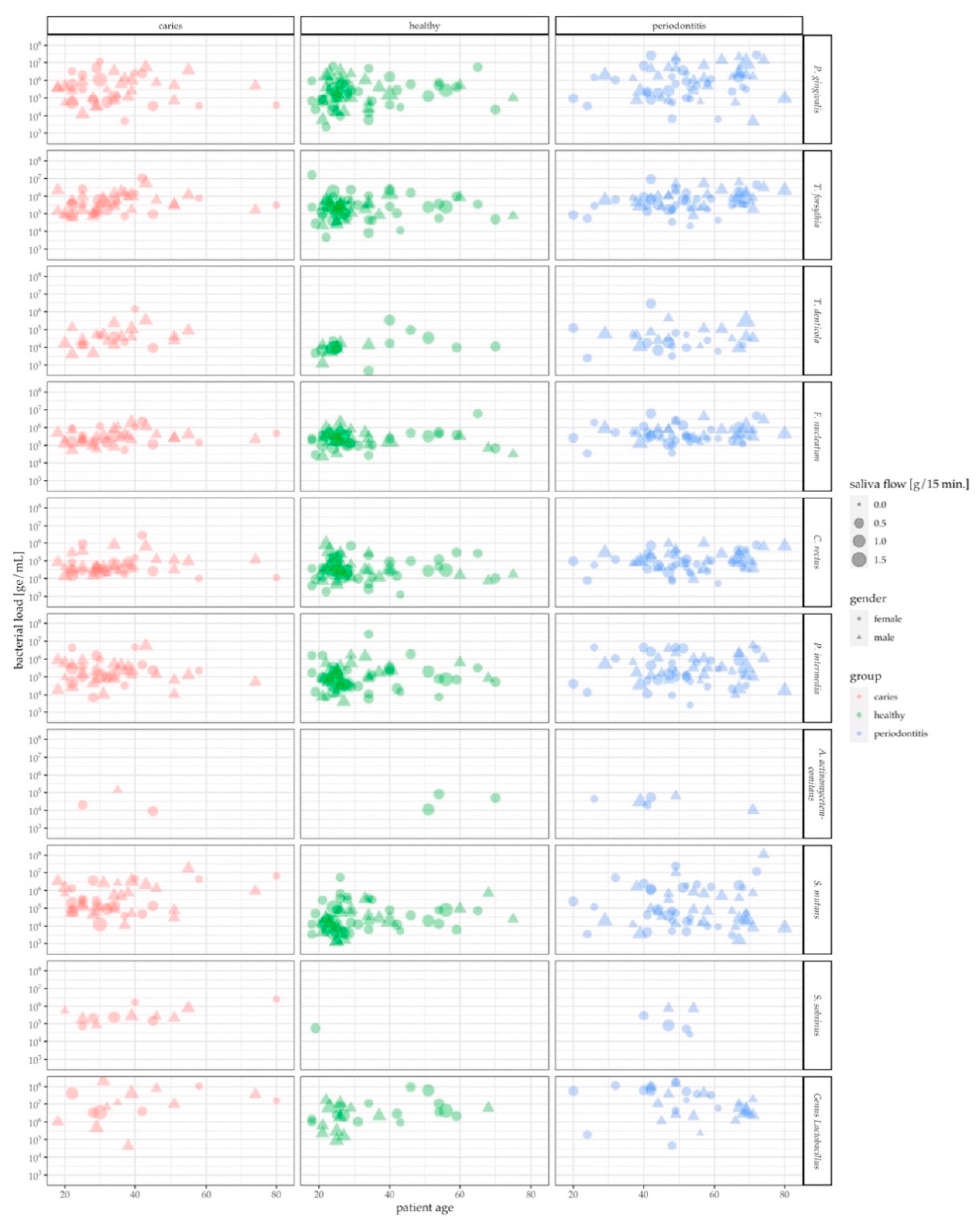
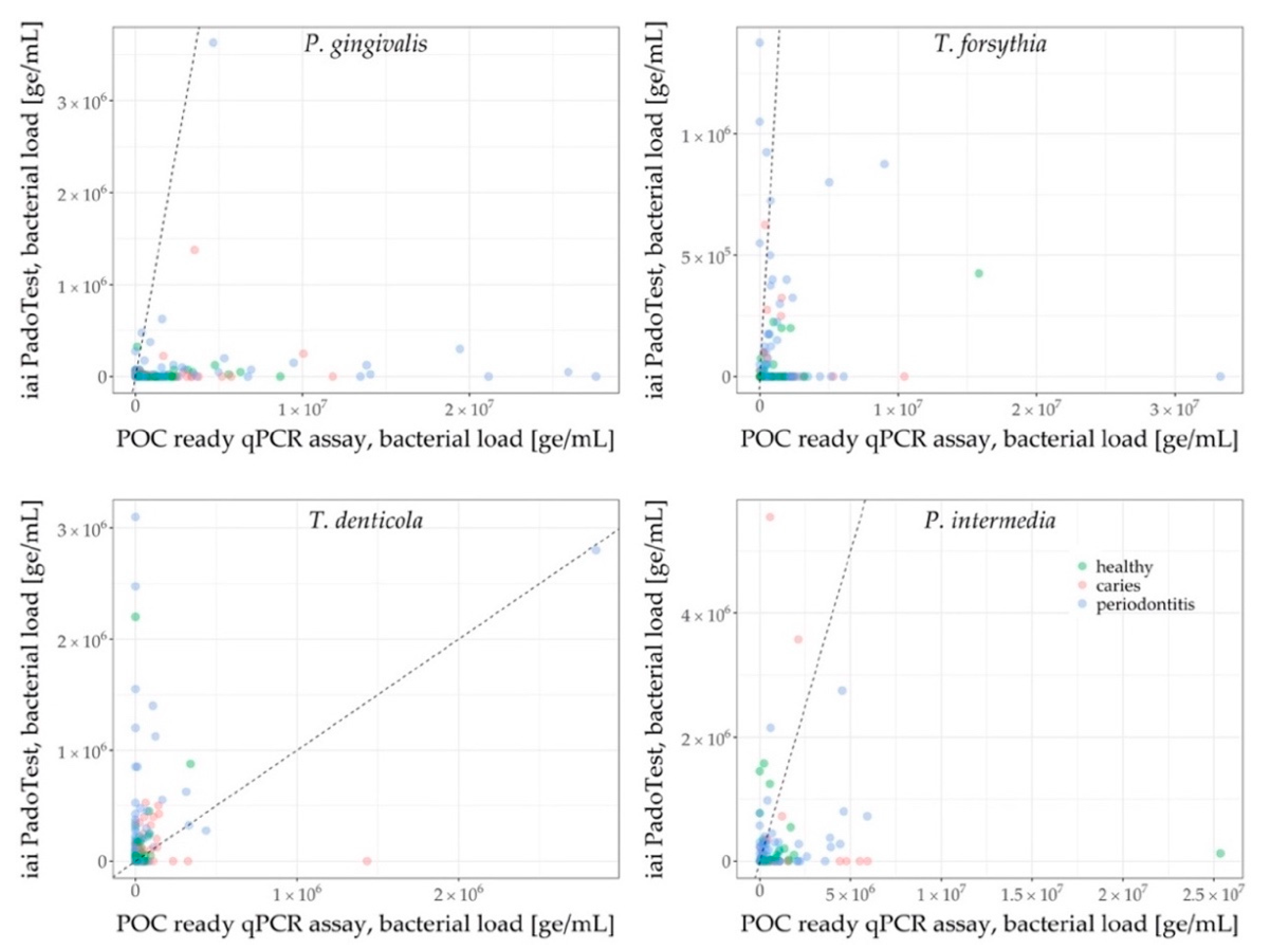
3.2. Characteristics of the qPCR Data
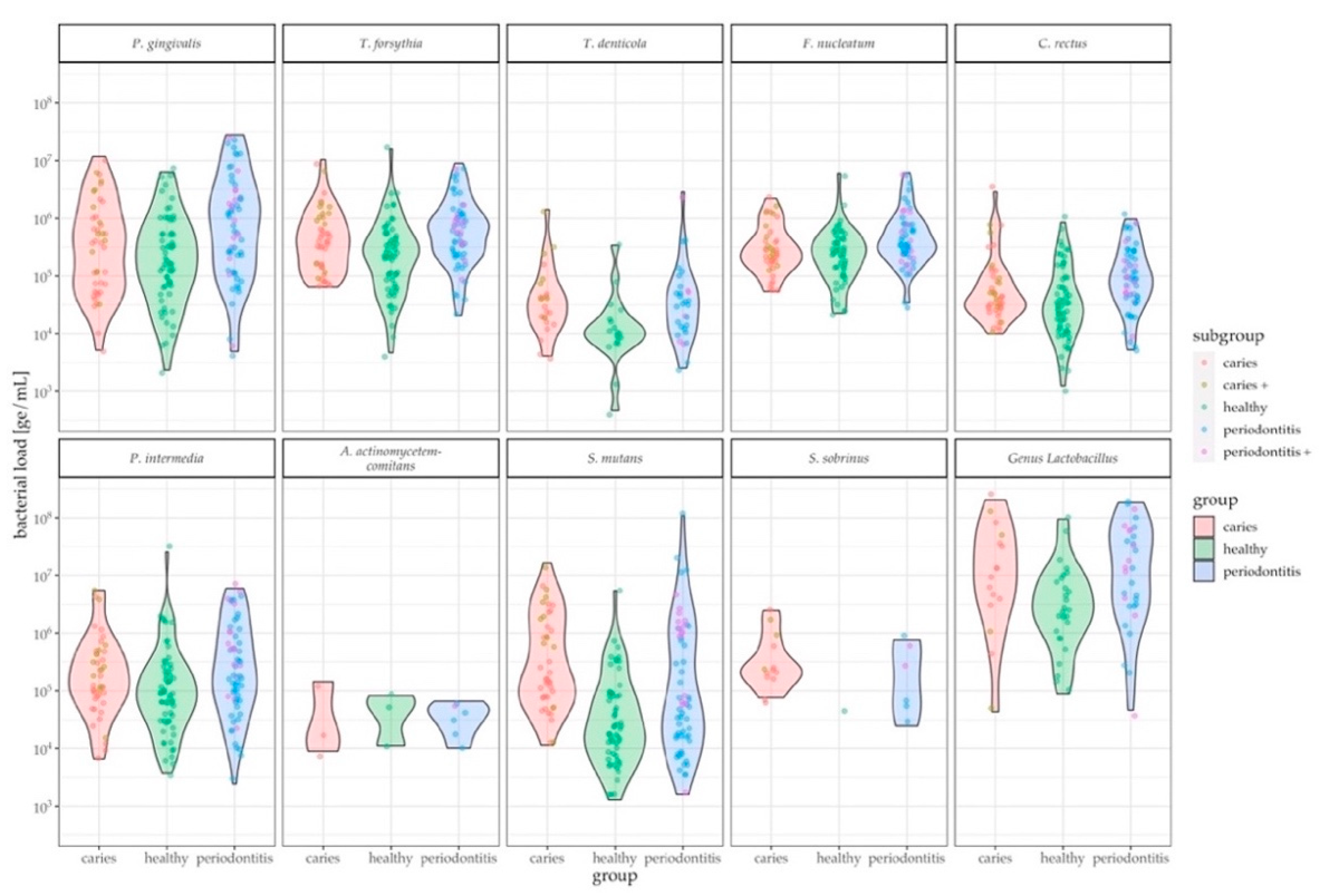
3.3. Assessment of Oral Health Status by Pathogen Levels in Saliva
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- OECD Health Statistics. Focus on, Spending on Primary Care: First Estimates; OECD: Paris, France, 2018. [Google Scholar]
- GBD 2017 Disease and Injury Incidence and Prevalence Collaborators. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1789–1858. [Google Scholar] [CrossRef]
- Belstrøm, D.; Fiehn, N.E.; Nielsen, C.H.; Klepac-Ceraj, V.; Paster, B.J.; Twetman, S.; Holmstrup, P. Differentiation of salivary bacterial profiles of subjects with periodontitis and dental caries. J. Oral Microbiol. 2015, 7, 27429. [Google Scholar] [CrossRef] [PubMed]
- Iwano, Y.; Sugano, N.; Matsumoto, K.; Nishihara, R.; Iizuka, T.; Yoshinuma, N.; Ito, K. Salivary microbial levels in relation to periodontal status and caries development. J. Periodontal Res. 2010, 45, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Liu, X.; Li, J.; Aprecio, R.M.; Zhang, W.; Li, Y. Real-time PCR quantification of six periodontal pathogens in saliva samples from healthy young adults. Clin. Oral Investig. 2015, 19, 937–946. [Google Scholar] [CrossRef]
- Kilian, M.; Chapple, I.L.; Hannig, M.; Marsh, P.D.; Meuric, V.; Pedersen, A.M.; Tonetti, M.S.; Wade, W.G.; Zaura, E. The oral microbiome—An update for oral healthcare professionals. Br. Dent. J. 2016, 221, 657–666. [Google Scholar] [CrossRef]
- de Araújo Nobre, M.; Maló, P. Prevalence of periodontitis, dental caries, and peri-implant pathology and their relation with systemic status and smoking habits: Results of an open-cohort study with 22009 patients in a private rehabilitation center. J. Dent. 2017, 67, 36–42. [Google Scholar] [CrossRef]
- Zarco, M.F.; Vess, T.J.; Ginsburg, G.S. The oral microbiome in health and disease and the potential impact on personalized dental medicine. Oral Dis. 2012, 18, 109–120. [Google Scholar] [CrossRef]
- Streckfus, C.F.; Bigler, L.R. Saliva as a diagnostic fluid. Oral Dis. 2002, 8, 69–76. [Google Scholar] [CrossRef]
- Ramseier, C.A.; Kinney, J.S.; Herr, A.E.; Braun, T.; Sugai, J.V.; Shelburne, C.A.; Rayburn, L.A.; Tran, H.M.; Singh, A.K.; Giannobile, W.V. Identification of pathogen and host-response markers correlated with periodontal disease. J. Periodontol. 2009, 80, 436–446. [Google Scholar] [CrossRef]
- Gomez, F.A. The future of microfluidic point-of-care diagnostic devices. Bioanalysis 2013, 5, 1–3. [Google Scholar] [CrossRef]
- Mombelli, A.; Schmid, J.; Walter, C.; Wetzel, A. Qualitätsleitlinie Parodontologie. Swiss Dent. J. 2014, 124, 261–267. [Google Scholar]
- Dige, I.; Schlafer, S.; Nyvad, B. Difference in initial dental biofilm accumulation between night and day. Acta Odontol. Scand. 2012, 70, 441–447. [Google Scholar] [CrossRef] [PubMed]
- Proctor, G.B. The physiology of salivary secretion. Periodontol. 2000 2016, 70, 11–25. [Google Scholar] [CrossRef]
- Navazesh, M.; Kumar, S.K.; University of Southern California School of Dentistry. Measuring salivary flow: Challenges and opportunities. J. Am. Dent. Assoc. 2008, 139, 35S–40S. [Google Scholar] [CrossRef] [PubMed]
- Kaur, M.; Kachlany, S.C. Aggregatibacter actinomycetemcomitans leukotoxin (LtxA.; Leukothera) induces cofilin dephosphorylation and actin depolymerization during killing of malignant monocytes. Microbiology 2014, 160, 2443–2452. [Google Scholar] [CrossRef]
- Vega, B.A.; Belinka, B.A.; Kachlany, S.C. Aggregatibacter actinomycetemcomitans Leukotoxin (LtxA.; Leukothera®): Mechanisms of Action and Therapeutic Applications. Toxins 2019, 11, 489. [Google Scholar] [CrossRef] [PubMed]
- Holt, S.C.; Ebersole, J.L. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: The “red complex”, a prototype polybacterial pathogenic consortium in periodontitis. Periodontology 2000 2005, 38, 72–122. [Google Scholar] [CrossRef]
- Dewhirst, F.E.; Chen, T.; Izard, J.; Paster, B.J.; Tanner, A.C.; Yu, W.H.; Lakshmanan, A.; Wade, W.G. The human oral microbiome. J. Bacteriol. 2010, 192, 5002–5017. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Deng, J.; Jiang, X. Advances in Reagents Storage and Release in Self-Contained Point-of-Care Devices. Adv. Mater. Technol. 2019, 4, 1800625. [Google Scholar] [CrossRef]
- Stumpf, F.; Schwemmer, F.; Hutzenlaub, T.; Baumann, D.; Strohmeier, O.; Dingemanns, G.; Simons, G.; Sager, C.; Plobner, L.; von Stetten, F.; et al. LabDisk with complete reagent prestorage for sample-to-answer nucleic acid based detection of respiratory pathogens verified with influenza A H3N2 virus. Lab Chip 2016, 16, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Luu-The, V.; Paquet, N.; Calvo, E.; Cumps, J. Improved real-time RT-PCR method for high-throughput measurements using second derivative calculation and double correction. Biotechniques 2005, 38, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Socransky, S.S.; Haffajee, A.D. Periodontal microbial ecology. Periodontology 2000 2005, 38, 135–187. [Google Scholar] [CrossRef] [PubMed]
- Belibasakis, G.N.; Schmidlin, P.R.; Sahrmann, P. Molecular microbiological evaluation of subgingival biofilm sampling by paper point and curette. APMIS 2014, 122, 347–352. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2015. [Google Scholar]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Pohlert, T. The Pairwise Multiple Comparison of Mean Ranks Package (PMCMR). R Package (2014). Available online: http://CRAN.R-project.org/package=PMCMR (accessed on 15 April 2020).
- Ammann, T.W.; Bostanci, N.; Belibasakis, G.N.; Thurnheer, T. Validation of a quantitative real-time PCR assay and comparison with fluorescence microscopy and selective agar plate counting for species-specific quantification of an in vitro subgingival biofilm model. J. Periodontal Res. 2013, 48, 517–526. [Google Scholar] [CrossRef]
- Asikainen, S.; Doğan, B.; Turgut, Z.; Paster, B.J.; Bodur, A.; Oscarsson, J. Specified species in gingival crevicular fluid predict bacterial diversity. PLoS ONE 2010, 5, e13589. [Google Scholar] [CrossRef]
- Salminen, A.; Kopra, K.A.; Hyvärinen, K.; Paju, S.; Mäntylä, P.; Buhlin, K.; Nieminen, M.S.; Sinisalo, J.; Pussinen, P.J. Quantitative PCR analysis of salivary pathogen burden in periodontitis. Front. Cell. Infect. Microbiol. 2015, 5, 69. [Google Scholar] [CrossRef]
- Choi, H.; Kim, E.; Kang, J.; Kim, H.J.; Lee, J.Y.; Choi, J.; Joo, J.Y. Real-time PCR quantification of 9 periodontal pathogens in saliva samples from periodontally healthy Korean young adults. J. Periodontal. Implant. Sci. 2018, 48, 261–271. [Google Scholar] [CrossRef]
- Mitsakakis, K.; Stumpf, F.; Strohmeier, O.; Klein, V.; Mark, D.; Von Stetten, F.; Peham, J.R.; Herz, C.; Tawakoli, P.N.; Wegehaupt, F.; et al. Chair/bedside diagnosis of oral and respiratory tract infections, and identification of antibiotic resistances for personalised monitoring and treatment. Stud. Health Technol. Inform. 2016, 224, 61–66. [Google Scholar]
- Yi, H.; Schumann, P.; Sohn, K.; Chun, J. Serinicoccus marinus gen. nov., sp. nov., a novel actinomycete with L-ornithine and L-serine in the peptidoglycan. Int. J. Syst. Evol. Microbiol. 2004, 54, 1585–1589. [Google Scholar] [CrossRef]
- Hoceini, A.; Klouche Khelil, N.; Ben-Yelles, I.; Mesli, A.; Ziouani, S.; Ghellai, L.; Aissaoui, N.; Arab, M. Caries-related factors and bacterial composition of supragingival plaques in caries free and caries active Algerian adults. Asian Pac. J. Trop. Biomed. 2016, 6, 720–726. [Google Scholar] [CrossRef]
- Khan, S.A.; Kong, E.F.; Meiller, T.F.; Jabra-Rizk, M.A. Periodontal Diseases: Bug Induced, Host Promoted. PLoS Pathog. 2015, 11, e1004952. [Google Scholar] [CrossRef] [PubMed]
- Yadaf, K.; Prakash, S. Dental Caries: A Microbial Approach. J. Clin. Infect. Dis. Pract. 2017, 2. [Google Scholar] [CrossRef]
- Ito, T.; Yasuda, M.; Kaneko, H.; Sasaki, H.; Kato, T.; Yajima, Y. Clinical evaluation of salivary periodontal pathogen levels by real-time polymerase chain reaction in patients before dental implant treatment. Clin. Oral Implant. Res. 2014, 25, 977–982. [Google Scholar] [CrossRef]
- Nørskov-Lauritsen, N.; Claesson, R.; Birkeholm Jensen, A.; Åberg, C.H.; Haubek, D. Aggregatibacter Actinomycetemcomitans: Clinical Significance of a Pathobiont Subjected to Ample Changes in Classification and Nomenclature. Pathogens 2019, 8, 243. [Google Scholar] [CrossRef]
- Claesson, R.; Höglund-Åberg, C.; Haubek, D.; Johansson, A. Age-related prevalence and characteristics of Aggregatibacter actinomycetemcomitans in periodontitis patients living in Sweden. J. Oral Microbiol. 2017, 9, 1334504. [Google Scholar] [CrossRef]
- Gross, E.L.; Beall, C.J.; Kutsch, S.R.; Firestone, N.D.; Leys, E.J.; Griffen, A.L. Beyond Streptococcus mutans: Dental caries onset linked to multiple species by 16S rRNA community analysis. PLoS ONE 2012, 7, e47722. [Google Scholar] [CrossRef]
- Preshaw, P.M. Detection and diagnosis of periodontal conditions amenable to prevention. BMC Oral Health 2015, 15 (Suppl. 1), S5. [Google Scholar] [CrossRef]

| Organism | DNA Sequence (5′-3′) | Strand on Template | Modification at 5′ | Modification at 3′ | Purification | Reference Material |
|---|---|---|---|---|---|---|
| Porphyromonas gingivalis | TTGCTAAGGTTGATGGCGAC | + | Desalted | DSMZ 20709 | ||
| ACAAGTGTATGCGGTTTTAGTCC | - | Desalted | ||||
| CGCGTATGCAACTTGCCTTA | + | 6-FAM | BHQ-1 | HPLC | ||
| Treponema denticola | ATTGGGACTGAGATACGGCC | + | Desalted | DSMZ 14222 | ||
| AGAAGCATTCCCTCTTCTTCTT | - | Desalted | ||||
| CCGTGTGAATGAAGAAGGCC | + | 6-FAM | BHQ-1 | HPLC | ||
| Tannerella forsythia | TGTACCTTGTGAATAAGCATCGG | + | Desalted | ATCC 43037 | ||
| CGGACTTAACAGCCCACCTA | - | Desalted | ||||
| CGGTAATACGGAGGATGCGA | + | 6-FAM | BHQ-1 | HPLC | ||
| Fusobacterium nucleatum | AACTTCGATTTGGGTGGCG | + | Desalted | DSMZ 15643 | ||
| AGCTTTCATAATTCTAGGATGCCC | - | Desalted | ||||
| CCTCACAGCTAGGGACAACA | + | 6-FAM | BHQ-1 | HPLC | ||
| Prevotella intermedia | GAACTGGCGGACTTGAGTG | + | Desalted | DSMZ 20706 | ||
| AGTAACACTCCCGTACGCTG | - | Desalted | ||||
| CGGAATTCATGGTGTAGCGG | + | 6-FAM | BHQ-1 | HPLC | ||
| Campylobacter rectus | AGCAAATCTATAAAATACGTCCCAGT | + | Desalted | DSMZ 3260 | ||
| CCGGTTTGGTATTTGGGCTTC | - | Desalted | ||||
| TACGTTCCCGGGTCTTGTAC | + | 6-FAM | BHQ-1 | HPLC | ||
| Aggregatibacter actinomycetemcomitans | GCTGATACTGCAACGAAAGC | + | Desalted | DSMZ 8324 | ||
| CAAGCATTCTCGCACGATCA | - | Desalted | ||||
| CGGGGCTTTCTACTACGGGA | + | 6-FAM | BHQ-1 | HPLC | ||
| Streptococcus mutans | CCAGAAAGGGACGGCTAACT | + | Desalted | DSMZ 20523 | ||
| GCCTTTTACTCCAGACTTTCCT | - | Desalted | ||||
| TATTGGGCGTAAAGGGAGCG | + | 6-FAM | BHQ-1 | HPLC | ||
| Streptococcus sobrinus | CCAAAATTCCGCAGAGTCGC | + | Desalted | DSMZ 20742 | ||
| CCTTCAAAGCACCAGGGACA | - | Desalted | ||||
| TGCAGGTCAAACAACGGATTCC | + | 6-FAM | BHQ-1 | HPLC | ||
| Oral Lactobacilli | GTGCAGAAGAGGASAGTGGA | + | Desalted | DSMZ 4905 | ||
| ATCCTGTTCGCTACCCATGC | - | Desalted | ||||
| ATGGAAGAACACCAGTGGCG | + | 6-FAM | BHQ-1 | HPLC | ||
| Serinicoccus marinus | CGCAGAGATGTGGTTTCCCT | + | Desalted | DSMZ 15273 | ||
| TCCCATGAGTCCCCAACCA | - | Desalted | ||||
| AGCGCAACCCTCGTTCCATGT | + | LightCycler Red 610 | BHQ-1 | HPLC |
| Healthy | Caries | Periodontitis | p-Value | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Panel | Strain | Median | IQR | Median | IQR | Median | IQR | Healthy-Caries | Healthy-Periodontitis | Caries-Periodontitis |
| POC compatible qPCR Assay | P. gingivalis | 193,889 | 537,489 | 334,152 | 1,007,053 | 948,848 | 2,519,962 | 0.232 | <0.001 *** | 0.064 |
| T. forsythia | 244,480 | 404,683 | 360,430 | 806,900 | 635,674 | 839,909 | 0.053 | <0.001 *** | 0.154 | |
| T. denticola | 11,025 | 9815 | 36,482 | 44,387 | 30,683 | 51,061 | 0.007 ** | 0.011 * | 0.638 | |
| F. nucleatum | 239,745 | 316,713 | 252,436 | 321,205 | 366,993 | 541,370 | 0.319 | 0.004 ** | 0.081 | |
| C. rectus | 29,311 | 64,630 | 41,116 | 60,922 | 82,206 | 195,432 | 0.068 | <0.001 *** | 0.059 | |
| P. intermedia | 83,001 | 192,885 | 146,802 | 376,529 | 185,607 | 989,823 | 0.107 | 0.012 * | 0.472 | |
| A. actinomycetemcomitans | 49,966 | 35,481 | 20,173 | 67,154 | 37,932 | 28,284 | 1.000 | 1.000 | 1.000 | |
| S. mutans | 22,218 | 72,897 | 194,821 | 1,305,829 | 68,350 | 871,276 | <0.001 *** | 0.004 ** | 0.004 ** | |
| S. sobrinus | 55,417 | - | 231,463 | 380,989 | 185,138 | 569,481 | 0.510 | 0.680 | 0.680 | |
| Genus Lactobacilli | 2,478,206 | 6,090,513 | 11,086,039 | 38,167,335 | 11,247,675 | 53,562,778 | 0.065 | 0.021* | 0.894 | |
| iai PadoTest | A. actinomycetemcomitans | - | - | - | - | - | - | - | - | - |
| T. forsythia | 87,500 | 150,000 | 450,000 | 175,000 | 300,000 | 525,000 | 0.140 | 0.050 | 0.510 | |
| P. gingivalis | 50,000 | 25,000 | 25,000 | 12,500 | 75,000 | 112,500 | 0.235 | 0.011 * | 0.011 * | |
| T. denticola | 125,000 | 150,000 | 100,000 | 93,750 | 200,000 | 362,500 | 0.432 | 0.130 | 0.016 * | |
| P. intermedia | 162,500 | 556,250 | 125,000 | 287,500 | 275,000 | 206,250 | 0.820 | 0.820 | 0.400 | |
| F. alocis | 75,000 | 150,000 | 75,000 | 118,750 | 100,000 | 275,000 | 1.000 | 1.000 | 1.000 | |
| Universal Primer | 671,500,000 | 1,136,250,000 | 873,000,000 | 1,317,000,000 | 744,000,000 | 1,055,500,000 | 0.350 | 0.720 | 0.720 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paqué, P.N.; Herz, C.; Jenzer, J.S.; Wiedemeier, D.B.; Attin, T.; Bostanci, N.; Belibasakis, G.N.; Bao, K.; Körner, P.; Fritz, T.; et al. Microbial Analysis of Saliva to Identify Oral Diseases Using a Point-of-Care Compatible qPCR Assay. J. Clin. Med. 2020, 9, 2945. https://doi.org/10.3390/jcm9092945
Paqué PN, Herz C, Jenzer JS, Wiedemeier DB, Attin T, Bostanci N, Belibasakis GN, Bao K, Körner P, Fritz T, et al. Microbial Analysis of Saliva to Identify Oral Diseases Using a Point-of-Care Compatible qPCR Assay. Journal of Clinical Medicine. 2020; 9(9):2945. https://doi.org/10.3390/jcm9092945
Chicago/Turabian StylePaqué, Pune N., Christopher Herz, Joël S. Jenzer, Daniel B. Wiedemeier, Thomas Attin, Nagihan Bostanci, Georgios N. Belibasakis, Kai Bao, Philipp Körner, Tanja Fritz, and et al. 2020. "Microbial Analysis of Saliva to Identify Oral Diseases Using a Point-of-Care Compatible qPCR Assay" Journal of Clinical Medicine 9, no. 9: 2945. https://doi.org/10.3390/jcm9092945
APA StylePaqué, P. N., Herz, C., Jenzer, J. S., Wiedemeier, D. B., Attin, T., Bostanci, N., Belibasakis, G. N., Bao, K., Körner, P., Fritz, T., Prinz, J., Schmidlin, P. R., Thurnheer, T., Wegehaupt, F. J., Mitsakakis, K., & Peham, J. R. (2020). Microbial Analysis of Saliva to Identify Oral Diseases Using a Point-of-Care Compatible qPCR Assay. Journal of Clinical Medicine, 9(9), 2945. https://doi.org/10.3390/jcm9092945










