Abstract
Acute myeloid leukemia (AML) is a highly heterogeneous hematopoietic malignancy characterized by excessive proliferation and accumulation of immature myeloid blasts in the bone marrow. AML has a very poor 5-year survival rate of just 16% in the UK; hence, more efficacious, tolerable, and targeted therapy is required. Persistent leukemia stem cell (LSC) populations underlie patient relapse and development of resistance to therapy. Identification of critical oncogenic signaling pathways in AML LSC may provide new avenues for novel therapeutic strategies. The phosphatidylinositol-3-kinase (PI3K)/Akt and the mammalian target of rapamycin (mTOR) signaling pathway, is often hyperactivated in AML, required to sustain the oncogenic potential of LSCs. Growing evidence suggests that targeting key components of this pathway may represent an effective treatment to kill AML LSCs. Despite this, accruing significant body of scientific knowledge, PI3K/Akt/mTOR inhibitors have not translated into clinical practice. In this article, we review the laboratory-based evidence of the critical role of PI3K/Akt/mTOR pathway in AML, and outcomes from current clinical studies using PI3K/Akt/mTOR inhibitors. Based on these results, we discuss the putative mechanisms of resistance to PI3K/Akt/mTOR inhibition, offering rationale for potential candidate combination therapies incorporating PI3K/Akt/mTOR inhibitors for precision medicine in AML.
Keywords:
AML; LSC; PI3K/Akt; mTOR; targeted therapy; drug resistance; combination treatment strategy 1. Introduction
Acute myeloid leukemia (AML) is a highly heterogeneous hematopoietic malignancy, characterized by excessive proliferation and accumulation of immature myeloid blasts in the bone marrow []. While reductions of bulk malignant cells can be achieved in the majority of patients by standard chemotherapy consisting of cell cycle active drugs, such as cytarabine and anthracyclines, approximately two-thirds of patients relapse after the induction therapy, highlighting an unmet need for a more targeted therapeutic approach []. A rare population of therapy-resistant cells are believed to be the origin of relapse, termed leukemia stem cells (LSCs), also referred to as leukemia-initiating cells (LICs) [,,]. These cells acquire enhanced self-renewal capacity and exhibit a block in differentiation. Thus, eliminating LSCs may prevent relapse, ultimately improving the currently poor outcome of AML. Designing LSC-targeted therapies is, however, challenging as LSCs share similar characteristics with normal hematopoietic stem cells (HSCs), making it difficult to kill LSCs whilst sparing HSCs [,]. In AML, dysregulated pathways affect the cellular functions of LSCs, which may represent putative targets to overcome resistance to therapy in AML [,].
The phosphatidylinositol-3-kinase (PI3K)/Akt and the mammalian target of rapamycin (mTOR) signaling pathway emerges as a promising therapeutic candidate to sensitize LSCs to chemotherapy. It plays an important role in both normal and malignant hematopoiesis; components of this pathway govern the expression of genes and proteins essential for cell proliferation, differentiation, and survival. Constitutive activation of PI3K/Akt/mTOR pathway is detected in 50–80% of AML patients, associated with decreased overall survival (OS) [,,]. Mutations in receptor tyrosine kinases (RTKs) or GTPases are the major causes leading to upregulation of the PI3K/Akt/mTOR pathway in AML []. One important mechanism leading to deregulation of PI3K/Akt/mTOR signaling is mutation of fms-like tyrosine kinase 3 (FLT3). Among them, internal tandem duplication (ITD) of FLT3 gene (FLT3-ITD) is one of the most frequent mutations in normal karyotype AML (approximately 25%). In recent clinical studies, few patients display prolonged remissions with RTK inhibitors, such as FLT3 inhibitors, highlighting the need for novel and/or partner targeted therapies [,]. Targeting the PI3K/Akt/mTOR pathway may be an option for FLT3-ITD AML patients.
Hyperactivation of PI3K/Akt/mTOR has also been associated with attenuated sensitivity to chemotherapy. Several studies have demonstrated that PI3K/Akt/mTOR inhibition may preferentially target LSCs. For example, the PI3K/Akt/mTOR pathway may regulate LSC survival through nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB). The pro-inflammatory transcription factor NF-κB, has been found to be aberrantly activated in LSCs but is not expressed in normal human CD34+ progenitor cells [,,]. NF-κB is known to mediate chemoresistance by upregulation of anti-apoptotic genes, which enable cells to increase proliferation and evade apoptosis [,,]. Targeting NF-κB may be selective for LSCs and/or sensitize LSCs to chemotherapy. Notably, NF-κB is a downstream target of PI3K/Akt/mTOR, and this signaling cascade can trigger NF-κB activation, which suggests a common survival pathway for LSCs. Treatment of AML patient samples with PI3K inhibitor LY294002 displayed inhibited Akt phosphorylation and NF-κB DNA-binding activity []. Furthermore, PI3K/Akt/mTOR inhibition induced apoptosis in primary AML cells and potentiated response to cytotoxic chemotherapy, while sparing normal HSC function, [,,,]. The LSC population was targeted by PI3K-directed therapies demonstrated by reduced engraftment ability of these cells in nonobese diabetic/severe combined immunodeficiency (NOD/SCID) mice [,]. PI3K/Akt/mTOR inhibitors may additionally potentiate LSC kill by synergizing with LSC-directed therapies. An essential feature of quiescent AML-LSCs is that they have relative lower production of reactive oxygen species (ROS) compared with bulk cells []. These ROS-low LSCs were shown to aberrantly overexpress Bcl-2, making them more susceptible to eradication by small-molecule Bcl-2 inhibitors like venetoclax. The therapeutic potential of venetoclax could be enhanced by PI3K/Akt/mTOR inhibition through Mcl-1-dependent mechanisms, which is a well-known determinant of resistance to venetoclax [].
Growing evidence signposts PI3K as a druggable target for AML; indeed, there has been very productive development of small-molecule inhibitors targeting the PI3K/Akt/mTOR pathway. While PI3K/Akt/mTOR inhibitors have been effective treating other hematological malignancies, such as chronic lymphoblastic leukemia (CLL) and follicular lymphoma (FL), in AML, the clinical potential of PI3K/Akt/mTOR inhibitors has not yet been fully elucidated [,,]. Clinical studies using PI3K/Akt/mTOR inhibitors as monotherapy have shown limited therapeutic efficacy, likely due to compensatory activation of other survival pathways []. Therefore, dissecting these relevant clinical findings and understanding how other signaling pathways impinge on PI3K/Akt/mTOR signaling pathway activity may provide us new clues as to how to effectively inhibit this pathway with potential candidate combination strategies to eradicate LSCs and so cure AML.
2. The PI3K/Akt/mTOR Signaling Pathway
2.1. Regulation of the PI3K/Akt/mTOR Pathway in Normal Hematopoiesis
The PI3K family consists of three distinct classes of PI3Ks (I-III), of which class I is implicated in regulation of hematopoiesis. Class I PI3K can be further divided into class IA and class IB enzymes, both of which are activated by cell surface receptors. Class IA PI3K can be activated by RTKs, G protein-coupled receptors (GPCRs), and oncoproteins such as the small G protein Ras, whereas class IB PI3K can be activated by GPCRs only [,]. Class IA PI3Ks form heterodimers between one of three catalytic subunits (p110α, p110β, or p110δ) and a regulatory adaptor molecule (p85α (or its splice variants p55α and p50α), p85β or p55γ) [,]. Each pair shares some overlap whilst maintaining distinct function. In contrast to the heterogeneity of class IA, a single class IB isoform has been described that associates catalytic subunit p110γ with regulatory adaptor molecule p101 or p84 [,]. While catalytic subunits p110α and p110β are consistently expressed in a broad range of tissues, p110γ and p110δ are specifically enriched within the hematopoietic system—preferentially in leukocytes [].
In response to extracellular stimuli (e.g., hormones, growth factors, and cytokines) and the subsequent activation of RTKs, class IA PI3K is recruited to the plasma membrane via interaction of p85 with adaptor proteins, such as insulin receptor substrate (IRS) 1/2 or growth factor receptor-bound protein 2-associated binding protein 2 (GAB2) that bind to the regulatory p85 subunit of PI3K [,]. The class IB p110γ is activated by GPCRs through direct interaction of its regulatory adaptor molecule with Gβγ subunit of trimeric G proteins []. The activated p110 catalytic subunit facilitates the phosphorylation of phosphatidylinositol-4,5-phosphate (PIP2) to generate phosphatidylinositol-3,4,5-phosphate (PIP3). PIP3 recruits phosphoinositide-dependent kinase 1 (PDK1) and Akt/protein kinase B (PKB) to the plasma membrane where PDK1 phosphorylates Akt at Threonine(T)308 residue within the activation loop of the kinase domain to initiate the activation of Akt [,] (Figure 1).
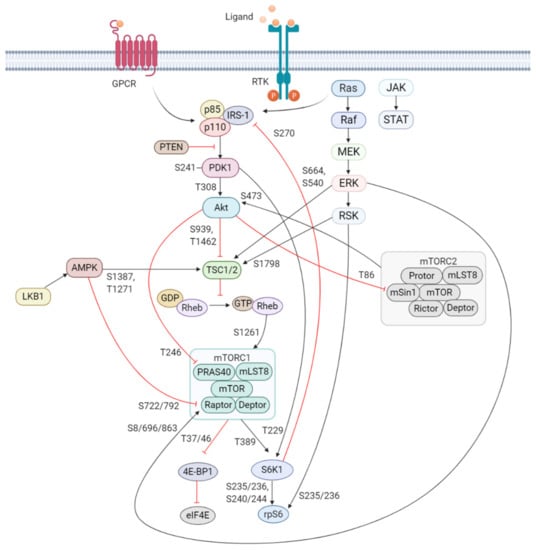
Figure 1.
Schematic overview of the activation and regulation of the PI3K/Akt/mTOR signaling pathway. Activation of PI3K is stimulated by binding of an extracellular ligand (e.g., hormones, growth factors, and cytokines) to a cell surface receptor such as the receptor tyrosine kinase (RTK) in the plasma membrane. Activated RTK recruits adaptor proteins, which bind to the regulatory p85 subunit of PI3K and subsequently activate the catalytic subunits for full PI3K activation. PI3K is also activated by G protein-coupled receptors (GPCR) or small GTPase Ras, which bind PI3K directly. Activated PI3K catalyzes the phosphorylation of phosphatidylinositol-4,5-phosphate (PIP2) to generate phosphatidylinositol-3,4,5-phosphate (PIP3). PIP3 recruits phosphoinositide-dependent kinase 1 (PDK1) and Akt to the plasma membrane inducing Akt phosphorylation by PDK1 at T308. Akt activation is completed by phosphorylation at S473 by mTOR complex 2 (mTORC2). The mTOR complex includes two distinct protein complexes, mTORC1 and mTORC2. mTORC1 comprises of mTOR, proline-rich Akt substrate 40 kDa (PRAS40), regulatory-associated protein of mTOR (Raptor), mammalian lethal with Sec13 protein 8 (mLST8, also known as GβL), and DEP-domain-containing mTOR-interacting protein (Deptor) []. mTORC2 comprises of mTOR, mLST8, Deptor, protein observed with Rictor-1 (Protor), rapamycin-insensitive companion of mTOR (Rictor), and mammalian stress-activated protein kinase interacting protein (mSin1) []. Akt indirectly activates mTORC1 by phosphorylation and inhibition of tuberous sclerosis complex 2 (TSC2) at S939 and T1462, releasing the inhibitory effects of this complex on Ras-related GTPase Rheb, an activator of mTORC1. Akt also directly controls activation of mTORC1 in a TSC2-independent manner via phosphorylation of PRAS40 at T246. The extracellular signal-regulated kinase (ERK)/90 kDa ribosomal S6 kinase (RSK) and liver kinase B1/AMP-activated protein kinase (LKB1/AMPK) signaling pathways impinge on several nodes of the PI3K/Akt/mTOR pathway and can modulate mTORC1 activity. Both ERK and RSK modulate mTORC1 activity by phosphorylation of TSC2 at S664 and S540 (ERK) and S1798 (RSK). ERK1/2 can also control mTORC1 activation by phosphorylation of Raptor at S8, S696, and S863. Master metabolic regulator AMPK inhibits mTORC1 activity in two different pathways, the first by phosphorylation of TSC2 at T1271 and S1387 and the second by phosphorylation of Raptor at S722 and S792. Activated mTORC1 promotes cap-dependent mRNA translation via phosphorylation of eukaryotic translation initiation factor 4E (eIF4E)-binding protein 1 (4E-BP1) at T37 and T46, which is a priming event required for subsequent phosphorylation of several carboxy-terminal serum-sensitive sites to release 4E-BP1 from eIF4E. Ribosomal protein S6 kinase beta-1 (S6K1) is a downstream target of mTORC1, activated by phosphorylation at T389 by mTORC1 as well as T229 phosphorylation mediated by PDK1. S6K1 in turn activates ribosomal protein S6 (rpS6), which is dispensable for cell growth and protein synthesis. RSK can also directly activate rpS6 via phosphorylation at S235 and S236. The black arrows represent positive regulation (activation), whereas the red blunt-ended lines indicate negative regulation (inhibition). IRS-1 = insulin receptor substrate 1, PTEN = phosphatase and tensin homolog, GDP = guanosine diphosphate, GTP = guanosine triphosphate, JAK = Janus kinase, STAT = signal transducer and activator of transcription. Created with BioRender.com.
Akt is a highly conserved serine/threonine kinase that has multiple diverse functions. Full Akt activation, in addition to PDK1-mediated phosphorylation, requires phosphorylation at Serine(S)473 residue in the regulatory domain, by mTOR complex 2 (mTORC2), integrin-linked kinase (ILK), PDK1 or members of the PI3K-related kinase (PIKK) family, such as DNA-dependent protein kinase (DNA-PK) [,]. Notably, Akt can activate mTORC2 through a positive feedback loop by direct phosphorylation of mTORC2 component mammalian stress-activated protein kinase interacting protein (mSin1) at T86 [,]. Activated Akt can phosphorylate a wide spectrum of protein substrates, including forkhead box class O (FoxO), glycogen synthase kinase-3 (GSK3) α/β, and Bcl-2 associated agonist of cell death (BAD), maintaining cell cycling, survival, metabolism, cell growth and other essential cellular functions (Figure 2) [].
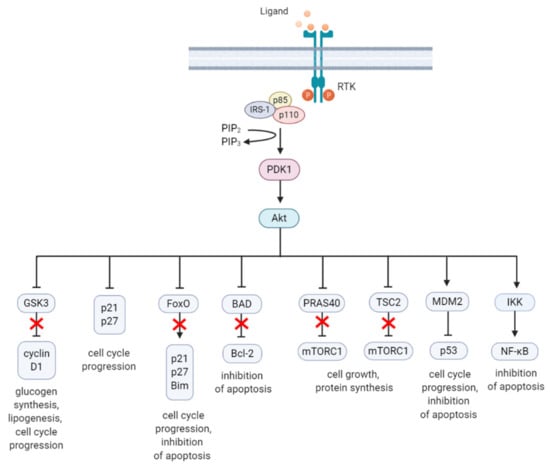
Figure 2.
A summary diagram of Akt downstream target molecules. (From left to right) Fully activated Akt controls numerous effectors implicated in cell growth, proliferation, differentiation, metabolism, and survival, of which some are highlighted. Akt regulates G1/S cell cycle progression by phosphorylation and inactivation of GSK3/cyclin D1, p21, and p27. Akt was found to regulate cell metabolism by mediating lipogenesis and glucose uptake through phosphorylation and inhibition of GSK3, which inhibits glycogen synthesis. Akt controls apoptosis by phosphorylation and inhibition of FoxO and pro-apoptotic Bcl-2 family member BAD. Akt promotes cell growth by activation of mTORC1 though phosphorylation of PRAS40, which prevents its inhibition of mTORC1. Akt can also induce mTORC1 activation through phosphorylation and inhibition of TSC2, relieving the inhibitory effects of the TSC1/TSC2 complex on mTORC1. Akt enhances MDM2-mediated ubiquitination and proteasomal-dependent degradation of p53. Akt can inhibit apoptosis and promote cell survival by activating NF-κB. The arrows represent positive regulation (induction/activation), whereas the blunt-ended lines indicate negative regulation (inhibition/inactivation). The red cross represents inhibition caused by Akt-mediated negative regulation. RTK = receptor tyrosine kinase, IRS-1 = insulin receptor substrate 1, PIP2 = phosphatidylinositol-4,5-phosphate, PIP3 = phosphatidylinositol-3,4,5-phosphate, PDK1 = phosphoinositide-dependent kinase-1, MDM2 = mouse double minute 2 homolog, GSK3 = glycogen synthase kinase 3, TSC2 = tuberous sclerosis complex 2, mTORC1 = mTOR complex 1, FoxO = forkhead box O, Bim = Bcl-2-like protein 11, BAD = BCL-2 associated agonist of cell death, Bcl-2 = B-cell lymphoma 2, IKK = IκB kinase, NF-κB = nuclear factor kappa-light-chain-enhancer of activated B cells. Created with BioRender.com.
Regulation of PI3K activity is negatively controlled by the tumor suppressor phosphatase and tensin homolog (PTEN) and Src homology domain-containing inositol phosphatase (SHIP) to maintain normal hematopoiesis. PTEN is a lipid phosphatase that hampers PI3K signaling through dephosphorylation of the lipid signaling intermediate PIP3 []. Loss of function of PTEN through mutations, genetic silencing, or epigenetic mechanism is implicated in the pathology of multiple human malignancies and can lead to aberrant PI3K/Akt/mTOR signaling []. SHIP is predominantly expressed in hematopoietic cells and hydrolyzes PIP3 to generate PI(3,4)P2 []. Mutation of the SHIP gene was detected in a low percentage of AML and acute lymphoblastic leukemia (ALL) patients [].
One of the key downstream targets of the Akt is mTOR, which positively regulates cell growth and proliferation by promoting protein synthesis and inhibition of autophagy [,]. The identification of this serine/threonine kinase stems from the discovery of the natural product rapamycin, originally extracted from the soil bacterium Streptomyces hygroscopicus []. mTOR participates in two functionally distinct multiprotein complexes, mTORC1 and mTORC2, of which only mTORC1 is sensitive to inhibition by rapamycin []. Akt indirectly activates mTORC1 by phosphorylation of tuberous sclerosis complex 2 (TSC2) at S939 and T1462 [,]. TSC2 functions with TSC1 forming a heterodimeric complex which blocks the activation of Ras homolog enriched in brain (Rheb). Akt phosphorylation of TSC2 inhibits the GTPase-activating protein (GAP) activity of this complex and in turn permits Rheb to activate mTORC1. Akt can also activate mTORC1 by a TSC2-independent mechanism which involves phosphorylation of proline-rich Akt substrate 40kDa (PRAS40), a component of mTORC1. Phosphorylation of PRAS40 at T246 results in dissociation of PRAS40 from mTORC1 and attenuates the inhibitory effect of PRAS40 on mTORC1 activity []. Upon activation, mTORC1 is phosphorylated on several residues (T2446, S2448, and S2481), but no function has been ascribed to any phosphorylation site [,,]. S1261 was identified as a site-specific phosphorylation site of mTORC1 that in response to insulin signals via the PI3K/TSC2/Rheb axis regulating mTORC1 function in an amino acid-dependent and rapamycin-insensitive mechanism.
The highly conserved protein kinase mTOR is a central hub of nutrient signaling and cell growth and integrates multiple intracellular signals []. With respect to the mTOR signaling pathway, the TSC1/TSC2 complex has emerged as a sensor and integrator of multiple signaling pathways to modulate mTORC1 activity [] (Figure 1). One important signaling pathway that negatively regulates mTORC1 activity is the liver kinase B1 (LKB1)/ AMP-activated protein kinase (AMPK) pathway []. AMPK is a cellular energy sensor activated in various conditions that deplete cellular energy, such as nutrient deprivation or hypoxia [,]. Phosphorylation of AMPK at T172 in the activation loop is required for its kinase activity and is mediated by LKB1, the upstream serine/threonine kinase of AMPK []. AMPK inhibits mTORC1 in two different ways, i.e., phosphorylation of TSC2 at T1227 and S1345; and phosphorylation of Raptor at S722/792, which is a component of mTORC1 [,]. In addition to AMPK, the extracellular signal-regulated kinase (ERK)/ p90 ribosomal S6 kinase (RSK) pathway also modulates mTORC1 activity. ERK/RSK is one of the other main signaling networks activated in parallel with PI3K by RTKs to control survival, differentiation, proliferation, and metabolism []. Both ERK and RSK promote mTORC1 activity by phosphorylation of TSC2 at ERK S664 and S540, and RSK S1798 [,]. In addition, ERK1/2 contributes to Ras-dependent activation of mTORC1 through phosphorylation of Raptor at S8, S696, and S863 [].
Upon activation, mTORC1 phosphorylates its main downstream targets eukaryotic initiation factor-4E (eIF4E) -binding protein 1 (4E-BP1) and rapamycin-sensitive ribosomal protein S6 kinase beta-1/p70 ribosomal S6 kinase (S6K1), involved in the translation of mRNAs. 4E-BP1 inhibits the initiation of cap-dependent translation by binding and inactivating eIF4E. This binding is reversible, and mTORC1 phosphorylation of 4E-BP1 at T37/46 relieves 4E-BP1 from eIF4E []. Released eIF4E assembles at the 5′ end of mRNA, which facilitates the recruitment of the ribosome and subsequent initiation of translation [].
S6K1 is the other main target of mTORC1 implicated in the regulation of cell growth. Activation of S6K1 requires phosphorylation at T229 and T389, of which T229 is phosphorylated by PDK1 and T389 by mTORC1 [,,]. Activated S6K1 activates 40S ribosomal protein (rp) S6 that represents the most extensively studied substrate of S6K1. rpS6 becomes phosphorylated on several serine residues []. While S6K1 phosphorylates rpS6 on all phosphorylation sites (S235/236 and S240/244), RSK phosphorylates rpS6 exclusively on S235/236 in an mTOR-independent mechanism, suggesting that ERK/RSK pathway contributes to rpS6 phosphorylation upon mitogen stimulation []. mTORC1-stimulated S6K1 mediates an important negative feedback regulation of PI3K through phosphorylation of IRS-1. As such, S6K1 phosphorylates IRS-1 proteins at several serine residues (S270, S307, S636, and S1101) of which S270 was found to be required for S6K1/IRS-1 interaction and subsequent phosphorylation of the other S6K1-specific residues []. Phosphorylation of IRS-1 induces its protein degradation and insulin resistance, thereby inhibiting the insulin-like growth factor 1 (IGF-1) -mediated PI3K activation.
2.2. Constitutive PI3K/Akt/mTOR Activation in AML
Regulated PI3K/Akt/mTOR signaling is critical for normal hematopoiesis, with deregulation of PI3K/Akt/mTOR activity linked to depletion of HSC pool []. For example, Pten deletion in adult mice HSCs activated the PI3K/Akt/mTOR pathway and promoted HSC proliferation and depletion through induced expression of p16Ink4a and p53, and leukemogenesis [,,]. These effects were mostly mediated by mTOR as rapamycin was able to suppress leukemogenesis and restore normal HSC function []. Myristoylated Akt1 (myr-Akt) was introduced into HSCs via retroviral transduction of bone marrow cells and subsequent transplantation, to mimic constitutively active Akt, which is frequently observed in AML []. Results revealed that myr-Akt contributes to myeloproliferative disorders (MPD), and T-cell lymphoma with high frequency, and AML with a lower penetrance. HSCs in the myr-Akt1 mice displayed transient expansion of immature myeloid cells in the bone marrow and spleen, and increased cycling, associated with impaired engraftment. The importance of mTOR signaling as a mediator of Akt was demonstrated with rapamycin. Rapamycin rescued cobblestone formation in myr-Akt–transduced bone marrow cells in vitro and increased survival of myr-Akt mice. TSC1 is also potentially involved in leukemogenesis. Deletion of TSC1 in HSCs leads to activation of mTOR signaling, causes rapid HSC cycling and elevated levels of ROS, and impaired HSC self-renewal []. Importantly, treatment with a ROS antagonist in vivo demonstrated that the TSC1/mTOR axis is important to maintain HSC quiescence and function by suppressing ROS. These findings indicate that mTOR is an important mediator of PI3K regulation in HSCs. Furthermore, FoxO transcription factors are functionally redundant in HSC homeostasis through regulation of HSC response to physiologic oxidative stress, quiescence, and survival [,,]. Mice engineered with conditional knockout alleles of Foxo1, Foxo3, and/or Foxo4 displayed increased cell cycling and apoptosis of HSC, and a marked increase in ROS levels.
The PI3K/Akt/mTOR signaling pathway is frequently hyperactivated in AML cells and potentially contributes to uncontrolled growth, proliferation, differentiation, metabolism, and survival [,]. The PI3K/Akt/mTOR pathway is also important for the regulation of the AML-LSC population, demonstrated in mouse models with genetic alterations of key PI3K/Akt/mTOR signaling genes. Rheb1 is overexpressed in AML patients, which was associated with reduced survival in comparison to patients with lower Rheb1 expression [,,]. Deletion of Rheb1 induced apoptosis and enhanced cell cycle arrest in LSCs, and prolonged survival of MLL-AF9-induced leukemic mice, suggesting that the mTORC1 pathway may be required for LSC maintenance []. PDK1 is overexpressed in over 40% of AML patients and is required for Akt activation []. Deletion of Pdk1 in MLL-AF9-induced mice resulted in a reduction of LSCs and upregulated the expression of apoptosis inducers, such as BAX and Tp53 [,]. Mice transplanted with MLL-AF9-positive LSCs were also shown dependent on S6K1 for LSC maintenance []. Loss of S6K1 improved survival of MLL-AF9-induced leukemic mice, which was associated with reduced Akt and 4E-BP1 phosphorylation. Furthermore, the PI3K/Akt/mTOR pathway is also implemented in the crosstalk between LSCs and the stromal cells associated with its niche that promotes the drug-resistant phenotype of LSCs. Several reports have demonstrated that pharmacological inhibition of PI3K/Akt/mTOR signaling may effectively target leukemic cells within the bone marrow niche [,,].
Dysregulation of the PI3K/Akt/mTOR pathway is often the result of loss or inactivation of tumor suppressors, mutation or amplification of PI3K, as well as activation of RTKs or oncoproteins upstream of PI3K (Figure 3) [,]. About 50–80% of patients with AML display constitutive PI3K/Akt/mTOR activation, and this was associated with significant poorer OS []. Poor prognosis in AML patients with constitutive PI3K/Akt/mTOR signaling could be related to the fact that this pathway is associated with chemoresistance, which contributes to the short-term survival in AML [,,]. However, no correlation was shown to exist between PI3K/Akt/mTOR activity and a particular AML subtype, cytogenetic abnormality, or etiology of the disease [,].
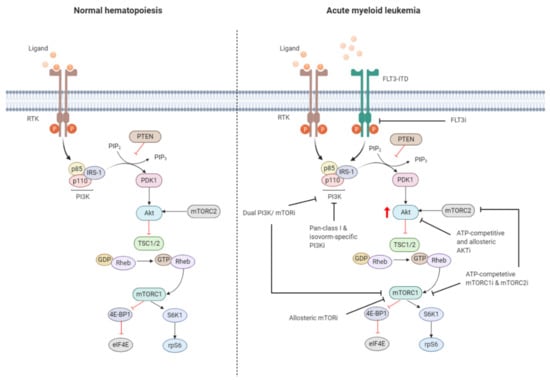
Figure 3.
Targeting the PI3K/Akt/mTOR pathway in AML. The PI3K/Akt/mTOR pathway is commonly dysregulated in AML caused by mutations in membrane-bound proteins such as receptor tyrosine kinases (RTKs) and small GTPase Ras. Activating mutations in fms-like tyrosine kinase 3 (FLT3), such as the FLT3-internal tandem duplication (FLT3-ITD), are an important mechanism leading to dysregulation of PI3K/Akt/mTOR signaling. The ITD mutation causes ligand-independent activation of the FLT3 receptor, leading to constitutive activation of the PI3K/Akt/mTOR pathway. Numerous small-molecule inhibitors of this pathway include FLT3 inhibitors (FLT3i), dual PI3K/mTORi, allosteric mTORi, pan-class I and isoform-specific PI3Ki, ATP-competitive and allosteric Akti, and ATP-competitive mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2) inhibitors (mTORC1i and mTORC2i). The red arrow indicates elevated Akt phosphorylation, whereas the red blunt-ended lines represent negative regulation (inhibition). IRS-1 = insulin receptor substrate 1, PIP2 = phosphatidylinositol-4,5-phosphate, PIP3 = phosphatidylinositol-3,4,5-phosphate, PTEN = phosphatase and tensin homolog, PDK1 = phosphoinositide-dependent kinase-1, TSC 1/2 = tuberous sclerosis complex 1/2, ATP = adenosine triphosphate, GDP = guanosine diphosphate, GTP = guanosine triphosphate, Rheb = Ras homolog enriched in brain, 4E-BP1 = eukaryotic translation initiation factor 4E (eIF4E)-binding protein 1, S6K1 = ribosomal protein S6 kinase beta-1, rpS6 = ribosomal protein S6. The black blunt-ended lines indicate the main targets for therapeutic intervention. Created with BioRender.com.
Exploring mechanisms of constitutive PI3K/Akt/mTOR activation in AML identified mutations of RTKs (e.g., FLT3-ITD, c-KIT) or GTPases (KRAS, NRAS) as well as autocrine IGF-1/IGF-1R signaling responsible for dysregulation [,]. Aberrant PI3K/Akt/mTOR signaling activation is often associated with enhanced Akt phosphorylation, mediated by phosphorylation at S473 by PDK1 and T308 by mTORC2. The OS of AML patients presenting with Akt phosphorylation at these sites was found to be significantly shorter in several studies [,]. Furthermore, mTORC1 is activated in most AML patients, indicated by enhanced phosphorylation of its main downstream substrates 4E-BP1, S6K1, and rpS6 [,]. However, activation of mTORC1 and its downstream target may also occur independently of PI3K/Akt though parallel signaling pathways [,,]. It is therefore important to dissect how PI3K/Akt/mTOR signaling converges with other signaling pathways, which may have clinical implications for selection of drugs targeting different signaling molecules.
About 30% of AML patients with normal karyotype present with an activating FLT3 receptor mutation, most often as FLT3-ITD, and is the major intercessory of PI3K/Akt/mTOR pathway dysregulation. FLT3 is a member of the class III RTK family and is important for the maintenance of hematopoietic homeostasis [,,]. FLT3-ITD exhibits ligand-independent constitutive tyrosine kinase activity and activates signaling pathways including PI3K/Akt/mTOR []. However, regulatory p85 subunit of PI3K does not bind to the FLT3 receptor, nor is it tyrosine phosphorylated after FLT3 ligand stimulation. Instead, p85 associated with SH2 domain-containing protein tyrosine phosphatase-2 (SHP-2) and SHIP, in murine Ba/F3 cells stably transfected with human FLT3-ITD []. FLT3-ITD expression in Ba/F3 cells was associated with constitutive activation of Akt and concomitant phosphorylation of FoxO3a [,]. FoxO3a has an important role in apoptosis and cell cycle regulation []. FLT3-ITD was shown to constitutively activate Akt, and concomitantly phosphorylate FoxO3a, suppressing the expression of FoxO3a target genes encoding for p27 and pro-apoptotic Bcl-2 family member, Bim [,]. FLT3-ITD negatively regulates FoxO3a, thereby suppressing FoxO3a-mediated apoptosis and bypassing the G1 cell cycle blockade.
2.3. Targeting the PI3K/Akt/mTOR Signaling Pathway in AML
Preclinical evidence underlines the significant role of PI3K/Akt/mTOR signaling in leukemia initiation and maintenance. There is considerable interest targeting PI3K/Akt/mTOR signaling for AML treatment, which has resulted into the rapid development of small molecule compounds that target either a single or multiple kinase (Figure 3). PI3K-Targeting molecules can be divided into isoform-specific PI3K inhibitors and ATP-competitive pan-PI3K inhibitors. The PI3K p110δ catalytic subunit is consistently expressed at a high level in AML blasts, making it an attractive therapeutic target for AML []. Idelalisib (also referred to as CAL-101), for example, is a p110δ inhibitor that is currently under Phase 3 clinical investigation for the treatment of B-cell malignancies []. Treatment of AML cells with idelalisib inhibited ribosomal RNA (rRNA) synthesis and cell proliferation by suppressing Akt phosphorylation with a greater effect observed in cells expressing higher levels of p110δ []. Pan-PI3K inhibitors target all isoforms of PI3K and may exert broader anti-leukemic effects but at the expense of higher toxicity. mTOR inhibitors include both ATP-pocket and allosteric mTOR binding drugs, e.g., rapalogs, such as everolimus and temsirolimus. Both drugs derive from the natural macrolide rapamycin and act by associating with immunophillin FK506 binding protein 12 (FKBP12), which in turn binds and inhibits mTORC1, although after lengthy exposure they inhibit also mTORC2 []. mTOR inhibitors represent the first class of PI3K/Akt/mTOR-directed therapies and yielded promising anti-proliferative effects without inhibition of normal CD34+ cells in preclinical settings []. The anti-leukemic effects were associated with reduced phosphorylation of S6K1 and 4E-BP1 and could be enhanced in combination with conventional cytotoxic drugs []. An important drawback of inhibiting mTORC1 is the increased phosphorylation of Akt. Dual PI3K and mTOR inhibitors block both the upstream and downstream targets of Akt, thereby circumventing the increased PI3K and Akt signaling subsequent to mTORC1 inhibition []. Dual PI3K/mTOR inhibitor dactolisib efficiently blocked PI3K and mTORC1 signaling and mTORC2 activity []. Furthermore, dactolisib inhibited protein translation in AML cells, reducing cell growth and inducing apoptosis without affecting survival of normal CD34+ cells. A small number of Akt inhibitors have been developed, but they are rarely evaluated in preclinical or clinical settings as the development of Akt inhibitors has long been hampered by high structural similarity of the Akt catalytic domain to that of other kinases of the AGC kinase family (named after the representative protein kinase A, G, and C families) [,].
3. Crosstalk of the PI3K/Akt/mTOR Signaling Pathway with Other Signaling Pathways in AML
We have demonstrated the complexity of the PI3K/Akt/mTOR signaling pathway resulting from several feedback loops and crosstalk with other signaling pathways that are able to modulate PI3K/Akt/mTOR signaling. In this section we will particularly focus on FLT3-ITD AML which is the major driver of dysregulation of PI3K/Akt/mTOR signaling. FLT3-ITD has been shown to induce, in addition to PI3K/Akt/mTOR, other downstream effectors, including MEK/ERK/RSK1 and Janus kinase (JAK)/signal transducer and activator of transcription (STAT) pathways, FoxO3a phosphorylation, and ROS production [,,]. These effectors interact with the PI3K/Akt/mTOR pathway, elucidation of which may reveal possible mechanisms of resistance to PI3K-directed therapies. Recent studies have stressed that FLT3-ITD induces genomic instability through ROS production, which potentially contributes to chemoresistance leading to disease relapse [,]. The PI3K/Akt/mTOR signaling pathway is linked to both ROS production and the DNA damage response (DDR) pathway, strengthening the premise of further exploiting this pathway as a potential therapeutic target [,,].
The Ras/Raf/MEK/ERK and PI3K/Akt/mTOR signaling pathways strongly regulate each other and share overlapping downstream effectors to regulate multiple cellular functions. Several mechanisms of integration of these two pathways have been identified, including negative feedback loops, cross-inhibition, cross-activation, and pathway convergence, which have been described in the previous section []. Briefly, the Ras/ERK pathway cross-activates PI3K/Akt/mTOR signaling by regulating PI3K, TSC2, and mTORC1. Ras-GTP can directly bind and allosterically activate PI3K []. Activated Ras/ERK pathway can positively regulate mTORC1 activity indirectly through phosphorylation of TSC2 or directly through PI3K-independent phosphorylation of proline-directed residues within Raptor [,,]. Furthermore, Ras/ERK and PI3K/Akt/mTOR pathways co-regulate downstream effectors, including FoxO and c-Myc transcription factors, BAD, and GSK3 [,,,,].
The JAK/STAT pathway is a core oncogenic signaling pathway activated by a plethora of cytokines and hormones and has a critical role in hematopoietic malignancies []. Deregulation of the JAK/STAT pathway, in particular STAT5, is frequently reported in AML and is indispensable for leukemia cell maintenance and survival [,]. Persistent STAT5 activity is driven by oncogenic FLT3-ITD independent of JAKs and is associated with resistance to PI3K/Akt/mTOR inhibitors [,,]. This resistance mechanism involved the increased expression of PIM kinases. PIM kinases are oncogenic serine/threonine kinases overexpressed in several malignancies that promote cellular functions, including cell cycle progression, proliferation, survival, and drug resistance [,]. PIM-1 is transcriptionally upregulated by constitutively active FLT3 and contributes to FLT3-ITD-mediated survival []. Furthermore, PIM-1 phosphorylates and stabilizes FLT3, promoting FLT3 signaling in a positive feedback loop []. It has been reported that PIM kinases are expressed downstream of STAT5 activation in FLT3-ITD AML and are implicated in the regulation of mTORC1 to promote cap-dependent translation in parallel with the PI3K/Akt pathway [,]. PIM and Akt share common substrates of the apoptosis machinery and cellular metabolism which may account for PIM kinases’ ability to act as a resistance mechanism []. For instance, it has been reported that PIM-1 elevates RTKs upon inhibition of Akt, leading to drug resistance [,]. Upregulation of PIM-1 mediated by FLT3-ITD enhanced survival through protection of the mTOR/4E-BP1/myeloid cell leukemia 1 (Mcl-1) pathway, which was abrogated by inhibition of the STAT5/PIM kinase axis [,,,,].
Both PI3K/Akt and STAT5 signaling converge to control ROS production, which is induced by FLT3-ITD. ROS are a heterogeneous group of molecules and radicals primarily produced in the electron transport chain (ETC) of the mitochondria, in particular from Complex I and III [,]. ROS have a critical role in the regulation of normal hematopoiesis and is commonly elevated in a wide range of human cancers, including AML [,]. Activating mutations of FLT3 has been shown to increase ROS production, causing DNA damage and affecting the DDR. The DNA damage is repaired by error-prone alternative non-homologous end-joining (alt-NHEJ) pathway, which promotes genomic instability and leukemogenesis []. Elevated ROS in FLT3-ITD AML is attributable to constitutive activation of nicotinamide adenine dinucleotide phosphate oxidases (NOX). [,,]. Another proposed mechanism of FLT3-ITD-driven ROS production involves interaction of STAT5 with RAC1, a small GTPase protein, which is an essential component of NADPH oxidase [,]. Interestingly, ROS can increase phosphorylation of STAT5 via JAK, which acts as a feed-forward loop []. It was demonstrated that inhibition of FLT3-ITD not only decreased tyrosine phosphorylation of STAT5 but also reduced RAC1 activity and its binding to NOX [].
Aberrant PI3K/Akt/mTOR signaling drives ROS production through different molecular mechanisms, including the activation of NOXs [,,]. Wortmannin (inhibitor for PI3Ks and PI3K-related enzymes) treatment or Akt1 knockout impaired translocation of NOX subunits and reduced production of ROS [,]. The PI3K/Akt/mTOR pathway is also involved in the regulation of the gene expression of endogenous antioxidant synthesis. ROS production is tightly regulated by the redox-sensitive signaling system Kelch-like ECH-associated protein 1 (Keap1)/nuclear factor erythroid 2-related factor 2 (Nrf2)/antioxidant response elements (ARE). Under physiological conditions, Nrf2 is sequestered in the cytosol via its association with Keap1. When ROS levels increase, the Nrf2 dissociates from Keap1 and enters the nucleus where it forms a heterodimer with a small musculoaponeurotic fibrosarcoma (MAF) protein to activate ARE-dependent gene expression of antioxidative proteins [,]. The PI3K/Akt/mTOR signaling pathway contributes to Nrf2-mediated regulation of antioxidative proteins as the nuclear translocation of Nrf2 requires PI3K activation []. PI3K inhibition using wortmannin and LY294002 was shown to inhibit Nrf2 activity []. As FLT3-ITD induces phosphorylation of FoxO3a (which requires the presence of Akt phosphorylation sites), it seems apparent that FLT3-ITD promotes ROS production at least in part through dysregulation of the PI3K/Akt/mTOR pathway [,]. Paradoxically, excessive ROS production can promote activation of PI3K/Akt/mTOR signaling through oxidative inhibition of PTEN, causing an increase in cellular PIP3 levels and activation of Akt [,,].
4. Clinical Implications of PI3K/Akt/mTOR Inhibitors in AML
There is considerable interest in small-molecule drugs targeting the PI3K/Akt/mTOR pathway, of which some have been approved by the United States Food and Drug Administration (FDA) for several human cancers (Table 1). However, despite extensive pre-clinical and clinical research of these drugs either as a monotherapy or in conjunction with conventional cytotoxic chemotherapeutics, they have not yet been successfully translated into clinical practice for the treatment of AML. Here, we review PI3K/Akt/mTOR inhibitors subjected to clinical evaluation for AML therapy, summarized in Table 1 and Table 2. The response criteria for AML are summarized in Table 3.

Table 1.
PI3K and mTOR inhibitors approved by FDA for human cancers.

Table 2.
Phase 1/2 clinical trials using PI3K/Akt/mTOR inhibitors.

Table 3.
Response criteria in AML.
4.1. Buparlisib
Buparlisib (BKM120; NVP-BKM120) is a highly selective pan-PI3K inhibitor that has shown promising activity against solid tumors as well as lymphoid malignancies. Buparlisib binds to all class IA p110 catalytic subunits and inhibits PI3K in an ATP-competitive manner [,]. Several reports have demonstrated that buparlisib inhibits cell proliferation, diminishes metabolic activity, and exerts cytotoxic effects on solid tumors and hematological malignancies, including AML, by selective inhibition of Akt activity [,,,]. The mechanism by which buparlisib impaired viability was associated with the induction of p21-mediated G2/M cell cycle arrest and reduced expression of NF-κB anti-apoptotic proteins []. In AML, buparlisib showed in vivo effectiveness in, and prolonged survival of, xenotransplant mouse models []. In addition, it has been demonstrated that combination with other small-molecule drugs, such as the glycolitic modulator dichloroacetate (DCA), proteasome inhibitor bortezomib, and c-Myc inhibitor 10058-F4, profoundly enhanced the cytotoxic effect of buparlisib in AML cell lines and/or primary samples [,].
Buparlisib was clinically evaluated by Ragon et al. in an open label, non-randomized Phase 1 dose escalation study in patients with relapsed/refractory AML (n = 12), ALL (n = 1) or mixed phenotype acute leukemia (MPAL) (n = 1) []. In the AML population, cytogenetic analysis detected diploid karyotype (n = 4), inversion 3 or 3q26 (n = 3), and other cytogenic alterations (n = 5). Molecular analysis revealed isocitrate dehydrogenase (IDH) and RAS mutation (n = 2), FLT3 D835 mutation (n = 1), NRAS mutation (n = 1), KRAS and NRAS mutation (n = 1), tumor protein 53 (TP53) mutation (n = 1), and nucleophosmin 1 (NPM1), NRAS, IDH1, DNA methyltransferase 3A (DNMT3A) mutation (n = 1). Three patients experienced clinically significant confusion, supported by earlier studies reporting that buparlisib crosses the blood-brain barrier and can cause neurotoxicity. Protein profiling by Western blot revealed that buparlisib was able to block PI3K/Akt/mTOR signaling, observed by decreased p-S6K1 and p-FoxO3a, and reverse phase protein array (RPPA) analysis showed significant downregulation of PRAS40. However, direct inhibition of PI3K led to Akt induction, pointing towards a compensatory feedback mechanism. These findings confirm that buparlisib can effectively inhibit and target the downstream PI3K/Akt/mTOR pathway, but such biological suppression did not translate into an improved clinical response in this patient population. Overall, only one patient (with inversion 3) had stable disease receiving maximum tolerated dose of BKM120 for a period of 82 days. Notably, three patients (21.4%) with 3q26 abnormalities had the longest OS. As patients carrying 3q26 gene rearrangements present up-regulation of PI3K/Akt/mTOR signaling and here displayed significantly improved OS, patients with 3q26 aberrations may be more sensitive to pan PI3K inhibitors.
4.2. Gedatolisib
Gedatolisib (PF-05212384; PKI-587) is a highly potent, selective and ATP-competitive dual inhibitor of PI3Kα, PIK3γ and mTOR for clinical development []. In vivo, gedatolisib has demonstrated antitumor activity in solid tumor xenograft models and has been clinically evaluated in patients with advanced cancers [,,,,]. Gedatolisib was able to inhibit cell viability in solid tumor models and effectively targeted and inhibited PI3K/Akt/mTOR signaling measured by decreased p-Akt and p-rpS6 expression [,]. However, many cells were resistant following gedatolisib treatment, which was attributed to over-activation of the MEK/ERK pathway through suppression of mTORC2 [,].
The clinical activity and tolerance of gedatolisib for AML therapy was evaluated in an open, prospective, single arm, multicentric Phase 2 study published by Vargaftig et al. []. Twelve patients were enrolled of which 10 were retained for analysis. All patients were adults diagnosed with relapsed AML (first relapse (n = 5), second relapse (n = 5)) and symptomatic uncontrolled AML. One patient had therapy-related AML (prostate adenocarcinoma). Overall, only one patient completed the treatment, and this study was terminated as no objective response was detected in any of the patients. Transcriptomic analysis revealed that gedatolisib treatment weakly affected gene expression pattern, of which four genes found to be upregulated were associated with natural killer (NK) cell immunity. This suggests that gedatolisib might affect immune cells. Current research is focusing on understanding mechanisms of resistance to gedatolisib in solid tumors.
4.3. Idelalisib
Idelalisib (CAL-101; GS-1101) is a highly selective inhibitor of PI3Kδ that has entered clinical trials for the treatment of hematologic malignancies. In a Phase 1 clinical trial, idelalisib was evaluated in relapsed/refractory CLL, non-Hodgkin lymphoma (NHL), AML, and multiple myeloma (MM). The results of this clinical study has been presented at several conferences and has been published in several reports [,,,]. Although it has been demonstrated that idelalisib has an acceptable safety profile and rapidly induced durable disease responses in CLL, NHL, and mantle cell lymphoma (MCL), the outcomes in AML have not yet been published. In one report, the interim results of the study were discussed in which seven AML patients were included []. The report suggests that idelalisib was not effective in AML.
4.4. Dactolisib
Dactolisib (BEZ235; NVP-BEZ235) is a dual ATP-competitive pan-class PI3K and mTOR inhibitor that has been clinically evaluated alone or in combination therapy in Phase 1/2 studies in solid tumors and hematological malignancies [,,]. Several reports have demonstrated that dactolisib exerts anti-proliferative and cytotoxic effects in vitro. Dactolisib was also shown to induce autophagy and enhance chemosensitivity [,,]. In AML, dactolisib showed similar biological effects in vitro and inhibited the colony-forming capability of LSCs [,]. Results obtained from clinical investigation of dactolisib for advanced cancers reported frequent dose-limiting toxicities accounting for adverse effects. Several Phase 2 studies reported poor tolerance of dactolisib with only modest clinical activity, which led to treatment discontinuation [,].
A Phase 1 study by Wunderle et al. evaluated the safety and efficacy of BEZ235 among 22 patients with relapsed or refractory AML (n = 11), B-cell precursor ALL (n = 9), T-cell ALL (n = 1), and chronic myeloid leukemia (CML) in myeloid blast phase (n = 9). Clinical responses were observed in four patients (18.2%): complete hematological and molecular remission or hematological improvement in three ALL patients and stable disease of four months in one AML patient. Notably, no activating mutations of PIK3CA, PTEN, or AKT were detected in any of the patients []. Detection of p-Akt, p-S6, and p-4E-BP1 by western blot and flow cytometry did not correlate with response. Similar to previous clinical findings, dactolisib induced adverse effects such as gastrointestinal implications, pneumonia, and sepsis. Due to disease progression, 19 patients (86.4%) were withdrawn from the study []. Together, these data raise the question whether inhibiting both PI3K and mTOR is a viable therapeutic strategy and whether we can do better.
4.5. MK-2206
MK-2206 is a highly specific allosteric pan-Akt inhibitor for all three isoforms of human Akt that has entered clinical trials for solid tumors as well as hematological malignancies. In vitro, MK-2206 induced apoptosis and enhanced the cytotoxicity of chemotherapy []. Although it was reported that MK-2206 effectively suppressed phosphorylation of Akt, MK-2206 monotherapy showed limited clinical activity, due to incomplete target inhibition at tolerable doses, paving the way for combination strategies [,,].
A Phase 2 study by Konopleva et al. evaluated the clinical efficacy and tolerability of MK-2206 in 19 patients, of which 18 were retained for analysis []. All patients were adults with relapsed/refractory AML requiring second salvage therapy. No activating mutations of upstream RTK were detected. In preclinical studies, MK-2206 inhibited cell growth and induced apoptosis in AML cell lines and primary samples. In clinical studies, among 18 evaluable patients, only one patient (5.6%) achieved CR with incomplete platelet count recovery. The remaining 17 patients (94.4%) were withdrawn from the study due to disease progression. Proteomic analysis by RPPA following MK-2206 treatment revealed upregulation of a subset of pro-survival proteins, suggesting that other approaches to block Akt signaling should be exploited for AML.
4.6. Afuresertib
Afuresertib (GSK21110183) is a reversible, ATP-competitive pan-Akt inhibitor that has entered clinical trials, both as monotherapy and combination with other agents, for solid tumors and MM [,,]. In vitro, afuresertib induced apoptosis and G1 phase arrest and effectively suppressed phosphorylation of multiple Akt substrates, including FoxO1, GSK3β, mTOR, and p70. Spencer et al. evaluated the maximum tolerated dose (MTD), safety, pharmacokinetics, and clinical activity of afuresertib in an open-label Phase 1 study for advanced hematological malignancies []. A total of 73 patents were included with MM (n = 34), NHL (n = 13), AML (n = 9), Hodgkin disease (n = 8), CLL (n = 7), ALL (n = 1), and Langerhan’s cell histiocytosis (n = 1). Among the 73 patients, 26 were evaluated for the dose escalation (part 1) and 47 patients in the expansion cohort (part 2). Disease inclusion criteria for part 2 were CLL, CML, ALL, AML, MM, or NHL. Clinical activity was observed primarily in MM patients and in patients with lymphomas. Overall, three patients (9%) achieved partial remission (PR), three patients (9%) achieved minimal response (MR), and some patients achieved prolonged disease stabilization. Among the nine AML patients, treatment failure was observed in five patients, and the other four did not have response assessed. Afuresertib was safe and generally well tolerated, but effects related to the gastrointestinal tract or fatigue were noted, suggesting that Akt inhibition with afuresertib may prove beneficial for the treatment of MM.
4.7. Uprosertib
Uprosertib (GSK2141795) is an ATP-competitive pan-Akt kinase inhibitor that has entered clinical trials alone or in combination with MEK inhibitor trametinib for the treatment of solid tumors and hematologic malignancies [,]. The efficacy and safety of uprosertib in combination with trametinib in patients with RAS-mutated relapsed/refractory AML was evaluated in a single-arm, open label, multi-institution Phase 2 study published by Ragon et al. []. In total, 23 patients were enrolled diagnosed with de novo AML (n = 9) or post-myelodysplastic syndromes (MDS)/myeloproliferative neoplasms (MPN)/chronic myelomonocytic leukemia (CMML) AML (n = 14). Molecular analysis confirmed KRAS mutations (n = 5), NRAS mutations (n = 15), or both NRAS/KRAS (n = 3). Concomitant mutations were identified in eight patients, including FLT3 (n = 1), IDH1/2 (n = 2), runt-related transcription factor 1 (RUNX1) (n = 1), DNMT3A (n = 3), additional sex combs like-1 (ASXL1) (n = 1), NPM1 (n = 1), CCAAT/enhancer-binding protein alpha (CEBPA) (n = 1), and protein tyrosine phosphatase non-receptor type 11 (PTPN11) (n = 2). RPPA and complementary flow cytometry analysis was performed to determine the effect of treatment on activation of PI3K/Akt/mTOR, MAPK, and other signaling pathways. RPPA analysis revealed that, in contrast to preclinical studies, no significant downregulation of PI3K/Akt/mTOR and MAPK signaling was observed following treatment with the drug combination. Phospho-flow analysis on the other hand demonstrated significant inhibition of p-ERK and a trend towards significant inhibition of p-rpS6 in peripheral blood blasts and CD34+ peripheral blood blasts. However, inhibition of MEK and Akt did not yield improved clinical efficacy. Overall, no patient obtained complete remission (CR) or CR with incomplete recovery of platelets (CRp), and the study was discontinued due to disease progression, dose-limiting toxicity (DLT), adverse effects determined unrelated to therapy, and death. Most toxicities were mild and included diarrhea, maculopapular rash, mucositis, and nausea/vomiting. Minor hematologic improvement and unconfirmed minor response was observed in seven patients. Ragon et al. suggest that this dual targeting approach could prove to be an effective treatment strategy in combination with cytotoxic, targeted, or epigenetic therapies to overcome possible compensatory signaling.
4.8. Sirolimus
Sirolimus, also known as rapamycin, is a naturally occurring macrolide compound that interacts with FKBP12. This complex binds to the FKBP rapamycin binding (FRB) domain located in the carboxyl terminus of mTOR and inhibits its kinase activity. In vitro, sirolimus inhibits cancer cell growth, promotes G1 cell cycle arrest, and enhances cytotoxic effects in combination with other agents [,,,]. Sirolimus is clinically often used in renal transplantations and for the treatment of a rare lung disease called lymphangioleiomyomatosis (LAM) but has also been investigated for the treatment of solid tumors and hematological malignancies [,,,,].
Kasner et al. evaluated the clinical potential of sirolimus for AML therapy in two sequential clinical trials in identical patient populations (NCT00780104 and NCT01184898). In total, 52 patients were enrolled, with 51 undergoing clinical evaluation. Among the 51 patients clinically evaluated, the majority had abnormal cytogenetics (56%), and FLT3-ITD was detected in seven patients (14%). The pilot study provided evidence that intracellular flow cytometry can be applied to monitor the biochemical efficacy of drugs targeting the PI3K/Akt/mTOR signaling pathway. Results revealed heterogeneity of rpS6 phosphorylation among particular patient populations, suggesting that evaluation of drug sensitivity ex vivo or in early therapy may prove useful in predicting response to molecular targeted therapies [,]. In the final clinical and pharmacodynamics study, the clinical response of sirolimus in combination with mitoxantrone/etoposide/cytarabine (MEC) chemotherapy in a larger study cohort was evaluated. Overall, 24 patients responded (47%) with 18 CR (35%), one CRp, and five PRs. Sirolimus combination with MEC was tolerable in patients with high risk AML; however, the combination therapy did not enhance clinical responses in AML and was limited to patients presenting baseline mTORC1 activation as measured by baseline rpS6 phosphorylation. [,].
4.9. Everolimus
Everolimus (RAD001), a derivative of rapamycin, is an allosteric inhibitor of mTORC1 showing promise for several cancer types. Everolimus was granted FDA approval for progressive, well-differentiated non-functional, neuroendocrine tumors of gastrointestinal or lung origin with unresectable, locally advanced, or metastatic disease []. Everolimus interacts with FKBP12, affecting downstream effectors of mTORC1 and ultimately inhibiting cell proliferation [,]. However, rapamycin-induced targeting of mTORC1 has not been effective inhibiting PI3K/Akt/mTOR activity, which may relate to a negative feedback loop promoting paradoxical hyperactivation of Akt, insensitivity to rapamycin, and IGF-1-mediated feedback loops [,].
A Phase 1b study by Park et al. evaluated the efficacy and tolerance of everolimus combined with conventional chemotherapy (daunorubicin plus cytarabine) in 28 younger (<65 years old) AML patients at first relapse []. Karyotyping revealed that six patients (21.4%) had poor cytogenetic abnormalities, 19 patients (67.9%) intermediate risk, and three (10.7%) good prognostic cytogenetic indicators. Mutational analysis identified FLT3-ITD or NPM1 mutation in each of two patients and co-existence of FLT3-ITD and NPM mutation in three patients (10.7%). Everolimus strongly inhibited mTORC1 signaling measured by decreased p-p70S6K at T389. Clinical outcomes revealed that 19 patients (67.9%) achieved CR. Notably, three out of four patients with FLT3-ITD achieved CR. Overall, this study showed that a weekly dose of everolimus (70mg) in combination with conventional therapy was tolerable and had acceptable toxicity in younger AML patients at relapse.
Another Phase 1b study by Tiong et al. evaluated the efficacy and tolerability of everolimus in combination with low dose cytarabine (LDAC) in elderly AML []. Twenty-four AML patients diagnosed with de novo AML (n = 15) or secondary AML (n = 9) were included. Mutational analysis by multiplexed mass spectrometry detected mutation of NRAS/KRAS (n = 4), NPM1 (n = 3), IDH1/2 (n = 3), DNMT3A (n = 2), FLT3 D835 (n = 2), MPL (n = 1), and FLT3-ITD (n = 1). Overall response rate (ORR) was achieved in six patients (25%) with four CR or CR with incomplete hematological recovery (CRi) (16.7%), one PR (4.2%), and one morphologic leukemia-free state (4.2%). Furthermore, two patients (8.3%) had >50% reduction in marrow leukemic blasts without blood count recovery. However, there was no clear association between mutational profile and response despite two responding patients carrying a RAS mutation. The combination of everolimus with LDAC neither revealed an efficacy signal nor potentially increased toxicity. The authors suggested combining mTOR-directed therapies with other novel agents for the treatment of AML.
5. Clinical Strategies to Overcome Resistance to PI3K/Akt/mTOR Inhibitors in AML
In the past, PI3K/Akt/mTOR inhibitors have been tried mostly combined with cytotoxic chemotherapeutics to improve efficacy and reduce drug-related toxicity. However, combination with targeted therapies may be more effective and cooperatively inhibit PI3K/Akt/mTOR signaling while preventing escape via alternative pathways []. Several new, targeted agents are currently under clinical investigation for AML therapy, including drugs targeting epigenetic regulators, kinases, monoclonal antibodies, Bcl-2, and metabolism (Figure 4). Here, we will provide the rationale for clinically investigating potential drug combinations using PI3K/Akt/mTOR inhibitors as a novel targeted therapeutic strategy for AML.
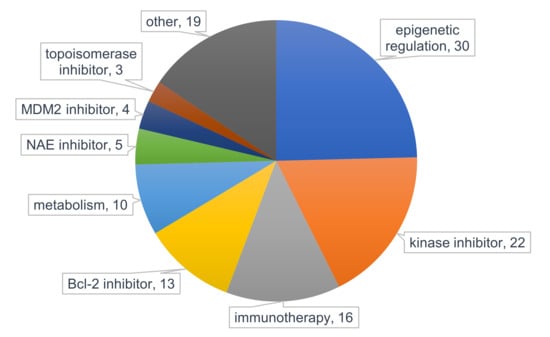
Figure 4.
Pie chart presenting the classification of interventions for AML therapy evaluated in clinical trials. Data was obtained from ClinicalTrials.gov and filtered for “recruiting” and “active, not recruiting” status in adult and elderly AML and study phase (1b-3) with start date from 1 January 2018. Numbers present frequency within 88 studies. * other = frequency <3 include dihydroorotate dehydrogenase (DHODH) inhibitor, proteasome inhibitor, p53 activator, hedgehog signaling inhibitor, CDK inhibitor, E-selectin inhibitor, nuclein export inhibitor, menin-MLL binding interaction inhibitor, CXCR4 antagonist, proteasome inhibitor, corticosteroid, thrombopoietin receptor agonist, and isoprenyl transferase inhibitor. MDM2 = mouse double minute 2 homolog, NAE = NEDD8-activating enzyme, Bcl-2 = B-cell lymphoma 2.
5.1. PI3K/Akt/mTOR Inhibitors in Combination with Epigenetic Targeting
The importance of epigenetics in AML pathology was demonstrated by the identification of recurrent mutations in genes encoding epigenetic regulators, such as DNA methyltransferase 3A (DNMT3A), Tet methylcytosine dioxygenase 2 (TET2), and IDH1/2 []. In addition to mutations in epigenetic regulators, oncogenic fusion proteins can dysregulate epigenetic modifications, driving abnormal transcriptional programs in AML [,]. Several targeted epigenetic therapies have been explored for AML, for example, hypomethylating agents (HMA), histone deacetylase (HDAC) inhibitors, and IDH1/2 inhibitors. However, as monotherapy these inhibitors do not seem to be effective, and treatment failure is common [,,,]. To potentiate the efficacy of epigenetic therapy, combination strategies with cytotoxic chemotherapy or other targeted drugs are currently being extensively studied.
Epigenetic modifiers participate in the PI3K/Akt/mTOR pathway, contributing to the oncogenicity of PI3K in cancer. The cooperation of the PI3K/Akt/mTOR pathway with epigenetic reprogramming involves modulation of DNA methylation and histone methylation to promote transcriptional competence [,]. Because of this collaboration, combination strategies with PI3K/Akt/mTOR inhibitors and epigenetic therapies have been exploited. There is emerging evidence demonstrating synergism when these classes of agents are combined, involving Akt inactivation, FoxO3a-mediated upregulation of pro-apoptotic targets Bim and Puma, and Mcl-1 downregulation [,,,]. PI3K/Akt/mTOR inhibitors may potentiate epigenetic therapies inducing LSC death by enhanced activation of ROS, to damage DNA [,]. In addition to enhanced anti-leukemic effects, there is preclinical evidence suggesting that DNMT or HDAC inhibitors can overcome acquired resistance to PI3K/Akt/mTOR inhibitors [,].
To our knowledge, there are two clinical studies that have evaluated the combination of mTOR inhibitors with a hypomethylating agent, reported by Liesveld et al. and Tan et al. Liesveld et al. evaluated decitabine followed by sirolimus in an open label, single arm, Phase 1 dose escalation study in relapsed/refractory AML []. Decitabine is a DNA-hypomethylating agent that has been reported to markedly improve CR rates compared to cytarabine but failed to significantly improve OS rates [,]. Thirteen patients were subjected to therapy of which 12 were included for safety evaluation. Complex cytogenetics were present in most patients and only one patient carried the FLT3-ITD mutation at diagnosis. Overall, the administration of decitabine followed by sirolimus was well tolerated and among the evaluable patients, but only one patient achieved CR. Four patients (31%) demonstrated disease progression, 5 (38%) had stable blast percentage, and 4 (31%) demonstrated reduced blast percentage after one cycle. Usually multiple cycles are required to achieve clinical response, but this was not possible in many of the patients due to persistent cytopenias and need for more palliative approaches. Furthermore, the response to inhibition of PI3K/Akt/mTOR signaling was inconsistent, reflecting on the heterogeneous nature of AML. More potent PI3K/Akt/mTOR inhibitors may be more effective in these studies.
Tan et al. evaluated the maximum tolerated dose (MTD) and efficacy of everolimus, a more potent mTOR inhibitor than sirolimus, in combination with azacitidine subcutaneously in an open-label, Phase 1b/2 study in relapsed/refractory AML. Forty patients with relapsed (n = 27), primary refractory (n = 11) or elderly patients unfit for intensive chemotherapy (n = 2) were enrolled, of which 37 were retained for evaluation of toxicity []. Similar to Liesveld et al., azacitidine and everolimus were given sequentially to maximize mTOR inhibitory effect and avoid potential mTOR-induced cell cycle inhibition which may attenuate azacitidine activity. Mutational analysis revealed IDH1/2 mutations in 15/35 (43%) and FLT3-ITD mutation in 7/37 (19%) patients. Notably, three patients with relapsed FLT3-ITD had >80% bone marrow blast reductions following sequential therapy, of which two underwent allogeneic stem cell transplantation but subsequently relapsed and died. Overall, the combination of everolimus and azacitidine was well tolerated with an ORR of 22.5% and mean survival of 8.5 months. Collectively, these findings support the clinical investigation of PI3K/Akt/mTOR inhibitors in combination with targeted epigenetic therapies in AML patients.
5.2. PI3K/Akt/mTOR Inhibitors in Combination with Bcl-2 Inhibitors
Bcl-2, encoded by the human BCL2 gene is the founding member of the Bcl-2 family of proteins which are divided into pro-apoptotic and anti-apoptotic members. The anti-apoptotic proteins include Bcl-2, Bcl-XL, Bcl-w, Mcl-1, and A1 that share three or four sequence homologies in the Bcl-2 homology (BH) domain. These proteins can directly interact with pro-apoptotic BH3-only proteins Bim, Puma, Bad, Bid, Bik, Bmf, Hrk, and Noxa, which share homology in the BH3 domain []. Apoptotic stimuli upregulate BH3-only proteins Bax and Bak that lead to permeabilization of the mitochondrial outer membrane and release cytochrome c into the cytosol for activation of caspases that dismantle cellular structures [,,].
Bcl-2 is often overexpressed in AML patients, both at diagnosis as well as at relapse and can render resistance to cytotoxic chemotherapeutics [,]. A major finding by Lagadinou et al. identified high levels of Bcl-2 as a feature of LSCs to maintain mitochondrial integrity for survival []. LSCs were characterized by low levels of ROS and displayed overexpression of Bcl-2. Importantly, it was demonstrated that small molecule Bcl-2 inhibitors reduced oxidative phosphorylation and selectively killed quiescent LSCs. Thus, targeting Bcl-2 may provide a promising therapeutic strategy to effectively target chemoresistant LSC population. BH3 mimetic venetoclax (ABT-199) is a specific inhibitor of Bcl-2 and has been approved by the FDA for the treatment of CLL. Venetoclax binds to Bcl-2 and prevents pro-apoptotic proteins Bak and Bax from binding to Bcl-2. As venetoclax showed potential both in vitro and xenograft models, its efficacy was evaluated for AML [,]. Konopleva et al. evaluated the efficacy of venetoclax as monotherapy in a Phase 2 study in 32 patients with high-risk relapsed/refractory AML or unfit for intensive chemotherapy []. Although venetoclax had acceptable tolerability, the ORR was 19%, of whom 6% achieved CR and 13% achieved CRi. Unfortunately, the clinical response of venetoclax was short-lived as all patients relapsed, suggesting the rapid development of venetoclax resistance. Myoblast dependence on Mcl-1 and Bcl-xL were identified as key players in the intrinsic resistance to venetoclax in AML [,]. In contrast to CLL, which is relatively homogenously dependent on Bcl-2, AML seems more heterogeneous and depends on multiple pro-apoptotic proteins [,]. Significant efforts have been made to overcome this resistance by combining venetoclax with DNA damaging agents such as daunorubicin and cytarabine, which reduce Mcl-1 levels and enhance venetoclax activity [,]. Hypomethylating agent azacitidine was also shown to reduce Mcl-1 protein levels, and synergistically induced apoptosis in AML cells []. The FDA approved venetoclax in combination with HMAs or LDAC for the treatment of AML patients who are previously untreated and older or unfit for chemotherapy [,,].
In addition to cytotoxic chemotherapy and hypomethylating agents, PI3K/Akt/mTOR inhibitors may also overcome venetoclax resistance and potentiate LSC targeting. The combination of PI3K/Akt/mTOR inhibitors with Bcl-2 inhibitors has been examined in several preclinical cancer models [,]. Results demonstrated that PI3K/Akt/mTOR inhibitors downregulate Mcl-1, in part through GSK3 activation, and upregulate Bim, Bad, Bax and Bak. Although PI3K/Akt/mTOR inhibition downregulated Mcl-1, it did not trigger apoptosis, presumably by increased Bim binding to Bcl-2 and Bcl-XL. Co-targeting Bcl-2 released Bim from Bcl-2 and Bcl-xL and induced Bax and Bak-dependent apoptosis in AML LSCs, whilst sparing normal hematopoietic progenitor cells [,,,,]. These data support the rationale for the evaluation of the combination of PI3K/Akt/mTOR inhibitors and Bcl-2 inhibitors in AML. Currently, there is one study exploring the combination of pan-PI3K inhibitor copalisib and venetoclax in relapsed/refractory NHL (NCT03886649) [].
5.3. PI3K/Akt/mTOR Inhibitors in Combination with Kinase Inhibitors
Previously, we discussed other signaling pathways that can integrate with the PI3K/Akt/mTOR pathway, which can potentially reduce the activity of PI3K/Akt/mTOR inhibitors. Parallel pathway inhibition may improve sensitivity to PI3K/Akt/mTOR inhibitors and enhance anti-leukemic effects in AML cells. For example, upregulation of PIM was shown to cause resistance to PI3K/Akt/mTOR inhibitors in AML through a mechanism that involves modulation of mTORC1 activity [,]. While pan-PIM inhibitor AZD1897 showed limited activity as monotherapy in AML cell lines and primary samples, including FLT3-ITD+ cells, combination of AZD1897 with Akt inhibitor AZD5363 displayed synergistic anti-leukemic activity []. This was associated with enhanced inhibition of mTORC1 signaling activity and Mcl-1 levels. Importantly, this combination strategy affected both bulk cells and LSC subsets.
There are two studies in which PIM inhibitors and PI3K inhibitors have been evaluated as combination therapy for hematological malignancies. Novartis Pharmaceuticals evaluated the safety and effectiveness of pan-PIM inhibitor LGH447 and PI3Kα inhibitor BYL719 in patients with relapsed/refractory MM in a Phase 1b/2 study, but the trial never made it to Phase 2 (NCT02144038). Incyte Corporation is currently examining the combination of NCB053914 (pan-PIM inhibitor) with INCB050465 (PI3Kδ inhibitor) in relapsed/refractory diffuse large B-cell lymphoma (DLBCL) in a Phase 1b study (NCT03688152) [].
Co-Targeting the PI3K/Akt/mTOR and MAPK/ERK pathways is another combination strategy that has been evaluated. In AML, the FLT3-ITD mutation is an important mechanism that feeds growth signals into main oncogenic signaling pathways such as PI3K/Akt/mTOR and MAPK/ERK. As crosstalk exists between PI3K/Akt/mTOR and MAPK/ERK pathways, inactivation of both pathways may interrupt oncogenic signals in AML [,]. This may be particularly beneficial in RAS mutated cases. In fact, several reports have shown in vitro enhanced induction of apoptosis when PI3K/Akt/mTOR inhibitors were combined with MAPK/ERK inhibitors []. Inhibition of MEK suppressed p-Erk level but caused feedback activation of the PI3K/Akt/mTOR pathway defined by increased p-mTOR and Mcl-1 levels which could be abrogated by co-targeting PI3K/Akt/mTOR []. The MEK inhibitor also resulted in increased expression of Bim and Bcl-2, which could not be abrogated by adding the PI3K/Akt/mTOR inhibitor. It has been suggested that the increased Bim was bound to Bcl-2, which prevented induction of cell death, and that inhibition of Bcl-2 might further sensitize AML cells to apoptotic cell death by the combination of the MEK and PI3K/Akt/mTOR inhibitors []. Indeed, simultaneous inhibition of PI3K, mTOR, and MAPK/ERK synergistically induced cell death in AML cells, which was further enhanced by the addition of venetoclax, while sparing the normal hematopoietic progenitor cells [,]. This triple-drug combination may potentiate the efficacy of venetoclax to target quiescent LSCs. There is one Phase 1b study that examined the MTD for the combination of PI3Kα inhibitor BYL719 and allosteric MEK1/2 inhibitor MEK162 in patients with advanced solid tumors and in adult patients with AML or high risk MDS carrying RAS or BRAF mutations (NCT01449058). However, the preliminary safety and efficacy findings warrant further exploration as adverse effects were common and stable disease lasting >6 weeks was noted as best response for 31% of patients [].
Furthermore, dual inhibition of FLT3 activation and downstream targeting of PI3K/Akt/mTOR may have synergistic activity in FLT3 mutated cases. Patients with FLT3-ITD tend to have a high risk of relapse and shorter OS compared to patients without the mutation []. In the last few years, several FLT3 tyrosine kinase inhibitors have been developed, which can be divided into first-generation pan-kinase inhibitors such as sorafenib and midostaurin, and next generation inhibitors such as quizartinib and gilteritinib, which are more potent and specific for FLT3 []. Although FLT3 inhibitors were able to induce response in AML patients with FLT3 mutations, these responses were often not durable, and resistance developed rapidly [,]. FLT3 inhibitors are currently evaluated in combination with chemotherapy or hypomethylating agents in various settings, to overcome resistance and prolong OS, respectively [,,]. The PI3K/Akt/mTOR pathway is a promising target to overcome resistance to FLT3 inhibitors. Upregulation of PI3K/Akt/mTOR pathway was shown in FLT3 inhibitor-resistant FLT3-ITD+ AML cells despite inhibition of FLT3 activation by FLT3 inhibitors, suggesting that the resistant cells become FLT3 independent [,]. Inhibition of PI3K/Akt/mTOR in vitro improved sensitivity to FLT3 inhibition and enhanced inhibition of growth and apoptosis. PI3K/Akt/mTOR inhibitors may also potentially suppress FLT3-ITD-induced LSC survival. It has been reported that FLT3-ITD upregulates Mcl-1 to promote survival of AML LSCs, which was abrogated by the tyrosine kinase inhibitor midostaurin (PKC-412) that targets multiple receptors (including FLT3 and c-Kit) []. As PI3K/Akt/mTOR inhibitors can induce inhibition of Mcl-1, combination of PI3K/Akt/mTOR inhibitors with FLT3 inhibitors may potentiate inhibition of LSC survival. Currently, there is one Phase 1 study evaluating the combination of mTOR inhibitor everolimus with midostaurin in patients with relapsed/refractory or poor prognosis AML or MDS (NCT00819546).
5.4. PI3K/Akt/mTOR Inhibitors in Combination with DNA Repair Inhibitors
In a previous section, we briefly highlighted the importance of increased ROS in AML, which can promote genomic instability and contributes to chemotherapy resistance. In particular, there is increasing evidence associating FLT3 mutations with DNA damage, specifically through increased ROS, resulting in double strand breaks (DSBs) and impaired DNA repair [,]. Targeting DNA damage repair mechanisms may provide a novel therapeutic opportunity in AML that may reduce repair errors and genomic instability []. Genomic stability is maintained by the DDR, a highly orchestrated signaling cascade that results from the induction and detection of DNA damage. At the heart of the DDR are members of the PIKK family comprising ataxia telangiectasia mutated (ATM), ataxia telangiectasia and Rad3-related protein (ATR), and DNA-PK, which upon sensing of DNA damage, in particular DSBs, phosphorylate a large number of substrates to effectively repair DNA []. Although DSB repair in normal mammalian cells is primarily dominated by homologous recombination (HR) and DNA-PK-dependent NHEJ, FLT3-ITD directs DSB repair towards an error-prone, alt-NHEJ pathway, which is mediated by poly(ADP-ribose) polymerase-1 (PARP-1) and DNA Lig IIIα [,].
In this regard, PARP inhibitors present a putative therapeutic strategy for AML. PARP1, an important protein of the DDR pathway, senses single strand breaks (SSBs) and has a crucial role for base excision repair (BER). Inhibition of PARP1 leads to accumulation of SSBs which then become DSBs that make cells more dependent on HR [,]. However, in cells with defective HR, such as those with BRCA deficiency, PARP1 inhibition induced synthetic lethality [,]. This concept of synthetic lethality using PARP inhibitors has been effectively examined in BRCA1-defective solid tumors, but was also exploited for the treatment hematological malignancies like AML [,,]. Indeed, several in vitro studies in AML reported that PARP inhibitors induced cell cycle arrest and apoptosis, and combination with chemotherapy may induce synthetic lethality [,,].
There is ongoing research evaluating the combination of PARP inhibitors with other agents to extend the therapeutic potential of these inhibitors beyond tumors with mutations of BRCA or other HR deficiencies. As the PI3K/Akt/mTOR pathway contributes to the DDR, combination of PI3K/Akt/mTOR inhibitors with PARP inhibitors was exploited in several solid tumor models [,]. Inhibition of the PI3K/Akt/mTOR pathway in ovarian, breast, and prostate cancers revealed impaired expression of BRCA1/2 and reduced cellular capacity to conduct HR [,,,]. Consistent with the reduction in the expression of BRCA1/2, PI3K/Akt/mTOR inhibitors sensitized cells to PARP inhibition and synergistically suppressed cell growth. Although these findings support the rationale for the combination of PI3K/Akt/mTOR inhibitors with PARP inhibitors, this combination strategy is yet to be evaluated in AML.
6. Conclusions and Future Directions
The PI3K/Akt/mTOR pathway is central to a broad range of cellular regulatory processes, such as proliferation, differentiation, and survival. Dysregulation of this pathway is common in AML and is often caused by mutations in membrane-bound proteins—in particular FLT3-ITD. Constitutive activation of PI3K/Akt/mTOR induced by such activating mutations is associated with poor OS and chemoresistance. As the PI3K/Akt/mTOR pathway is considered a putative target for AML, significant efforts have been made to develop small-molecule drugs that showed promise in preclinical settings. However, the initial clinical results using PI3K/Akt/mTOR inhibitors as monotherapy have not been successful in clinical practice due to drug-related toxicity, incomplete pathway inhibition, and heterogeneous constitutive activation of the pathway. Therefore, PI3K/Akt/mTOR inhibitors may be more effective in combination with other therapeutic agents. Several clinical studies have exploited the combination of PI3K/Akt/mTOR inhibitors with induction chemotherapy, and although this drug combination strategy was generally well tolerated with acceptable toxicities, only modest improvement of clinical outcome was achieved. Notably, activation of parallel pathways that cooperate with PI3K/Akt/mTOR was reported, and therapeutic targeting of the PI3K/Akt/mTOR pathway was restricted to only a subset of patients. Given the limited clinical effects of PI3K/Akt/mTOR inhibitors as monotherapy or in combination with induction chemotherapy, it appears more promising to evaluate the combination of PI3K/Akt/mTOR inhibitors with other agents, e.g., novel targeted therapies or pathway inhibitors. In that regard, we have discussed how other signaling pathways impinge on the PI3K/Akt/mTOR signaling pathway activity and provided rationale for combining PI3K/Akt/mTOR inhibitors with novel targeted agents that have emerged as potential therapeutic candidates for AML. These combination treatment strategies may effectively inhibit the PI3K/Akt/mTOR pathway and kill AML cells. It is however important to identify the appropriate subgroups of patients that would benefit from such drug combination strategies.
Author Contributions
S.D. and H.A. wrote the manuscript. G.H., H.G.J., S.M., and X.H. conceived the framework, provided guidance, reviewed and edited the draft. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by AIRCRAFT [2016/19186]; the Wellcome Trust [105614/Z/14/Z]; Leukaemia UK [2016/JGF/0005]; the Howat Foundation and Friends of Paul O’Gorman. S.D. is a recipient of University of Modena and Reggio Emilia PhD studentship.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Saultz, J.N.; Garzon, R. Acute Myeloid Leukemia: A Concise Review. J. Clin. Med. 2016, 5, 33. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, M.; Wang, F.; Loghavi, S.; Bueso-Ramos, C.; Gumbs, C.; Little, L.; Song, X.; Zhang, J.; Kadia, T.; Borthakur, G.; et al. Late relapse in acute myeloid leukemia (AML): Clonal evolution or therapy-related leukemia? Blood Cancer J. 2019, 9, 7. [Google Scholar] [CrossRef]
- Hanekamp, D.; Cloos, J.; Schuurhuis, G.J. Leukemic stem cells: Identification and clinical application. Int. J. Hematol. 2017, 105, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, D.; Dick, J.E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997, 3, 730–737. [Google Scholar] [CrossRef] [PubMed]
- Lapidot, T.; Sirard, C.; Vormoor, J.; Murdoch, B.; Hoang, T.; Caceres-Cortes, J.; Minden, M.; Paterson, B.; Caligiuri, M.A.; Dick, J.E. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature 1994, 367, 645–648. [Google Scholar] [CrossRef]
- Riether, C.; Schürch, C.M.; Ochsenbein, A.F. Regulation of hematopoietic and leukemic stem cells by the immune system. Cell Death Differ. 2015, 22, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Tabe, Y.; Konopleva, M. Advances in understanding the leukaemia microenvironment. Br. J. Haematol. 2014, 164, 767–778. [Google Scholar] [CrossRef]
- Zhou, H.-S.; Carter, B.Z.; Andreeff, M. Bone marrow niche-mediated survival of leukemia stem cells in acute myeloid leukemia: Yin and Yang. Cancer Biol. Med. 2016, 13, 248–259. [Google Scholar] [CrossRef]
- Wang, X.; Huang, S.; Chen, J.-L. Understanding of leukemic stem cells and their clinical implications. Mol. Cancer 2017, 16, 2. [Google Scholar] [CrossRef]
- Park, S.; Chapuis, N.; Tamburini, J.; Bardet, V.; Cornillet-Lefebvre, P.; Willems, L.; Green, A.; Mayeux, P.; Lacombe, C.; Bouscary, D. Role of the PI3K/AKT and mTOR signaling pathways in acute myeloid leukemia. Haematologica 2010, 95, 819–828. [Google Scholar] [CrossRef]
- Martelli, A.M.; Evangelisti, C.; Chiarini, F.; Grimaldi, C.; Cappellini, A.; Ognibene, A.; McCubrey, J.A. The emerging role of the phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin signaling network in normal myelopoiesis and leukemogenesis. Biochim. Biophys. Acta 2010, 1803, 991–1002. [Google Scholar] [CrossRef] [PubMed]
- Bertacchini, J.; Guida, M.; Accordi, B.; Mediani, L.; Martelli, A.M.; Barozzi, P.; Petricoin, E.; Liotta, L.; Milani, G.; Giordan, M.; et al. Feedbacks and adaptive capabilities of the PI3K/Akt/mTOR axis in acute myeloid leukemia revealed by pathway selective inhibition and phosphoproteome analysis. Leukemia 2014, 28, 2197–2205. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.-A.; Tien, H.-F. Genomic landscape in acute myeloid leukemia and its implications in risk classification and targeted therapies. J. Biomed. Sci. 2020, 27. [Google Scholar] [CrossRef] [PubMed]
- Daver, N.; Schlenk, R.F.; Russell, N.H.; Levis, M.J. Targeting FLT3 mutations in AML: Review of current knowledge and evidence. Leukemia 2019, 33, 299–312. [Google Scholar] [CrossRef]
- Zhou, F.; Ge, Z.; Chen, B. Quizartinib (AC220): A promising option for acute myeloid leukemia. Drug Des. Dev. Ther. 2019, 13, 1117–1125. [Google Scholar] [CrossRef]
- Guzman, M.L.; Neering, S.J.; Upchurch, D.; Grimes, B.; Howard, D.S.; Rizzieri, D.A.; Luger, S.M.; Jordan, C.T. Nuclear factor-kappaB is constitutively activated in primitive human acute myelogenous leukemia cells. Blood 2001, 98, 2301–2307. [Google Scholar] [CrossRef]
- Guzman, M.L.; Swiderski, C.F.; Howard, D.S.; Grimes, B.A.; Rossi, R.M.; Szilvassy, S.J.; Jordan, C.T. Preferential induction of apoptosis for primary human leukemic stem cells. Proc. Natl. Acad. Sci. USA 2002, 99, 16220–16225. [Google Scholar] [CrossRef]
- Zhou, J.; Ching, Y.Q.; Chng, W.-J. Aberrant nuclear factor-kappa B activity in acute myeloid leukemia: From molecular pathogenesis to therapeutic target. Oncotarget 2015, 6, 5490–5500. [Google Scholar] [CrossRef]
- Li, F.; Sethi, G. Targeting transcription factor NF-kappaB to overcome chemoresistance and radioresistance in cancer therapy. Biochim. Biophys. Acta 2010, 1805, 167–180. [Google Scholar] [CrossRef]
- Jacamo, R.; Chen, Y.; Wang, Z.; Ma, W.; Zhang, M.; Spaeth, E.L.; Wang, Y.; Battula, V.L.; Mak, P.Y.; Schallmoser, K.; et al. Reciprocal leukemia-stroma VCAM-1/VLA-4-dependent activation of NF-κB mediates chemoresistance. Blood 2014, 123, 2691–2702. [Google Scholar] [CrossRef]
- Zhou, J.; Chooi, J.-Y.; Ching, Y.Q.; Quah, J.Y.; Toh, S.H.-M.; Ng, Y.; Tan, T.Z.; Chng, W.-J. NF-κB promotes the stem-like properties of leukemia cells by activation of LIN28B. World J. Stem Cells 2018, 10, 34–42. [Google Scholar] [CrossRef]
- Birkenkamp, K.U.; Geugien, M.; Schepers, H.; Westra, J.; Lemmink, H.H.; Vellenga, E. Constitutive NF-kappaB DNA-binding activity in AML is frequently mediated by a Ras/PI3-K/PKB-dependent pathway. Leukemia 2004, 18, 103–112. [Google Scholar] [CrossRef]
- Xu, Q.; Simpson, S.-E.; Scialla, T.J.; Bagg, A.; Carroll, M. Survival of acute myeloid leukemia cells requires PI3 kinase activation. Blood 2003, 102, 972–980. [Google Scholar] [CrossRef]
- Xu, Q.; Thompson, J.E.; Carroll, M. mTOR regulates cell survival after etoposide treatment in primary AML cells. Blood 2005, 106, 4261–4268. [Google Scholar] [CrossRef] [PubMed]
- Bertacchini, J.; Frasson, C.; Chiarini, F.; D’Avella, D.; Accordi, B.; Anselmi, L.; Barozzi, P.; Forghieri, F.; Luppi, M.; Martelli, A.M.; et al. Dual inhibition of PI3K/mTOR signaling in chemoresistant AML primary cells. Adv. Biol. Regul. 2018, 68, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Grandage, V.L.; Gale, R.E.; Linch, D.C.; Khwaja, A. PI3-kinase/Akt is constitutively active in primary acute myeloid leukaemia cells and regulates survival and chemoresistance via NF-kappaB, Mapkinase and p53 pathways. Leukemia 2005, 19, 586–594. [Google Scholar] [CrossRef] [PubMed]
- Lagadinou, E.D.; Sach, A.; Callahan, K.; Rossi, R.M.; Neering, S.J.; Minhajuddin, M.; Ashton, J.M.; Pei, S.; Grose, V.; O’Dwyer, K.M.; et al. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell 2013, 12, 329–341. [Google Scholar] [CrossRef] [PubMed]
- Vachhani, P.; Bose, P.; Rahmani, M.; Grant, S. Rational combination of dual PI3K/mTOR blockade and Bcl-2/-xL inhibition in AML. Physiol. Genom. 2014, 46, 448–456. [Google Scholar] [CrossRef]
- Markham, A. Copanlisib: First Global Approval. Drugs 2017, 77, 2057–2062. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, D.A.; Sagrillo, F.S.; Fraga, C.A.M. Duvelisib: A 2018 Novel FDA-Approved Small Molecule Inhibiting Phosphoinositide. Pharmaceuticals 2019, 12, 69. [Google Scholar] [CrossRef]
- Miller, B.W.; Przepiorka, D.; de Claro, R.A.; Lee, K.; Nie, L.; Simpson, N.; Gudi, R.; Saber, H.; Shord, S.; Bullock, J.; et al. FDA approval: Idelalisib monotherapy for the treatment of patients with follicular lymphoma and small lymphocytic lymphoma. Clin. Cancer Res. 2015, 21, 1525–1529. [Google Scholar] [CrossRef] [PubMed]
- Nepstad, I.; Hatfield, K.J.; Gronningsaeter, I.S.; Reikvam, H. The PI3K-Akt-mTOR Signaling Pathway in Human Acute Myeloid Leukemia (AML) Cells. Int. J. Mol. Sci. 2020, 21, 2907. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Cheng, H.; Roberts, T.M.; Zhao, J.J. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat. Rev. Drug Discov. 2009, 8, 627–644. [Google Scholar] [CrossRef]
- Fruman, D.A.; Rommel, C. PI3K and cancer: Lessons, challenges and opportunities. Nat. Rev. Drug Discov. 2014, 13, 140–156. [Google Scholar] [CrossRef]
- Vanhaesebroeck, B.; Guillermet-Guibert, J.; Graupera, M.; Bilanges, B. The emerging mechanisms of isoform-specific PI3K signalling. Nat. Rev. Mol. Cell Biol. 2010, 11, 329–341. [Google Scholar] [CrossRef] [PubMed]
- Vadas, O.; Burke, J.E.; Zhang, X.; Berndt, A.; Williams, R.L. Structural basis for activation and inhibition of class I phosphoinositide 3-kinases. Sci. Signal. 2011, 4, re2. [Google Scholar] [CrossRef] [PubMed]
- Suire, S.; Coadwell, J.; Ferguson, G.J.; Davidson, K.; Hawkins, P.; Stephens, L. p84, a new Gbetagamma-activated regulatory subunit of the type IB phosphoinositide 3-kinase p110gamma. Curr. Biol. 2005, 15, 566–570. [Google Scholar] [CrossRef]
- Brock, C.; Schaefer, M.; Reusch, H.P.; Czupalla, C.; Michalke, M.; Spicher, K.; Schultz, G.; Nürnberg, B. Roles of Gβγ in membrane recruitment and activation of p110γ/p101 phosphoinositide 3-kinase γ. J. Cell Biol. 2003, 160, 89–99. [Google Scholar] [CrossRef]
- Kok, K.; Nock, G.E.; Verrall, E.A.G.; Mitchell, M.P.; Hommes, D.W.; Peppelenbosch, M.P.; Vanhaesebroeck, B. Regulation of p110delta PI 3-kinase gene expression. PLoS ONE 2009, 4, e5145. [Google Scholar] [CrossRef]
- Hemmings, B.A.; Restuccia, D.F. PI3K-PKB/Akt pathway. Cold Spring Harb. Perspect. Biol. 2012, 4, a011189. [Google Scholar] [CrossRef]
- Kiyatkin, A.; Aksamitiene, E.; Markevich, N.I.; Borisov, N.M.; Hoek, J.B.; Kholodenko, B.N. Scaffolding protein Grb2-associated binder 1 sustains epidermal growth factor-induced mitogenic and survival signaling by multiple positive feedback loops. J. Biol. Chem. 2006, 281, 19925–19938. [Google Scholar] [CrossRef] [PubMed]
- Alessi, D.R.; Andjelkovic, M.; Caudwell, B.; Cron, P.; Morrice, N.; Cohen, P.; Hemmings, B.A. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 1996, 15, 6541–6551. [Google Scholar] [CrossRef]
- Toker, A.; Marmiroli, S. Signaling Specificity in the Akt Pathway in Biology and Disease. Adv. Biol. Regul. 2014, 55, 28–38. [Google Scholar] [CrossRef]
- Laplante, M.; Sabatini, D.M. mTOR signaling at a glance. J. Cell Sci. 2009, 122, 3589–3594. [Google Scholar] [CrossRef]
- Pearce, L.R.; Huang, X.; Boudeau, J.; Pawłowski, R.; Wullschleger, S.; Deak, M.; Ibrahim, A.F.M.; Gourlay, R.; Magnuson, M.A.; Alessi, D.R. Identification of Protor as a novel Rictor-binding component of mTOR complex-2. Biochem. J. 2007, 405, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Turner, K.M.; Alfred Yung, W.K.; Chen, K.; Zhang, W. Role of AKT signaling in DNA repair and clinical response to cancer therapy. Neuro-oncology 2014, 16, 1313–1323. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Hung, M.-C. Physiological regulation of Akt activity and stability. Am. J. Transl. Res. 2010, 2, 19–42. [Google Scholar]
- Humphrey, S.J.; Yang, G.; Yang, P.; Fazakerley, D.J.; Stöckli, J.; Yang, J.Y.; James, D.E. Dynamic adipocyte phosphoproteome reveals that Akt directly regulates mTORC2. Cell Metab. 2013, 17, 1009–1020. [Google Scholar] [CrossRef]
- Yang, G.; Murashige, D.S.; Humphrey, S.J.; James, D.E. A Positive Feedback Loop between Akt and mTORC2 via SIN1 Phosphorylation. Cell Rep. 2015, 12, 937–943. [Google Scholar] [CrossRef]
- Manning, B.D.; Toker, A. AKT/PKB Signaling: Navigating the Network. Cell 2017, 169, 381–405. [Google Scholar] [CrossRef]
- Carracedo, A.; Pandolfi, P.P. The PTEN-PI3K pathway: Of feedbacks and cross-talks. Oncogene 2008, 27, 5527–5541. [Google Scholar] [CrossRef] [PubMed]
- Milella, M.; Falcone, I.; Conciatori, F.; Cesta Incani, U.; Del Curatolo, A.; Inzerilli, N.; Nuzzo, C.M.A.; Vaccaro, V.; Vari, S.; Cognetti, F.; et al. PTEN: Multiple Functions in Human Malignant Tumors. Front. Oncol. 2015, 5, 24. [Google Scholar] [CrossRef] [PubMed]
- Hazen, A.L.; Smith, M.J.; Desponts, C.; Winter, O.; Moser, K.; Kerr, W.G. SHIP is required for a functional hematopoietic stem cell niche. Blood 2009, 113, 2924–2933. [Google Scholar] [CrossRef]
- Luo, J.-M.; Liu, Z.-L.; Hao, H.-L.; Wang, F.-X.; Dong, Z.-R.; Ohno, R. Mutation analysis of SHIP gene in acute leukemia. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2004, 12, 420–426. [Google Scholar] [PubMed]
- Rabanal-Ruiz, Y.; Otten, E.G.; Korolchuk, V.I. mTORC1 as the main gateway to autophagy. Essays Biochem. 2017, 61, 565–584. [Google Scholar] [CrossRef]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef]
- Park, S.R.; Yoo, Y.J.; Ban, Y.-H.; Yoon, Y.J. Biosynthesis of rapamycin and its regulation: Past achievements and recent progress. J. Antibiot. 2010, 63, 434–441. [Google Scholar] [CrossRef]
- Gibbons, J.J.; Abraham, R.T.; Yu, K. Mammalian target of rapamycin: Discovery of rapamycin reveals a signaling pathway important for normal and cancer cell growth. Semin. Oncol. 2009, 36 (Suppl. S3), S3–S17. [Google Scholar] [CrossRef]
- Potter, C.J.; Pedraza, L.G.; Xu, T. Akt regulates growth by directly phosphorylating Tsc2. Nat. Cell Biol. 2002, 4, 658–665. [Google Scholar] [CrossRef]
- Manning, B.D.; Cantley, L.C. Rheb fills a GAP between TSC and TOR. Trends Biochem. Sci. 2003, 28, 573–576. [Google Scholar] [CrossRef]
- Kovacina, K.S.; Park, G.Y.; Bae, S.S.; Guzzetta, A.W.; Schaefer, E.; Birnbaum, M.J.; Roth, R.A. Identification of a proline-rich Akt substrate as a 14-3-3 binding partner. J. Biol. Chem. 2003, 278, 10189–10194. [Google Scholar] [CrossRef] [PubMed]
- Chiang, G.G.; Abraham, R.T. Phosphorylation of mammalian target of rapamycin (mTOR) at Ser-2448 is mediated by p70S6 kinase. J. Biol. Chem. 2005, 280, 25485–25490. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.W.Y.; Fryer, L.G.D.; Carling, D.; Shepherd, P.R. Thr2446 is a novel mammalian target of rapamycin (mTOR) phosphorylation site regulated by nutrient status. J. Biol. Chem. 2004, 279, 15719–15722. [Google Scholar] [CrossRef] [PubMed]
- Holz, M.K.; Blenis, J. Identification of S6 kinase 1 as a novel mammalian target of rapamycin (mTOR)-phosphorylating kinase. J. Biol. Chem. 2005, 280, 26089–26093. [Google Scholar] [CrossRef]
- Kim, J.; Guan, K.-L. mTOR as a central hub of nutrient signalling and cell growth. Nat. Cell Biol. 2019, 21, 63–71. [Google Scholar] [CrossRef]
- Huang, J.; Manning, B.D. The TSC1-TSC2 complex: A molecular switchboard controlling cell growth. Biochem. J. 2008, 412, 179–190. [Google Scholar] [CrossRef]
- Huang, X.; Wullschleger, S.; Shpiro, N.; McGuire, V.A.; Sakamoto, K.; Woods, Y.L.; McBurnie, W.; Fleming, S.; Alessi, D.R. Important role of the LKB1-AMPK pathway in suppressing tumorigenesis in PTEN-deficient mice. Biochem. J. 2008, 412, 211–221. [Google Scholar] [CrossRef]
- Hardie, D.G. New roles for the LKB1-->AMPK pathway. Curr. Opin. Cell Biol. 2005, 17, 167–173. [Google Scholar] [CrossRef]
- Dengler, F. Activation of AMPK under Hypoxia: Many Roads Leading to Rome. Int. J. Mol. Sci. 2020, 21, 2428. [Google Scholar] [CrossRef]
- Hawley, S.A.; Davison, M.; Woods, A.; Davies, S.P.; Beri, R.K.; Carling, D.; Hardie, D.G. Characterization of the AMP-activated protein kinase kinase from rat liver and identification of threonine 172 as the major site at which it phosphorylates AMP-activated protein kinase. J. Biol. Chem. 1996, 271, 27879–27887. [Google Scholar] [CrossRef]
- Gwinn, D.M.; Shackelford, D.B.; Egan, D.F.; Mihaylova, M.M.; Mery, A.; Vasquez, D.S.; Turk, B.E.; Shaw, R.J. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 2008, 30, 214–226. [Google Scholar] [CrossRef] [PubMed]
- Inoki, K.; Zhu, T.; Guan, K.-L. TSC2 mediates cellular energy response to control cell growth and survival. Cell 2003, 115, 577–590. [Google Scholar] [CrossRef]
- Mendoza, M.C.; Er, E.E.; Blenis, J. The Ras-ERK and PI3K-mTOR pathways: Cross-talk and compensation. Trends Biochem. Sci. 2011, 36, 320–328. [Google Scholar] [CrossRef]
- Ma, L.; Chen, Z.; Erdjument-Bromage, H.; Tempst, P.; Pandolfi, P.P. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell 2005, 121, 179–193. [Google Scholar] [CrossRef] [PubMed]
- Roux, P.P.; Ballif, B.A.; Anjum, R.; Gygi, S.P.; Blenis, J. Tumor-promoting phorbol esters and activated Ras inactivate the tuberous sclerosis tumor suppressor complex via p90 ribosomal S6 kinase. Proc. Natl. Acad. Sci. USA 2004, 101, 13489–13494. [Google Scholar] [CrossRef]
- Carriere, A.; Romeo, Y.; Acosta-Jaquez, H.A.; Moreau, J.; Bonneil, E.; Thibault, P.; Fingar, D.C.; Roux, P.P. ERK1/2 phosphorylate Raptor to promote Ras-dependent activation of mTOR complex 1 (mTORC1). J. Biol. Chem. 2011, 286, 567–577. [Google Scholar] [CrossRef]
- Gingras, A.C.; Gygi, S.P.; Raught, B.; Polakiewicz, R.D.; Abraham, R.T.; Hoekstra, M.F.; Aebersold, R.; Sonenberg, N. Regulation of 4E-BP1 phosphorylation: A novel two-step mechanism. Genes Dev. 1999, 13, 1422–1437. [Google Scholar] [CrossRef]
- Jackson, R.J.; Hellen, C.U.T.; Pestova, T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010, 11, 113–127. [Google Scholar] [CrossRef]
- Han, J.W.; Pearson, R.B.; Dennis, P.B.; Thomas, G. Rapamycin, wortmannin, and the methylxanthine SQ20006 inactivate p70s6k by inducing dephosphorylation of the same subset of sites. J. Biol. Chem. 1995, 270, 21396–21403. [Google Scholar] [CrossRef]
- Alessi, D.R.; Kozlowski, M.T.; Weng, Q.P.; Morrice, N.; Avruch, J. 3-Phosphoinositide-dependent protein kinase 1 (PDK1) phosphorylates and activates the p70 S6 kinase in vivo and in vitro. Curr. Biol. 1998, 8, 69–81. [Google Scholar] [CrossRef]
- Kim, D.-H.; Sarbassov, D.D.; Ali, S.M.; King, J.E.; Latek, R.R.; Erdjument-Bromage, H.; Tempst, P.; Sabatini, D.M. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 2002, 110, 163–175. [Google Scholar] [CrossRef]
- Ruvinsky, I.; Meyuhas, O. Ribosomal protein S6 phosphorylation: From protein synthesis to cell size. Trends Biochem. Sci. 2006, 31, 342–348. [Google Scholar] [CrossRef] [PubMed]
- Pende, M.; Um, S.H.; Mieulet, V.; Sticker, M.; Goss, V.L.; Mestan, J.; Mueller, M.; Fumagalli, S.; Kozma, S.C.; Thomas, G. S6K1(-/-)/S6K2(-/-) mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol. Cell. Biol. 2004, 24, 3112–3124. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Gao, Z.; Yin, J.; Quon, M.J.; Ye, J. S6K directly phosphorylates IRS-1 on Ser-270 to promote insulin resistance in response to TNF-(alpha) signaling through IKK2. J. Biol. Chem. 2008, 283, 35375–35382. [Google Scholar] [CrossRef] [PubMed]
- Kharas, M.G.; Okabe, R.; Ganis, J.J.; Gozo, M.; Khandan, T.; Paktinat, M.; Gilliland, D.G.; Gritsman, K. Constitutively active AKT depletes hematopoietic stem cells and induces leukemia in mice. Blood 2010, 115, 1406–1415. [Google Scholar] [CrossRef] [PubMed]
- Magee, J.A.; Ikenoue, T.; Nakada, D.; Lee, J.Y.; Guan, K.-L.; Morrison, S.J. Temporal changes in PTEN and mTORC2 regulation of hematopoietic stem cell self-renewal and leukemia suppression. Cell Stem Cell 2012, 11, 415–428. [Google Scholar] [CrossRef]
- Yilmaz, O.H.; Valdez, R.; Theisen, B.K.; Guo, W.; Ferguson, D.O.; Wu, H.; Morrison, S.J. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature 2006, 441, 475–482. [Google Scholar] [CrossRef]
- Lee, J.Y.; Nakada, D.; Yilmaz, O.H.; Tothova, Z.; Joseph, N.M.; Lim, M.S.; Gilliland, D.G.; Morrison, S.J. mTOR activation induces tumor suppressors that inhibit leukemogenesis and deplete hematopoietic stem cells after Pten deletion. Cell Stem Cell 2010, 7, 593–605. [Google Scholar] [CrossRef]
- Chen, C.; Liu, Y.; Liu, R.; Ikenoue, T.; Guan, K.-L.; Liu, Y.; Zheng, P. TSC-mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. J. Exp. Med. 2008, 205, 2397–2408. [Google Scholar] [CrossRef]
- Tothova, Z.; Gilliland, D.G. FoxO transcription factors and stem cell homeostasis: Insights from the hematopoietic system. Cell Stem Cell 2007, 1, 140–152. [Google Scholar] [CrossRef]
- Tothova, Z.; Kollipara, R.; Huntly, B.J.; Lee, B.H.; Castrillon, D.H.; Cullen, D.E.; McDowell, E.P.; Lazo-Kallanian, S.; Williams, I.R.; Sears, C.; et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell 2007, 128, 325–339. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, K.; Araki, K.Y.; Naka, K.; Arai, F.; Takubo, K.; Yamazaki, S.; Matsuoka, S.; Miyamoto, T.; Ito, K.; Ohmura, M.; et al. Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell 2007, 1, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Fransecky, L.; Mochmann, L.H.; Baldus, C.D. Outlook on PI3K/AKT/mTOR inhibition in acute leukemia. Mol. Cell. Ther. 2015, 3, 2. [Google Scholar] [CrossRef]
- Gao, Y.; Gao, J.; Li, M.; Zheng, Y.; Wang, Y.; Zhang, H.; Wang, W.; Chu, Y.; Wang, X.; Xu, M.; et al. Rheb1 promotes tumor progression through mTORC1 in MLL-AF9-initiated murine acute myeloid leukemia. J. Hematol. Oncol. 2016, 9, 36. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, J.; Kobayashi, M.; Ramdas, B.; Chatterjee, A.; Ma, P.; Mali, R.S.; Carlesso, N.; Liu, Y.; Plas, D.R.; Chan, R.J.; et al. S6K1 regulates hematopoietic stem cell self-renewal and leukemia maintenance. J. Clin. Investig. 2016, 126, 2621–2625. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, J.; Kapur, R. Role of mTORC1-S6K1 signaling pathway in regulation of hematopoietic stem cell and acute myeloid leukemia. Exp. Hematol. 2017, 50, 13–21. [Google Scholar] [CrossRef][Green Version]
- Zabkiewicz, J.; Pearn, L.; Hills, R.K.; Morgan, R.G.; Tonks, A.; Burnett, A.K.; Darley, R.L. The PDK1 master kinase is over-expressed in acute myeloid leukemia and promotes PKC-mediated survival of leukemic blasts. Haematologica 2014, 99, 858–864. [Google Scholar] [CrossRef]
- Peña-Blanco, A.; García-Sáez, A.J. Bax, Bak and beyond-mitochondrial performance in apoptosis. FEBS J. 2018, 285, 416–431. [Google Scholar] [CrossRef]
- Pan, R.; Ruvolo, V.; Mu, H.; Leverson, J.D.; Nichols, G.; Reed, J.C.; Konopleva, M.; Andreeff, M. Synthetic Lethality of Combined Bcl-2 Inhibition and p53 Activation in AML: Mechanisms and Superior Antileukemic Efficacy. Cancer Cell 2017, 32, 748–760. [Google Scholar] [CrossRef]
- Zeng, Z.; Shi, Y.X.; Tsao, T.; Qiu, Y.; Kornblau, S.M.; Baggerly, K.A.; Liu, W.; Jessen, K.; Liu, Y.; Kantarjian, H.; et al. Targeting of mTORC1/2 by the mTOR kinase inhibitor PP242 induces apoptosis in AML cells under conditions mimicking the bone marrow microenvironment. Blood 2012, 120, 2679–2689. [Google Scholar] [CrossRef]
- Reikvam, H.; Nepstad, I.; Bruserud, Ø.; Hatfield, K.J. Pharmacological targeting of the PI3K/mTOR pathway alters the release of angioregulatory mediators both from primary human acute myeloid leukemia cells and their neighboring stromal cells. Oncotarget 2013, 4, 830–843. [Google Scholar] [CrossRef] [PubMed]
- Stemke-Hale, K.; Gonzalez-Angulo, A.M.; Lluch, A.; Neve, R.M.; Kuo, W.-L.; Davies, M.; Carey, M.; Hu, Z.; Guan, Y.; Sahin, A.; et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008, 68, 6084–6091. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Nie, J.; Ma, X.; Wei, Y.; Peng, Y.; Wei, X. Targeting PI3K in cancer: Mechanisms and advances in clinical trials. Mol. Cancer 2019, 18, 26. [Google Scholar] [CrossRef] [PubMed]
- Gallay, N.; Dos Santos, C.; Cuzin, L.; Bousquet, M.; Simmonet Gouy, V.; Chaussade, C.; Attal, M.; Payrastre, B.; Demur, C.; Récher, C. The level of AKT phosphorylation on threonine 308 but not on serine 473 is associated with high-risk cytogenetics and predicts poor overall survival in acute myeloid leukaemia. Leukemia 2009, 23, 1029–1038. [Google Scholar] [CrossRef]
- Zhang, J.; Gu, Y.; Chen, B. Mechanisms of drug resistance in acute myeloid leukemia. Onco Targets Ther. 2019, 12, 1937–1945. [Google Scholar] [CrossRef]
- Min, Y.H.; Eom, J.I.; Cheong, J.W.; Maeng, H.O.; Kim, J.Y.; Jeung, H.K.; Lee, S.T.; Lee, M.H.; Hahn, J.S.; Ko, Y.W. Constitutive phosphorylation of Akt/PKB protein in acute myeloid leukemia: Its significance as a prognostic variable. Leukemia 2003, 17, 995–997. [Google Scholar] [CrossRef]
- Martelli, A.M.; Nyåkern, M.; Tabellini, G.; Bortul, R.; Tazzari, P.L.; Evangelisti, C.; Cocco, L. Phosphoinositide 3-kinase/Akt signaling pathway and its therapeutical implications for human acute myeloid leukemia. Leukemia 2006, 20, 911–928. [Google Scholar] [CrossRef]
- Nepstad, I.; Hatfield, K.J.; Aasebø, E.; Hernandez-Valladares, M.; Brenner, A.K.; Bartaula-Brevik, S.; Berven, F.; Selheim, F.; Skavland, J.; Gjertsen, B.T.; et al. Two acute myeloid leukemia patient subsets are identified based on the constitutive PI3K-Akt-mTOR signaling of their leukemic cells; a functional, proteomic, and transcriptomic comparison. Expert Opin. Ther. Targets 2018, 22, 639–653. [Google Scholar] [CrossRef]
- Chapuis, N.; Tamburini, J.; Cornillet-Lefebvre, P.; Gillot, L.; Bardet, V.; Willems, L.; Park, S.; Green, A.S.; Ifrah, N.; Dreyfus, F.; et al. Autocrine IGF-1/IGF-1R signaling is responsible for constitutive PI3K/Akt activation in acute myeloid leukemia: Therapeutic value of neutralizing anti-IGF-1R antibody. Haematologica 2010, 95, 415–423. [Google Scholar] [CrossRef]
- Brandts, C.H.; Sargin, B.; Rode, M.; Biermann, C.; Lindtner, B.; Schwäble, J.; Buerger, H.; Müller-Tidow, C.; Choudhary, C.; McMahon, M.; et al. Constitutive activation of Akt by Flt3 internal tandem duplications is necessary for increased survival, proliferation, and myeloid transformation. Cancer Res. 2005, 65, 9643–9650. [Google Scholar] [CrossRef]
- Tamburini, J.; Green, A.S.; Bardet, V.; Chapuis, N.; Park, S.; Willems, L.; Uzunov, M.; Ifrah, N.; Dreyfus, F.; Lacombe, C.; et al. Protein synthesis is resistant to rapamycin and constitutes a promising therapeutic target in acute myeloid leukemia. Blood 2009, 114, 1618–1627. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Jiang, B.; Zhang, Y. 4E-BP1, a multifactor regulated multifunctional protein. Cell Cycle 2016, 15, 781–786. [Google Scholar] [CrossRef] [PubMed]
- Chow, S.; Minden, M.D.; Hedley, D.W. Constitutive phosphorylation of the S6 ribosomal protein via mTOR and ERK signaling in the peripheral blasts of acute leukemia patients. Exp. Hematol. 2006, 34, 1183–1191. [Google Scholar] [CrossRef] [PubMed]
- Chu, S.H.; Heiser, D.; Li, L.; Kaplan, I.; Collector, M.; Huso, D.; Sharkis, S.J.; Civin, C.; Small, D. FLT3-ITD knockin impairs hematopoietic stem cell quiescence/homeostasis, leading to myeloproliferative neoplasm. Cell Stem Cell 2012, 11, 346–358. [Google Scholar] [CrossRef]
- Gilliland, D.G.; Griffin, J.D. The roles of FLT3 in hematopoiesis and leukemia. Blood 2002, 100, 1532–1542. [Google Scholar] [CrossRef]
- Kikushige, Y.; Yoshimoto, G.; Miyamoto, T.; Iino, T.; Mori, Y.; Iwasaki, H.; Niiro, H.; Takenaka, K.; Nagafuji, K.; Harada, M.; et al. Human Flt3 is expressed at the hematopoietic stem cell and the granulocyte/macrophage progenitor stages to maintain cell survival. J. Immunol. 2008, 180, 7358–7367. [Google Scholar] [CrossRef]
- Chen, W.; Drakos, E.; Grammatikakis, I.; Schlette, E.J.; Li, J.; Leventaki, V.; Staikou-Drakopoulou, E.; Patsouris, E.; Panayiotidis, P.; Medeiros, L.J.; et al. mTOR signaling is activated by FLT3 kinase and promotes survival of FLT3-mutated acute myeloid leukemia cells. Mol. Cancer 2010, 9, 292. [Google Scholar] [CrossRef]
- Zhang, S.; Broxmeyer, H.E. p85 subunit of PI3 kinase does not bind to human Flt3 receptor, but associates with SHP2, SHIP, and a tyrosine-phosphorylated 100-kDa protein in Flt3 ligand-stimulated hematopoietic cells. Biochem. Biophys. Res. Commun. 1999, 254, 440–445. [Google Scholar] [CrossRef]
- Scheijen, B.; Ngo, H.T.; Kang, H.; Griffin, J.D. FLT3 receptors with internal tandem duplications promote cell viability and proliferation by signaling through Foxo proteins. Oncogene 2004, 23, 3338–3349. [Google Scholar] [CrossRef]
- Zhang, X.; Tang, N.; Hadden, T.J.; Rishi, A.K. Akt, FoxO and regulation of apoptosis. Biochim. Biophys. Acta 2011, 1813, 1978–1986. [Google Scholar] [CrossRef]
- Santo, E.E.; Stroeken, P.; Sluis, P.V.; Koster, J.; Versteeg, R.; Westerhout, E.M. FOXO3a is a major target of inactivation by PI3K/AKT signaling in aggressive neuroblastoma. Cancer Res. 2013, 73, 2189–2198. [Google Scholar] [CrossRef] [PubMed]
- Sujobert, P.; Bardet, V.; Cornillet-Lefebvre, P.; Hayflick, J.S.; Prie, N.; Verdier, F.; Vanhaesebroeck, B.; Muller, O.; Pesce, F.; Ifrah, N.; et al. Essential role for the p110delta isoform in phosphoinositide 3-kinase activation and cell proliferation in acute myeloid leukemia. Blood 2005, 106, 1063–1066. [Google Scholar] [CrossRef] [PubMed]
- Gopal, A.; Graf, S. Idelalisib for the treatment of non-Hodgkin lymphoma. Expert Opin. Pharmacother. 2016, 17, 265–274. [Google Scholar] [CrossRef]
- Nguyen, L.X.T.; Sesay, A.; Mitchell, B.S. Effect of CAL-101, a PI3Kδ inhibitor, on ribosomal rna synthesis and cell proliferation in acute myeloid leukemia cells. Blood Cancer J. 2014, 4, e228. [Google Scholar] [CrossRef] [PubMed]
- Sabers, C.J.; Martin, M.M.; Brunn, G.J.; Williams, J.M.; Dumont, F.J.; Wiederrecht, G.; Abraham, R.T. Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J. Biol. Chem. 1995, 270, 815–822. [Google Scholar] [CrossRef]
- Carneiro, B.A.; Kaplan, J.B.; Altman, J.K.; Giles, F.J.; Platanias, L.C. Targeting mTOR signaling pathways and related negative feedback loops for the treatment of acute myeloid leukemia. Cancer Biol. Ther. 2015, 16, 648–656. [Google Scholar] [CrossRef]
- Chapuis, N.; Tamburini, J.; Green, A.S.; Vignon, C.; Bardet, V.; Neyret, A.; Pannetier, M.; Willems, L.; Park, S.; Macone, A.; et al. Dual Inhibition of PI3K and mTORC1/2 Signaling by NVP-BEZ235 as a New Therapeutic Strategy for Acute Myeloid Leukemia. Clin. Cancer Res. 2010, 16, 5424–5435. [Google Scholar] [CrossRef]
- Nitulescu, G.M.; Margina, D.; Juzenas, P.; Peng, Q.; Olaru, O.T.; Saloustros, E.; Fenga, C.; Spandidos, D.A.; Libra, M.; Tsatsakis, A.M. Akt inhibitors in cancer treatment: The long journey from drug discovery to clinical use (Review). Int. J. Oncol. 2016, 48, 869–885. [Google Scholar] [CrossRef]
- Hayakawa, F.; Towatari, M.; Kiyoi, H.; Tanimoto, M.; Kitamura, T.; Saito, H.; Naoe, T. Tandem-duplicated Flt3 constitutively activates STAT5 and MAP kinase and introduces autonomous cell growth in IL-3-dependent cell lines. Oncogene 2000, 19, 624–631. [Google Scholar] [CrossRef]
- Sallmyr, A.; Fan, J.; Datta, K.; Kim, K.-T.; Grosu, D.; Shapiro, P.; Small, D.; Rassool, F. Internal tandem duplication of FLT3 (FLT3/ITD) induces increased ROS production, DNA damage, and misrepair: Implications for poor prognosis in AML. Blood 2008, 111, 3173–3182. [Google Scholar] [CrossRef]
- Sallmyr, A.; Fan, J.; Rassool, F.V. Genomic instability in myeloid malignancies: Increased reactive oxygen species (ROS), DNA double strand breaks (DSBs) and error-prone repair. Cancer Lett. 2008, 270, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Rebechi, M.T.; Pratz, K.W. Genomic instability is a principle pathologic feature of FLT3 ITD kinase activity in acute myeloid leukemia leading to clonal evolution and disease progression. Leuk. Lymphoma 2017, 58, 2040–2050. [Google Scholar] [CrossRef]
- Ng, K.L.N.; Lynn, C.; Leung, T.W.Y.; So, E.C.W.; Leung, A.Y.H. Targeting DNA Damage and Repair in Acute Myeloid Leukemia Carrying Internal Tandem Duplication of Fms-like Tyrosine Kinase 3 (FLT3-ITD)-a Mechanistic Study. Blood 2019, 134, 1261. [Google Scholar] [CrossRef]
- Juvekar, A.; Hu, H.; Yadegarynia, S.; Lyssiotis, C.A.; Ullas, S.; Lien, E.C.; Bellinger, G.; Son, J.; Hok, R.C.; Seth, P.; et al. Phosphoinositide 3-kinase inhibitors induce DNA damage through nucleoside depletion. Proc. Natl. Acad. Sci. USA 2016, 113, E4338–E4347. [Google Scholar] [CrossRef]
- Shen, W.H.; Balajee, A.S.; Wang, J.; Wu, H.; Eng, C.; Pandolfi, P.P.; Yin, Y. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell 2007, 128, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Kodaki, T.; Woscholski, R.; Hallberg, B.; Rodriguez-Viciana, P.; Downward, J.; Parker, P.J. The activation of phosphatidylinositol 3-kinase by Ras. Curr. Biol. 1994, 4, 798–806. [Google Scholar] [CrossRef]
- She, Q.-B.; Solit, D.B.; Ye, Q.; O’Reilly, K.E.; Lobo, J.; Rosen, N. The BAD protein integrates survival signaling by EGFR/MAPK and PI3K/Akt kinase pathways in PTEN-deficient tumor cells. Cancer Cell 2005, 8, 287–297. [Google Scholar] [CrossRef]
- Yang, J.-Y.; Zong, C.S.; Xia, W.; Yamaguchi, H.; Ding, Q.; Xie, X.; Lang, J.-Y.; Lai, C.-C.; Chang, C.-J.; Huang, W.-C.; et al. ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat. Cell Biol. 2008, 10, 138–148. [Google Scholar] [CrossRef]
- Zhu, J.; Blenis, J.; Yuan, J. Activation of PI3K/Akt and MAPK pathways regulates Myc-mediated transcription by phosphorylating and promoting the degradation of Mad1. Proc. Natl. Acad. Sci. USA 2008, 105, 6584–6589. [Google Scholar] [CrossRef]
- Ding, Q.; Xia, W.; Liu, J.-C.; Yang, J.-Y.; Lee, D.-F.; Xia, J.; Bartholomeusz, G.; Li, Y.; Pan, Y.; Li, Z.; et al. Erk associates with and primes GSK-3beta for its inactivation resulting in upregulation of beta-catenin. Mol. Cell 2005, 19, 159–170. [Google Scholar] [CrossRef]
- McCubrey, J.A.; Rakus, D.; Gizak, A.; Steelman, L.S.; Abrams, S.L.; Lertpiriyapong, K.; Fitzgerald, T.L.; Yang, L.V.; Montalto, G.; Cervello, M.; et al. Effects of mutations in Wnt/β-catenin, hedgehog, Notch and PI3K pathways on GSK-3 activity-Diverse effects on cell growth, metabolism and cancer. Biochim. Biophys. Acta 2016, 1863, 2942–2976. [Google Scholar] [CrossRef] [PubMed]
- Vainchenker, W.; Constantinescu, S.N. JAK/STAT signaling in hematological malignancies. Oncogene 2013, 32, 2601–2613. [Google Scholar] [CrossRef] [PubMed]
- Birkenkamp, K.U.; Geugien, M.; Lemmink, H.H.; Kruijer, W.; Vellenga, E. Regulation of constitutive STAT5 phosphorylation in acute myeloid leukemia blasts. Leukemia 2001, 15, 1923–1931. [Google Scholar] [CrossRef] [PubMed]
- Wingelhofer, B.; Maurer, B.; Heyes, E.C.; Cumaraswamy, A.A.; Berger-Becvar, A.; de Araujo, E.D.; Orlova, A.; Freund, P.; Ruge, F.; Park, J.; et al. Pharmacologic inhibition of STAT5 in acute myeloid leukemia. Leukemia 2018, 32, 1135–1146. [Google Scholar] [CrossRef]
- Britschgi, A.; Andraos, R.; Brinkhaus, H.; Klebba, I.; Romanet, V.; Müller, U.; Murakami, M.; Radimerski, T.; Bentires-Alj, M. JAK2/STAT5 Inhibition Circumvents Resistance to PI3K/mTOR Blockade: A Rationale for Cotargeting These Pathways in Metastatic Breast Cancer. Cancer Cell 2012, 22, 796–811. [Google Scholar] [CrossRef]
- Spiekermann, K.; Bagrintseva, K.; Schwab, R.; Schmieja, K.; Hiddemann, W. Overexpression and constitutive activation of FLT3 induces STAT5 activation in primary acute myeloid leukemia blast cells. Clin. Cancer Res. 2003, 9, 2140–2150. [Google Scholar]
- Choudhary, C.; Brandts, C.; Schwable, J.; Tickenbrock, L.; Sargin, B.; Ueker, A.; Böhmer, F.-D.; Berdel, W.E.; Müller-Tidow, C.; Serve, H. Activation mechanisms of STAT5 by oncogenic Flt3-ITD. Blood 2007, 110, 370–374. [Google Scholar] [CrossRef]
- Green, A.S.; Maciel, T.T.; Hospital, M.-A.; Yin, C.; Mazed, F.; Townsend, E.C.; Pilorge, S.; Lambert, M.; Paubelle, E.; Jacquel, A.; et al. Pim kinases modulate resistance to FLT3 tyrosine kinase inhibitors in FLT3-ITD acute myeloid leukemia. Sci. Adv. 2015, 1, e1500221. [Google Scholar] [CrossRef]
- Keane, N.A.; Reidy, M.; Natoni, A.; Raab, M.S.; O’Dwyer, M. Targeting the Pim kinases in multiple myeloma. Blood Cancer J. 2015, 5, e325. [Google Scholar] [CrossRef]
- Kim, K.-T.; Baird, K.; Ahn, J.-Y.; Meltzer, P.; Lilly, M.; Levis, M.; Small, D. Pim-1 is up-regulated by constitutively activated FLT3 and plays a role in FLT3-mediated cell survival. Blood 2005, 105, 1759–1767. [Google Scholar] [CrossRef]
- Natarajan, K.; Xie, Y.; Burcu, M.; Linn, D.E.; Qiu, Y.; Baer, M.R. Pim-1 kinase phosphorylates and stabilizes 130 kDa FLT3 and promotes aberrant STAT5 signaling in acute myeloid leukemia with FLT3 internal tandem duplication. PLoS ONE 2013, 8, e74653. [Google Scholar] [CrossRef] [PubMed]
- Aziz, A.U.R.; Farid, S.; Qin, K.; Wang, H.; Liu, B. PIM Kinases and Their Relevance to the PI3K/AKT/mTOR Pathway in the Regulation of Ovarian Cancer. Biomolecules 2018, 8, 7. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Beharry, Z.M.; Harris, T.E.; Lilly, M.B.; Smith, C.D.; Mahajan, S.; Kraft, A.S. PIM1 protein kinase regulates PRAS40 phosphorylation and mTOR activity in FDCP1 cells. Cancer Biol. Ther. 2009, 8, 846–853. [Google Scholar] [CrossRef] [PubMed]
- Amaravadi, R.; Thompson, C.B. The survival kinases Akt and Pim as potential pharmacological targets. J. Clin. Investig. 2005, 115, 2618–2624. [Google Scholar] [CrossRef] [PubMed]
- Cen, B.; Mahajan, S.; Wang, W.; Kraft, A.S. Elevation of receptor tyrosine kinases by small molecule AKT inhibitors in prostate cancer is mediated by Pim-1. Cancer Res. 2013, 73, 3402–3411. [Google Scholar] [CrossRef]
- Song, J.H.; Padi, S.K.R.; Luevano, L.A.; Minden, M.D.; DeAngelo, D.J.; Hardiman, G.; Ball, L.E.; Warfel, N.A.; Kraft, A.S. Insulin receptor substrate 1 is a substrate of the Pim protein kinases. Oncotarget 2016, 7, 20152–20165. [Google Scholar] [CrossRef]
- Yoshimoto, G.; Miyamoto, T.; Jabbarzadeh-Tabrizi, S.; Iino, T.; Rocnik, J.L.; Kikushige, Y.; Mori, Y.; Shima, T.; Iwasaki, H.; Takenaka, K.; et al. FLT3-ITD up-regulates MCL-1 to promote survival of stem cells in acute myeloid leukemia via FLT3-ITD-specific STAT5 activation. Blood 2009, 114, 5034–5043. [Google Scholar] [CrossRef]
- Okada, K.; Nogami, A.; Ishida, S.; Akiyama, H.; Chen, C.; Umezawa, Y.; Miura, O. FLT3-ITD induces expression of Pim kinases through STAT5 to confer resistance to the PI3K/Akt pathway inhibitors on leukemic cells by enhancing the mTORC1/Mcl-1 pathway. Oncotarget 2018, 9, 8870–8886. [Google Scholar] [CrossRef]
- Nogami, A.; Oshikawa, G.; Okada, K.; Fukutake, S.; Umezawa, Y.; Nagao, T.; Kurosu, T.; Miura, O. FLT3-ITD confers resistance to the PI3K/Akt pathway inhibitors by protecting the mTOR/4EBP1/Mcl-1 pathway through STAT5 activation in acute myeloid leukemia. Oncotarget 2015, 6, 9189–9205. [Google Scholar] [CrossRef]
- Nogami, A.; Okada, K.; Ishida, S.; Akiyama, H.; Umezawa, Y.; Miura, O. Inhibition of the STAT5/Pim Kinase Axis Enhances Cytotoxic Effects of Proteasome Inhibitors on FLT3-ITD-Positive AML Cells by Cooperatively Inhibiting the mTORC1/4EBP1/S6K/Mcl-1 Pathway. Transl. Oncol. 2019, 12, 336–349. [Google Scholar] [CrossRef]
- Watanabe, D.; Nogami, A.; Okada, K.; Akiyama, H.; Umezawa, Y.; Miura, O. FLT3-ITD Activates RSK1 to Enhance Proliferation and Survival of AML Cells by Activating mTORC1 and eIF4B Cooperatively with PIM or PI3K and by Inhibiting Bad and BIM. Cancers 2019, 11, 1827. [Google Scholar] [CrossRef] [PubMed]
- Maranzana, E.; Barbero, G.; Falasca, A.I.; Lenaz, G.; Genova, M.L. Mitochondrial Respiratory Supercomplex Association Limits Production of Reactive Oxygen Species from Complex I. Antioxid. Redox Signal. 2013, 19, 1469–1480. [Google Scholar] [CrossRef] [PubMed]
- Bleier, L.; Dröse, S. Superoxide generation by complex III: From mechanistic rationales to functional consequences. Biochim. Biophys. Acta 2013, 1827, 1320–1331. [Google Scholar] [CrossRef] [PubMed]
- Tong, L.; Chuang, C.-C.; Wu, S.; Zuo, L. Reactive oxygen species in redox cancer therapy. Cancer Lett. 2015, 367, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Sillar, J.R.; Germon, Z.P.; DeIuliis, G.N.; Dun, M.D. The Role of Reactive Oxygen Species in Acute Myeloid Leukaemia. Int. J. Mol. Sci. 2019, 20, 6003. [Google Scholar] [CrossRef] [PubMed]
- Hole, P.S.; Zabkiewicz, J.; Munje, C.; Newton, Z.; Pearn, L.; White, P.; Marquez, N.; Hills, R.K.; Burnett, A.K.; Tonks, A.; et al. Overproduction of NOX-derived ROS in AML promotes proliferation and is associated with defective oxidative stress signaling. Blood 2013, 122, 3322–3330. [Google Scholar] [CrossRef]
- Jayavelu, A.K.; Moloney, J.N.; Böhmer, F.-D.; Cotter, T.G. NOX-driven ROS formation in cell transformation of FLT3-ITD-positive AML. Exp. Hematol. 2016, 44, 1113–1122. [Google Scholar] [CrossRef]
- Stanicka, J.; Russell, E.G.; Woolley, J.F.; Cotter, T.G. NADPH oxidase-generated hydrogen peroxide induces DNA damage in mutant FLT3-expressing leukemia cells. J. Biol. Chem. 2015, 290, 9348–9361. [Google Scholar] [CrossRef]
- Mi, T.; Wang, Z.; Bunting, K.D. The Cooperative Relationship between STAT5 and Reactive Oxygen Species in Leukemia: Mechanism and Therapeutic Potential. Cancers 2018, 10, 359. [Google Scholar] [CrossRef]
- Bourgeais, J.; Gouilleux-Gruart, V.; Gouilleux, F. Oxidative metabolism in cancer: A STAT affair? Jak-Stat 2013, 2, e25764. [Google Scholar] [CrossRef]
- Koundouros, N.; Poulogiannis, G. Phosphoinositide 3-Kinase/Akt Signaling and Redox Metabolism in Cancer. Front. Oncol. 2018, 8, 160. [Google Scholar] [CrossRef] [PubMed]
- Moloney, J.N.; Jayavelu, A.K.; Stanicka, J.; Roche, S.L.; O’Brien, R.L.; Scholl, S.; Böhmer, F.-D.; Cotter, T.G. Nuclear membrane-localised NOX4D generates pro-survival ROS in FLT3-ITD-expressing AML. Oncotarget 2017, 8, 105440–105457. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Browning, E.A.; Hong, N.; DeBolt, K.; Sorokina, E.M.; Liu, W.; Birnbaum, M.J.; Fisher, A.B. Membrane depolarization is the trigger for PI3K/Akt activation and leads to the generation of ROS. Am. J. Physiol. Heart Circ. Physiol. 2012, 302, H105–H114. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, A.; Wada, Y.; Kitagishi, Y.; Matsuda, S. Link between PI3K/AKT/PTEN Pathway and NOX Proteinin Diseases. Aging Dis. 2014, 5, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Zimta, A.-A.; Cenariu, D.; Irimie, A.; Magdo, L.; Nabavi, S.M.; Atanasov, A.G.; Berindan-Neagoe, I. The Role of Nrf2 Activity in Cancer Development and Progression. Cancers 2019, 11, 1755. [Google Scholar] [CrossRef]
- Bellezza, I.; Giambanco, I.; Minelli, A.; Donato, R. Nrf2-Keap1 signaling in oxidative and reductive stress. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 721–733. [Google Scholar] [CrossRef]
- Nakaso, K.; Yano, H.; Fukuhara, Y.; Takeshima, T.; Wada-Isoe, K.; Nakashima, K. PI3K is a key molecule in the Nrf2-mediated regulation of antioxidative proteins by hemin in human neuroblastoma cells. FEBS Lett. 2003, 546, 181–184. [Google Scholar] [CrossRef]
- Wang, L.; Chen, Y.; Sternberg, P.; Cai, J. Essential Roles of the PI3 Kinase/Akt Pathway in Regulating Nrf2-Dependent Antioxidant Functions in the RPE. Investig. Ophthalmol. Vis. Sci. 2008, 49, 1671–1678. [Google Scholar] [CrossRef]
- Vazquez, F.; Grossman, S.R.; Takahashi, Y.; Rokas, M.V.; Nakamura, N.; Sellers, W.R. Phosphorylation of the PTEN tail acts as an inhibitory switch by preventing its recruitment into a protein complex. J. Biol. Chem. 2001, 276, 48627–48630. [Google Scholar] [CrossRef]
- Leslie, N.R.; Bennett, D.; Lindsay, Y.E.; Stewart, H.; Gray, A.; Downes, C.P. Redox regulation of PI 3-kinase signalling via inactivation of PTEN. EMBO J. 2003, 22, 5501–5510. [Google Scholar] [CrossRef]
- Markham, A. Alpelisib: First Global Approval. Drugs 2019, 79, 1249–1253. [Google Scholar] [CrossRef] [PubMed]
- Hasskarl, J. Everolimus. Recent Results Cancer Res. 2018, 211, 101–123. [Google Scholar] [CrossRef] [PubMed]
- Kwitkowski, V.E.; Prowell, T.M.; Ibrahim, A.; Farrell, A.T.; Justice, R.; Mitchell, S.S.; Sridhara, R.; Pazdur, R. FDA approval summary: Temsirolimus as treatment for advanced renal cell carcinoma. Oncologist 2010, 15, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R. Properties of FDA-approved small molecule protein kinase inhibitors. Pharmacol. Res. 2019, 144, 19–50. [Google Scholar] [CrossRef]
- Maira, S.-M.; Pecchi, S.; Huang, A.; Burger, M.; Knapp, M.; Sterker, D.; Schnell, C.; Guthy, D.; Nagel, T.; Wiesmann, M.; et al. Identification and characterization of NVP-BKM120, an orally available pan-class I PI3-kinase inhibitor. Mol. Cancer Ther. 2012, 11, 317–328. [Google Scholar] [CrossRef]
- Burger, M.T.; Pecchi, S.; Wagman, A.; Ni, Z.-J.; Knapp, M.; Hendrickson, T.; Atallah, G.; Pfister, K.; Zhang, Y.; Bartulis, S.; et al. Identification of NVP-BKM120 as a Potent, Selective, Orally Bioavailable Class I PI3 Kinase Inhibitor for Treating Cancer. ACS Med. Chem. Lett. 2011, 2, 774–779. [Google Scholar] [CrossRef]
- Yang, S.; Li, X.; Guan, W.; Qian, M.; Yao, Z.; Yin, X.; Zhao, H. NVP-BKM120 inhibits colon cancer growth via FoxO3a-dependent PUMA induction. Oncotarget 2017, 8, 83052–83062. [Google Scholar] [CrossRef]
- Jin, L.; Jin, M.-H.; Nam, A.-R.; Park, J.-E.; Bang, J.-H.; Oh, D.-Y.; Bang, Y.-J. Anti-tumor effects of NVP-BKM120 alone or in combination with MEK162 in biliary tract cancer. Cancer Lett. 2017, 411, 162–170. [Google Scholar] [CrossRef]
- Yang, L.; Yang, G.; Ding, Y.; Huang, Y.; Liu, S.; Zhou, L.; Wei, W.; Wang, J.; Hu, G. Combined treatment with PI3K inhibitor BKM120 and PARP inhibitor olaparib is effective in inhibiting the gastric cancer cells with ARID1A deficiency. Oncol. Rep. 2018, 40, 479–487. [Google Scholar] [CrossRef]
- Allegretti, M.; Ricciardi, M.R.; Licchetta, R.; Mirabilii, S.; Orecchioni, S.; Reggiani, F.; Talarico, G.; Foà, R.; Bertolini, F.; Amadori, S.; et al. The pan-class I phosphatidyl-inositol-3 kinase inhibitor NVP-BKM120 demonstrates anti-leukemic activity in acute myeloid leukemia. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef]
- Sadeghi, S.; Esmaeili, S.; Pourbagheri-Sigaroodi, A.; Safaroghli-Azar, A.; Bashash, D. PI3K abrogation using pan PI3K inhibitor BKM120 give rise to a weighty anti-cancer effect on AML-derived KG-1 cells by inducing apoptosis and G2/M arrest. Turk. J. Haematol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Ragon, B.K.; Kantarjian, H.; Jabbour, E.; Ravandi, F.; Cortes, J.; Borthakur, G.; DeBose, L.; Zeng, Z.; Schneider, H.; Pemmaraju, N.; et al. Buparlisib, a PI3K inhibitor, demonstrates acceptable tolerability and preliminary activity in a phase I trial of patients with advanced leukemias. Am. J. Hematol. 2017, 92, 7–11. [Google Scholar] [CrossRef]
- Venkatesan, A.M.; Dehnhardt, C.M.; Delos Santos, E.; Chen, Z.; Dos Santos, O.; Ayral-Kaloustian, S.; Khafizova, G.; Brooijmans, N.; Mallon, R.; Hollander, I.; et al. Bis(morpholino-1,3,5-triazine) derivatives: Potent adenosine 5′-triphosphate competitive phosphatidylinositol-3-kinase/mammalian target of rapamycin inhibitors: Discovery of compound 26 (PKI-587), a highly efficacious dual inhibitor. J. Med. Chem. 2010, 53, 2636–2645. [Google Scholar] [CrossRef] [PubMed]
- Mallon, R.; Feldberg, L.R.; Lucas, J.; Chaudhary, I.; Dehnhardt, C.; Santos, E.D.; Chen, Z.; dos Santos, O.; Ayral-Kaloustian, S.; Venkatesan, A.; et al. Antitumor efficacy of PKI-587, a highly potent dual PI3K/mTOR kinase inhibitor. Clin. Cancer Res. 2011, 17, 3193–3203. [Google Scholar] [CrossRef] [PubMed]
- Wainberg, Z.A.; Alsina, M.; Soares, H.P.; Braña, I.; Britten, C.D.; Del Conte, G.; Ezeh, P.; Houk, B.; Kern, K.A.; Leong, S.; et al. A Multi-Arm Phase I Study of the PI3K/mTOR Inhibitors PF-04691502 and Gedatolisib (PF-05212384) plus Irinotecan or the MEK Inhibitor PD-0325901 in Advanced Cancer. Target Oncol. 2017, 12, 775–785. [Google Scholar] [CrossRef]
- Langdon, S.P.; Kay, C.; Um, I.H.; Dodds, M.; Muir, M.; Sellar, G.; Kan, J.; Gourley, C.; Harrison, D.J. Evaluation of the dual mTOR/PI3K inhibitors Gedatolisib (PF-05212384) and PF-04691502 against ovarian cancer xenograft models. Sci. Rep. 2019, 9, 18742. [Google Scholar] [CrossRef]
- Iezzi, A.; Caiola, E.; Broggini, M. Activity of Pan-Class I Isoform PI3K/mTOR Inhibitor PF-05212384 in Combination with Crizotinib in Ovarian Cancer Xenografts and PDX. Transl. Oncol. 2016, 9, 458–465. [Google Scholar] [CrossRef][Green Version]
- Del Campo, J.M.; Birrer, M.; Davis, C.; Fujiwara, K.; Gollerkeri, A.; Gore, M.; Houk, B.; Lau, S.; Poveda, A.; González-Martín, A.; et al. A randomized phase II non-comparative study of PF-04691502 and gedatolisib (PF-05212384) in patients with recurrent endometrial cancer. Gynecol. Oncol. 2016, 142, 62–69. [Google Scholar] [CrossRef]
- Soares, H.P.; Ming, M.; Mellon, M.; Young, S.H.; Han, L.; Sinnet-Smith, J.; Rozengurt, E. Dual PI3K/mTOR inhibitors induce rapid over-activation of the MEK/ERK pathway in human pancreatic cancer cells through suppression of mTORC2. Mol. Cancer Ther. 2015, 14, 1014–1023. [Google Scholar] [CrossRef]
- Shah, K.; Moharram, S.A.; Kazi, J.U. Acute leukemia cells resistant to PI3K/mTOR inhibition display upregulation of P2RY14 expression. Clin. Epigenetics 2018, 10, 83. [Google Scholar] [CrossRef]
- Vargaftig, J.; Farhat, H.; Ades, L.; Briaux, A.; Benoist, C.; Turbiez, I.; Vey, N.; Glaisner, S.; Callens, C. Phase 2 Trial of Single Agent Gedatolisib (PF-05212384), a Dual PI3K/mTOR Inhibitor, for Adverse Prognosis and Relapse/Refractory AML: Clinical and Transcriptomic Results. Blood 2018, 132, 5233. [Google Scholar] [CrossRef]
- Furman, R.R.; Byrd, J.C.; Brown, J.R.; Coutre, S.E.; Benson, D.M.; Wagner-Johnston, N.D.; Flinn, I.W.; Kahl, B.S.; Spurgeon, S.E.; Lannutti, B.; et al. CAL-101, An Isoform-Selective Inhibitor of Phosphatidylinositol 3-Kinase P110δ, Demonstrates Clinical Activity and Pharmacodynamic Effects In Patients with Relapsed or Refractory Chronic Lymphocytic Leukemia. Blood 2010, 116, 55. [Google Scholar] [CrossRef]
- Kahl, B.S.; Spurgeon, S.E.; Furman, R.R.; Flinn, I.W.; Coutre, S.E.; Brown, J.R.; Benson, D.M.; Byrd, J.C.; Peterman, S.; Cho, Y.; et al. A phase 1 study of the PI3Kδ inhibitor idelalisib in patients with relapsed/refractory mantle cell lymphoma (MCL). Blood 2014, 123, 3398–3405. [Google Scholar] [CrossRef] [PubMed]
- Flinn, I.W.; Kahl, B.S.; Leonard, J.P.; Furman, R.R.; Brown, J.R.; Byrd, J.C.; Wagner-Johnston, N.D.; Coutre, S.E.; Benson, D.M.; Peterman, S.; et al. Idelalisib, a selective inhibitor of phosphatidylinositol 3-kinase-δ, as therapy for previously treated indolent non-Hodgkin lymphoma. Blood 2014, 123, 3406–3413. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.R.; Byrd, J.C.; Coutre, S.E.; Benson, D.M.; Flinn, I.W.; Wagner-Johnston, N.D.; Spurgeon, S.E.; Kahl, B.S.; Bello, C.; Webb, H.K.; et al. Idelalisib, an inhibitor of phosphatidylinositol 3-kinase p110δ, for relapsed/refractory chronic lymphocytic leukemia. Blood 2014, 123, 3390–3397. [Google Scholar] [CrossRef] [PubMed]
- Flinn, I.W.; Byrd, J.C.; Furman, R.R.; Brown, J.R.; Benson, D.M.; Coutre, S.E.; Kahl, B.S.; Smith, B.D.; Wagner-Johnston, N.D.; Spurgeon, S.E.; et al. Evidence of Clinical Activity in a Phase 1 Study of CAL-101, An Oral P110Δ Isoform-Selective Inhibitor of Phosphatidylinositol 3-Kinase, in Patients with Relapsed or Refractory B-Cell Malignancies. Blood 2009, 114, 922. [Google Scholar] [CrossRef]
- Pongas, G.; Fojo, T. BEZ235: When Promising Science Meets Clinical Reality. Oncologist 2016, 21, 1033–1034. [Google Scholar] [CrossRef]
- Carlo, M.I.; Molina, A.M.; Lakhman, Y.; Patil, S.; Woo, K.; DeLuca, J.; Lee, C.-H.; Hsieh, J.J.; Feldman, D.R.; Motzer, R.J.; et al. A Phase Ib Study of BEZ235, a Dual Inhibitor of Phosphatidylinositol 3-Kinase (PI3K) and Mammalian Target of Rapamycin (mTOR), in Patients with Advanced Renal Cell Carcinoma. Oncologist 2016, 21, 787–788. [Google Scholar] [CrossRef]
- Salazar, R.; Garcia-Carbonero, R.; Libutti, S.K.; Hendifar, A.E.; Custodio, A.; Guimbaud, R.; Lombard-Bohas, C.; Ricci, S.; Klümpen, H.-J.; Capdevila, J.; et al. Phase II Study of BEZ235 versus Everolimus in Patients with Mammalian Target of Rapamycin Inhibitor-Naïve Advanced Pancreatic Neuroendocrine Tumors. Oncologist 2018, 23, 766-e90. [Google Scholar] [CrossRef]
- Huang, J.-C.; Cui, Z.-F.; Chen, S.-M.; Yang, L.-J.; Lian, H.-K.; Liu, B.; Su, Z.-H.; Liu, J.-S.; Wang, M.; Hu, Z.-B.; et al. NVP-BEZ235 synergizes cisplatin sensitivity in osteosarcoma. Oncotarget 2018, 9, 10483–10496. [Google Scholar] [CrossRef]
- Ma, Y.; Jin, Z.; Yu, K.; Liu, Q. NVP-BEZ235-induced autophagy as a potential therapeutic approach for multiple myeloma. Am. J. Transl. Res. 2019, 11, 87–105. [Google Scholar] [PubMed]
- Mitchell, R.; Hopcroft, L.E.M.; Baquero, P.; Allan, E.K.; Hewit, K.; James, D.; Hamilton, G.; Mukhopadhyay, A.; O’Prey, J.; Hair, A.; et al. Targeting BCR-ABL-Independent TKI Resistance in Chronic Myeloid Leukemia by mTOR and Autophagy Inhibition. J. Natl. Cancer Inst. 2018, 110, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.-Y.; Hu, L.-S.; Han, H.-J.; Song, C.-Y.; Huang, Y.-X.; Guo, K.-Y. [NVP-BEZ235 inhibits proliferation and colony-forming capability of CD34(+)CD38(-) human acute myeloid leukemia stem cells]. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2013, 21, 334–338. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Jiang, L.; Ln, X.-H.; Tseng, K.-F.; Liu, Y.; Zhang, X.; Dong, R.-H.; Lu, Z.-G.; Wang, X.-J. The PI3K/mTOR dual inhibitor BEZ235 suppresses proliferation and migration and reverses multidrug resistance in acute myeloid leukemia. Acta Pharmacol. Sin. 2017, 38, 382–391. [Google Scholar] [CrossRef] [PubMed]
- Wunderle, L.; Badura, S.; Lang, F.; Wolf, A.; Schleyer, E.; Serve, H.; Goekbuget, N.; Pfeifer, H.; Bug, G.; Ottmann, O.G. Safety and Efficacy of BEZ235, a Dual PI3-Kinase /mTOR Inhibitor, In Adult Patients with Relapsed or Refractory Acute Leukemia: Results of a Phase I Study. Blood 2013, 122, 2675. [Google Scholar] [CrossRef]
- Lu, J.-W.; Lin, Y.-M.; Lai, Y.-L.; Chen, C.-Y.; Hu, C.-Y.; Tien, H.-F.; Ou, D.-L.; Lin, L.-I. MK-2206 induces apoptosis of AML cells and enhances the cytotoxicity of cytarabine. Med. Oncol. 2015, 32, 206. [Google Scholar] [CrossRef]
- Xing, Y.; Lin, N.U.; Maurer, M.A.; Chen, H.; Mahvash, A.; Sahin, A.; Akcakanat, A.; Li, Y.; Abramson, V.; Litton, J.; et al. Phase II trial of AKT inhibitor MK-2206 in patients with advanced breast cancer who have tumors with PIK3CA or AKT mutations, and/or PTEN loss/PTEN mutation. Breast Cancer Res. 2019, 21, 78. [Google Scholar] [CrossRef]
- Ma, C.X.; Suman, V.; Goetz, M.P.; Northfelt, D.; Burkard, M.E.; Ademuyiwa, F.; Naughton, M.; Margenthaler, J.; Aft, R.; Gray, R.; et al. A Phase II Trial of Neoadjuvant MK-2206, an AKT Inhibitor, with Anastrozole in Clinical Stage II or III PIK3CA-Mutant ER-Positive and HER2-Negative Breast Cancer. Clin. Cancer Res. 2017, 23, 6823–6832. [Google Scholar] [CrossRef]
- Akcakanat, A.; Meric-Bernstam, F. MK-2206 window of opportunity study in breast cancer. Ann. Transl. Med. 2018, 6. [Google Scholar] [CrossRef]
- Konopleva, M.Y.; Walter, R.B.; Faderl, S.H.; Jabbour, E.J.; Zeng, Z.; Borthakur, G.; Huang, X.; Kadia, T.M.; Ruvolo, P.P.; Feliu, J.B.; et al. Preclinical and early clinical evaluation of the oral AKT inhibitor, MK-2206, for the treatment of acute myelogenous leukemia. Clin. Cancer Res. 2014, 20, 2226–2235. [Google Scholar] [CrossRef]
- Tolcher, A.W.; Patnaik, A.; Papadopoulos, K.P.; Rasco, D.W.; Becerra, C.R.; Allred, A.J.; Orford, K.; Aktan, G.; Ferron-Brady, G.; Ibrahim, N.; et al. Phase I study of the MEK inhibitor trametinib in combination with the AKT inhibitor afuresertib in patients with solid tumors and multiple myeloma. Cancer Chemother. Pharmacol. 2015, 75, 183–189. [Google Scholar] [CrossRef]
- Blagden, S.P.; Hamilton, A.L.; Mileshkin, L.; Wong, S.; Michael, A.; Hall, M.; Goh, J.C.; Lisyanskaya, A.S.; DeSilvio, M.; Frangou, E.; et al. Phase IB Dose Escalation and Expansion Study of AKT Inhibitor Afuresertib with Carboplatin and Paclitaxel in Recurrent Platinum-resistant Ovarian Cancer. Clin. Cancer Res. 2019, 25, 1472–1478. [Google Scholar] [CrossRef] [PubMed]
- Naymagon, L.; Abdul-Hay, M. Novel agents in the treatment of multiple myeloma: A review about the future. J. Hematol. Oncol. 2016, 9, 52. [Google Scholar] [CrossRef]
- Spencer, A.; Yoon, S.-S.; Harrison, S.J.; Morris, S.R.; Smith, D.A.; Brigandi, R.A.; Gauvin, J.; Kumar, R.; Opalinska, J.B.; Chen, C. The novel AKT inhibitor afuresertib shows favorable safety, pharmacokinetics, and clinical activity in multiple myeloma. Blood 2014, 124, 2190–2195. [Google Scholar] [CrossRef] [PubMed]
- Algazi, A.P.; Esteve-Puig, R.; Nosrati, A.; Hinds, B.; Hobbs-Muthukumar, A.; Nandoskar, P.; Ortiz-Urda, S.; Chapman, P.B.; Daud, A. Dual MEK/AKT inhibition with trametinib and GSK2141795 does not yield clinical benefit in metastatic NRAS-mutant and wild-type melanoma. Pigment Cell Melanoma Res. 2018, 31, 110–114. [Google Scholar] [CrossRef]
- Westin, S.N.; Sill, M.W.; Coleman, R.L.; Waggoner, S.; Moore, K.N.; Mathews, C.A.; Martin, L.P.; Modesitt, S.C.; Lee, S.; Ju, Z.; et al. Safety lead-in of the MEK inhibitor trametinib in combination with GSK2141795, an AKT inhibitor, in patients with recurrent endometrial cancer: An NRG Oncology/GOG study. Gynecol. Oncol. 2019, 155, 420–428. [Google Scholar] [CrossRef]
- Ragon, B.K.; Odenike, O.; Baer, M.R.; Stock, W.; Borthakur, G.; Patel, K.; Han, L.; Chen, H.; Ma, H.; Joseph, L.; et al. Oral MEK 1/2 Inhibitor Trametinib in Combination with AKT Inhibitor GSK2141795 in Patients With Acute Myeloid Leukemia With RAS Mutations: A Phase II Study. Clin. Lymphoma Myeloma Leuk. 2019, 19, 431–440.e13. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhao, R.; Tseng, K.; Li, K.; Lu, Z.; Liu, Y.; Han, K.; Gan, Z.; Lin, S.; Hu, H.; et al. Sirolimus induces apoptosis and reverses multidrug resistance in human osteosarcoma cells in vitro via increasing microRNA-34b expression. Acta Pharmacol. Sin. 2016, 37, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Imrali, A.; Mao, X.; Yeste-Velasco, M.; Shamash, J.; Lu, Y. Rapamycin inhibits prostate cancer cell growth through cyclin D1 and enhances the cytotoxic efficacy of cisplatin. Am. J. Cancer Res. 2016, 6, 1772–1784. [Google Scholar]
- Yao, C.; Liu, J.; Shao, L. Rapamycin inhibits the proliferation and apoptosis of gastric cancer cells by down regulating the expression of survivin. Hepatogastroenterology 2011, 58, 1075–1080. [Google Scholar]
- Wagner, F.; Henningsen, B.; Lederer, C.; Eichenmüller, M.; Gödeke, J.; Müller-Höcker, J.; von Schweinitz, D.; Kappler, R. Rapamycin blocks hepatoblastoma growth in vitro and in vivo implicating new treatment options in high-risk patients. Eur. J. Cancer 2012, 48, 2442–2450. [Google Scholar] [CrossRef] [PubMed]
- Kolos, J.M.; Voll, A.M.; Bauder, M.; Hausch, F. FKBP Ligands-Where We Are and Where to Go? Front. Pharmacol. 2018, 9, 1425. [Google Scholar] [CrossRef] [PubMed]
- Grignani, G.; Palmerini, E.; Ferraresi, V.; D’Ambrosio, L.; Bertulli, R.; Asaftei, S.D.; Tamburini, A.; Pignochino, Y.; Sangiolo, D.; Marchesi, E.; et al. Sorafenib and everolimus for patients with unresectable high-grade osteosarcoma progressing after standard treatment: A non-randomised phase 2 clinical trial. Lancet Oncol. 2015, 16, 98–107. [Google Scholar] [CrossRef]
- Colon-Otero, G.; Weroha, S.J.; Foster, N.R.; Haluska, P.; Hou, X.; Wahner-Hendrickson, A.E.; Jatoi, A.; Block, M.S.; Dinh, T.A.; Robertson, M.W.; et al. Phase 2 trial of everolimus and letrozole in relapsed estrogen receptor-positive high-grade ovarian cancers. Gynecol. Oncol. 2017, 146, 64–68. [Google Scholar] [CrossRef]
- Zhu, A.X.; Abrams, T.A.; Miksad, R.; Blaszkowsky, L.S.; Meyerhardt, J.A.; Zheng, H.; Muzikansky, A.; Clark, J.W.; Kwak, E.L.; Schrag, D.; et al. Phase 1/2 study of everolimus in advanced hepatocellular carcinoma. Cancer 2011, 117, 5094–5102. [Google Scholar] [CrossRef]
- Perl, A.E.; Kasner, M.T.; Shank, D.; Luger, S.M.; Carroll, M. Single-cell pharmacodynamic monitoring of S6 ribosomal protein phosphorylation in AML blasts during a clinical trial combining the mTOR inhibitor sirolimus and intensive chemotherapy. Clin. Cancer Res. 2012, 18, 1716–1725. [Google Scholar] [CrossRef]
- Kasner, M.; Luger, S.M.; Jeschke, G.R.; Mick, R.; Carroll, M.; Perl, A.E. Single-Cell Pharmacodynamic Monitoring of S6 Ribosomal Protein in AML Blasts During Trials Combining Sirolimus and Intensive Chemotherapy: Target Inhibition Enhances Response. Blood 2011, 118, 230. [Google Scholar] [CrossRef]
- Kasner, M.T.; Mick, R.; Jeschke, G.R.; Carabasi, M.; Filicko-O’Hara, J.; Flomenberg, N.; Frey, N.V.; Hexner, E.O.; Luger, S.M.; Loren, A.W.; et al. Sirolimus enhances remission induction in patients with high risk acute myeloid leukemia and mTORC1 target inhibition. Investig. New Drugs 2018, 36, 657–666. [Google Scholar] [CrossRef]
- Perl, A.E.; Kasner, M.T.; Tsai, D.E.; Vogl, D.T.; Loren, A.W.; Schuster, S.J.; Porter, D.L.; Stadtmauer, E.A.; Goldstein, S.C.; Frey, N.V.; et al. A phase I study of the mammalian target of rapamycin inhibitor sirolimus and MEC chemotherapy in relapsed and refractory acute myelogenous leukemia. Clin. Cancer Res. 2009, 15, 6732–6739. [Google Scholar] [CrossRef]
- Raimondo, L.; D’Amato, V.; Servetto, A.; Rosa, R.; Marciano, R.; Formisano, L.; Mauro, C.D.; Orsini, R.C.; Cascetta, P.; Ciciola, P.; et al. Everolimus induces Met inactivation by disrupting the FKBP12/Met complex. Oncotarget 2016, 7, 40073–40084. [Google Scholar] [CrossRef]
- Yang, H.; Rudge, D.G.; Koos, J.D.; Vaidialingam, B.; Yang, H.J.; Pavletich, N.P. mTOR kinase structure, mechanism and regulation. Nature 2013, 497, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Tamburini, J.; Chapuis, N.; Bardet, V.; Park, S.; Sujobert, P.; Willems, L.; Ifrah, N.; Dreyfus, F.; Mayeux, P.; Lacombe, C.; et al. Mammalian target of rapamycin (mTOR) inhibition activates phosphatidylinositol 3-kinase/Akt by up-regulating insulin-like growth factor-1 receptor signaling in acute myeloid leukemia: Rationale for therapeutic inhibition of both pathways. Blood 2008, 111, 379–382. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Harkavy, B.; Shen, N.; Grohar, P.; Helman, L.J. Rapamycin induces feedback activation of Akt signaling through an IGF-1R-dependent mechanism. Oncogene 2007, 26, 1932–1940. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Chapuis, N.; Saint Marcoux, F.; Recher, C.; Prebet, T.; Chevallier, P.; Cahn, J.-Y.; Leguay, T.; Bories, P.; Witz, F.; et al. A phase Ib GOELAMS study of the mTOR inhibitor RAD001 in association with chemotherapy for AML patients in first relapse. Leukemia 2013, 27, 1479–1486. [Google Scholar] [CrossRef] [PubMed]
- Tiong, I.S.; Tan, P.; McManus, J.; Cummings, N.; Sadawarte, S.; Catalano, J.; Hills, R.; Wei, A. Phase Ib study of the mTOR inhibitor everolimus with low dose cytarabine in elderly acute myeloid leukemia. Leuk. Lymphoma 2018, 59, 493–496. [Google Scholar] [CrossRef]
- Herschbein, L.; Liesveld, J.L. Dueling for dual inhibition: Means to enhance effectiveness of PI3K/Akt/mTOR inhibitors in AML. Blood Rev. 2018, 32, 235–248. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network; Ley, T.J.; Miller, C.; Ding, L.; Raphael, B.J.; Mungall, A.J.; Robertson, A.G.; Hoadley, K.; Triche, T.J.; Laird, P.W.; et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013, 368, 2059–2074. [Google Scholar] [CrossRef]
- Alcalay, M.; Meani, N.; Gelmetti, V.; Fantozzi, A.; Fagioli, M.; Orleth, A.; Riganelli, D.; Sebastiani, C.; Cappelli, E.; Casciari, C.; et al. Acute myeloid leukemia fusion proteins deregulate genes involved in stem cell maintenance and DNA repair. J. Clin. Investig. 2003, 112, 1751–1761. [Google Scholar] [CrossRef]
- Martens, J.H.A.; Stunnenberg, H.G. The molecular signature of oncofusion proteins in acute myeloid leukemia. FEBS Lett. 2010, 584, 2662–2669. [Google Scholar] [CrossRef]
- San José-Enériz, E.; Gimenez-Camino, N.; Agirre, X.; Prosper, F. HDAC Inhibitors in Acute Myeloid Leukemia. Cancers 2019, 11, 1794. [Google Scholar] [CrossRef]
- Gardin, C.; Dombret, H. Hypomethylating Agents as a Therapy for AML. Curr. Hematol. Malig. Rep. 2017, 12, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Bewersdorf, J.P.; Shallis, R.; Stahl, M.; Zeidan, A.M. Epigenetic therapy combinations in acute myeloid leukemia: What are the options? Ther. Adv. Hematol. 2019, 10. [Google Scholar] [CrossRef]
- Choe, S.; Wang, H.; DiNardo, C.D.; Stein, E.M.; de Botton, S.; Roboz, G.J.; Altman, J.K.; Mims, A.S.; Watts, J.M.; Pollyea, D.A.; et al. Molecular mechanisms mediating relapse following ivosidenib monotherapy in IDH1-mutant relapsed or refractory AML. Blood Adv. 2020, 4, 1894–1905. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Jiang, W.; Hou, P. Emerging role of PI3K/AKT in tumor-related epigenetic regulation. Semin. Cancer Biol. 2019, 59, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Spangle, J.M.; Roberts, T.M.; Zhao, J.J. The emerging role of PI3K/AKT-mediated epigenetic regulation in cancer. Biochim. Biophys. Acta Rev. Cancer 2017, 1868, 123–131. [Google Scholar] [CrossRef]
- Long, J.; Jia, M.-Y.; Fang, W.-Y.; Chen, X.-J.; Mu, L.-L.; Wang, Z.-Y.; Shen, Y.; Xiang, R.-F.; Wang, L.-N.; Wang, L.; et al. FLT3 inhibition upregulates HDAC8 via FOXO to inactivate p53 and promote maintenance of FLT3-ITD+ acute myeloid leukemia. Blood 2020, 135, 1472–1483. [Google Scholar] [CrossRef]
- Nishioka, C.; Ikezoe, T.; Yang, J.; Koeffler, H.P.; Yokoyama, A. Blockade of mTOR signaling potentiates the ability of histone deacetylase inhibitor to induce growth arrest and differentiation of acute myelogenous leukemia cells. Leukemia 2008, 22, 2159–2168. [Google Scholar] [CrossRef]
- Thépot, S.; Lainey, E.; Cluzeau, T.; Sébert, M.; Leroy, C.; Adès, L.; Tailler, M.; Galluzzi, L.; Baran-Marszak, F.; Roudot, H.; et al. Hypomethylating agents reactivate FOXO3A in acute myeloid leukemia. Cell Cycle 2011, 10, 2323–2330. [Google Scholar] [CrossRef]
- Rahmani, M.; Aust, M.M.; Benson, E.C.; Wallace, L.; Friedberg, J.; Grant, S. PI3K/mTOR inhibition markedly potentiates HDAC inhibitor activity in NHL cells through BIM- and MCL-1-dependent mechanisms in vitro and in vivo. Clin. Cancer Res. 2014, 20, 4849–4860. [Google Scholar] [CrossRef]
- Li, Y.; Chen, K.; Zhou, Y.; Xiao, Y.; Deng, M.; Jiang, Z.; Ye, W.; Wang, X.; Wei, X.; Li, J.; et al. A New Strategy to Target Acute Myeloid Leukemia Stem and Progenitor Cells Using Chidamide, a Histone Deacetylase Inhibitor. Curr. Cancer Drug Targets 2015, 15, 493–503. [Google Scholar] [CrossRef]
- Qian, X.; Li, Y.; Yu, Y.; Yang, F.; Deng, R.; Ji, J.; Jiao, L.; Li, X.; Wu, R.-Y.; Chen, W.-D.; et al. Inhibition of DNA methyltransferase as a novel therapeutic strategy to overcome acquired resistance to dual PI3K/mTOR inhibitors. Oncotarget 2015, 6, 5134–5146. [Google Scholar] [CrossRef]
- Gupta, M.; Ansell, S.M.; Novak, A.J.; Kumar, S.; Kaufmann, S.H.; Witzig, T.E. Inhibition of histone deacetylase overcomes rapamycin-mediated resistance in diffuse large B-cell lymphoma by inhibiting Akt signaling through mTORC2. Blood 2009, 114, 2926–2935. [Google Scholar] [CrossRef] [PubMed]
- Liesveld, J.L.; O’Dwyer, K.; Walker, A.; Becker, M.W.; Ifthikharuddin, J.J.; Mulford, D.; Chen, R.; Bechelli, J.; Rosell, K.; Minhajuddin, M.; et al. A phase I study of decitabine and rapamycin in relapsed/refractory AML. Leuk. Res. 2013, 37, 1622–1627. [Google Scholar] [CrossRef] [PubMed]
- Malik, P.; Cashen, A.F. Decitabine in the treatment of acute myeloid leukemia in elderly patients. Cancer Manag Res. 2014, 6, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Kantarjian, H.M.; Thomas, X.G.; Dmoszynska, A.; Wierzbowska, A.; Mazur, G.; Mayer, J.; Gau, J.-P.; Chou, W.-C.; Buckstein, R.; Cermak, J.; et al. Multicenter, randomized, open-label, phase III trial of decitabine versus patient choice, with physician advice, of either supportive care or low-dose cytarabine for the treatment of older patients with newly diagnosed acute myeloid leukemia. J. Clin. Oncol. 2012, 30, 2670–2677. [Google Scholar] [CrossRef]
- Tan, P.; Tiong, I.S.; Fleming, S.; Pomilio, G.; Cummings, N.; Droogleever, M.; McManus, J.; Schwarer, A.; Catalano, J.; Patil, S.; et al. The mTOR inhibitor everolimus in combination with azacitidine in patients with relapsed/refractory acute myeloid leukemia: A phase Ib/II study. Oncotarget 2017, 8, 52269–52280. [Google Scholar] [CrossRef]
- Huang, D.C.; Strasser, A. BH3-Only proteins-essential initiators of apoptotic cell death. Cell 2000, 103, 839–842. [Google Scholar] [CrossRef]
- Kuwana, T.; Newmeyer, D.D. Bcl-2-family proteins and the role of mitochondria in apoptosis. Curr. Opin. Cell Biol. 2003, 15, 691–699. [Google Scholar] [CrossRef]
- Hata, A.N.; Engelman, J.A.; Faber, A.C. The BCL-2 family: Key mediators of the apoptotic response to targeted anti-cancer therapeutics. Cancer Discov. 2015, 5, 475–487. [Google Scholar] [CrossRef]
- Willis, S.N.; Chen, L.; Dewson, G.; Wei, A.; Naik, E.; Fletcher, J.I.; Adams, J.M.; Huang, D.C.S. Proapoptotic Bak is sequestered by Mcl-1 and Bcl-xL, but not Bcl-2, until displaced by BH3-only proteins. Genes Dev. 2005, 19, 1294–1305. [Google Scholar] [CrossRef]
- Zhou, J.-D.; Zhang, T.-J.; Xu, Z.-J.; Gu, Y.; Ma, J.-C.; Li, X.-X.; Guo, H.; Wen, X.-M.; Zhang, W.; Yang, L.; et al. BCL2 overexpression: Clinical implication and biological insights in acute myeloid leukemia. Diagn. Pathol. 2019, 14, 68. [Google Scholar] [CrossRef] [PubMed]
- Andreeff, M.; Jiang, S.; Zhang, X.; Konopleva, M.; Estrov, Z.; Snell, V.E.; Xie, Z.; Okcu, M.F.; Sanchez-Williams, G.; Dong, J.; et al. Expression of Bcl-2-related genes in normal and AML progenitors: Changes induced by chemotherapy and retinoic acid. Leukemia 1999, 13, 1881–1892. [Google Scholar] [CrossRef] [PubMed]
- Pan, R.; Hogdal, L.J.; Benito, J.M.; Bucci, D.; Han, L.; Borthakur, G.; Cortes, J.; DeAngelo, D.J.; Debose, L.; Mu, H.; et al. Selective BCL-2 inhibition by ABT-199 causes on-target cell death in acute myeloid leukemia. Cancer Discov. 2014, 4, 362–375. [Google Scholar] [CrossRef]
- Souers, A.J.; Leverson, J.D.; Boghaert, E.R.; Ackler, S.L.; Catron, N.D.; Chen, J.; Dayton, B.D.; Ding, H.; Enschede, S.H.; Fairbrother, W.J.; et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat. Med. 2013, 19, 202–208. [Google Scholar] [CrossRef]
- Konopleva, M.; Pollyea, D.A.; Potluri, J.; Chyla, B.; Hogdal, L.; Busman, T.; McKeegan, E.; Salem, A.H.; Zhu, M.; Ricker, J.L.; et al. Efficacy and Biological Correlates of Response in a Phase II Study of Venetoclax Monotherapy in Patients with Acute Myelogenous Leukemia. Cancer Discov. 2016, 6, 1106–1117. [Google Scholar] [CrossRef] [PubMed]
- Bose, P.; Gandhi, V.; Konopleva, M. Pathways and mechanisms of venetoclax resistance. Leuk. Lymphoma 2017, 58. [Google Scholar] [CrossRef]
- Xiang, Z.; Luo, H.; Payton, J.E.; Cain, J.; Ley, T.J.; Opferman, J.T.; Tomasson, M.H. Mcl1 haploinsufficiency protects mice from Myc-induced acute myeloid leukemia. J. Clin. Investig. 2010, 120, 2109–2118. [Google Scholar] [CrossRef]
- Lin, K.H.; Winter, P.S.; Xie, A.; Roth, C.; Martz, C.A.; Stein, E.M.; Anderson, G.R.; Tingley, J.P.; Wood, K.C. Targeting MCL-1/BCL-XL Forestalls the Acquisition of Resistance to ABT-199 in Acute Myeloid Leukemia. Sci. Rep. 2016, 6, 27696. [Google Scholar] [CrossRef]
- Teh, T.-C.; Nguyen, N.-Y.; Moujalled, D.M.; Segal, D.; Pomilio, G.; Rijal, S.; Jabbour, A.; Cummins, K.; Lackovic, K.; Blombery, P.; et al. Enhancing venetoclax activity in acute myeloid leukemia by co-targeting MCL1. Leukemia 2018, 32, 303–312. [Google Scholar] [CrossRef]
- Konopleva, M.; Letai, A. BCL-2 inhibition in AML: An unexpected bonus? Blood 2018, 132, 1007–1012. [Google Scholar] [CrossRef]
- Tsao, T.; Shi, Y.; Kornblau, S.; Lu, H.; Konoplev, S.; Antony, A.; Ruvolo, V.; Qiu, Y.H.; Zhang, N.; Coombes, K.R.; et al. Concomitant inhibition of DNA methyltransferase and BCL-2 protein function synergistically induce mitochondrial apoptosis in acute myelogenous leukemia cells. Ann. Hematol. 2012, 91, 1861–1870. [Google Scholar] [CrossRef] [PubMed]
- DiNardo, C.D.; Pratz, K.W.; Letai, A.; Jonas, B.A.; Wei, A.H.; Thirman, M.; Arellano, M.; Frattini, M.G.; Kantarjian, H.; Popovic, R.; et al. Safety and preliminary efficacy of venetoclax with decitabine or azacitidine in elderly patients with previously untreated acute myeloid leukaemia: A non-randomised, open-label, phase 1b study. Lancet Oncol. 2018, 19, 216–228. [Google Scholar] [CrossRef]
- DiNardo, C.D.; Pratz, K.; Pullarkat, V.; Jonas, B.A.; Arellano, M.; Becker, P.S.; Frankfurt, O.; Konopleva, M.; Wei, A.H.; Kantarjian, H.M.; et al. Venetoclax combined with decitabine or azacitidine in treatment-naive, elderly patients with acute myeloid leukemia. Blood 2019, 133, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Wei, A.H.; Strickland, S.A.; Hou, J.-Z.; Fiedler, W.; Lin, T.L.; Walter, R.B.; Enjeti, A.; Tiong, I.S.; Savona, M.; Lee, S.; et al. Venetoclax Combined with Low-Dose Cytarabine for Previously Untreated Patients With Acute Myeloid Leukemia: Results From a Phase Ib/II Study. J. Clin. Oncol. 2019, 37, 1277–1284. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.M.; Chao, J.R.; Chen, W.; Kuo, M.L.; Yen, J.J.; Yang-Yen, H.F. The antiapoptotic gene mcl-1 is up-regulated by the phosphatidylinositol 3-kinase/Akt signaling pathway through a transcription factor complex containing CREB. Mol. Cell. Biol. 1999, 19, 6195–6206. [Google Scholar] [CrossRef] [PubMed]
- Pugazhenthi, S.; Nesterova, A.; Sable, C.; Heidenreich, K.A.; Boxer, L.M.; Heasley, L.E.; Reusch, J.E. Akt/protein kinase B up-regulates Bcl-2 expression through cAMP-response element-binding protein. J. Biol. Chem. 2000, 275, 10761–10766. [Google Scholar] [CrossRef]
- Su, Y.; Li, X.; Ma, J.; Zhao, J.; Liu, S.; Wang, G.; Edwards, H.; Taub, J.W.; Lin, H.; Ge, Y. Targeting PI3K, mTOR, ERK, and Bcl-2 signaling network shows superior antileukemic activity against AML ex vivo. Biochem. Pharmacol. 2018, 148, 13–26. [Google Scholar] [CrossRef]
- Rahmani, M.; Nkwocha, J.; Hawkins, E.; Pei, X.; Parker, R.E.; Kmieciak, M.; Leverson, J.D.; Sampath, D.; Ferreira-Gonzalez, A.; Grant, S. Cotargeting BCL-2 and PI3K Induces BAX-Dependent Mitochondrial Apoptosis in AML Cells. Cancer Res. 2018, 78, 3075–3086. [Google Scholar] [CrossRef]
- Bose, P.; Rahmani, M.; Grant, S. Coordinate PI3K pathway and Bcl-2 family disruption in AML. Oncotarget 2012, 3, 1499–1500. [Google Scholar] [CrossRef]
- Rahmani, M.; Aust, M.M.; Attkisson, E.; Williams, D.C.; Ferreira-Gonzalez, A.; Grant, S. Dual inhibition of Bcl-2 and Bcl-xL strikingly enhances PI3K inhibition-induced apoptosis in human myeloid leukemia cells through a GSK3- and Bim-dependent mechanism. Cancer Res. 2013, 73, 1340–1351. [Google Scholar] [CrossRef]
- Choudhary, G.S.; Al-Harbi, S.; Mazumder, S.; Hill, B.T.; Smith, M.R.; Bodo, J.; Hsi, E.D.; Almasan, A. MCL-1 and BCL-xL-dependent resistance to the BCL-2 inhibitor ABT-199 can be overcome by preventing PI3K/AKT/mTOR activation in lymphoid malignancies. Cell Death Dis. 2015, 6, e1593. [Google Scholar] [CrossRef] [PubMed]
- Song, J.H.; Singh, N.; Luevano, L.A.; Padi, S.K.R.; Okumura, K.; Olive, V.; Black, S.M.; Warfel, N.A.; Goodrich, D.W.; Kraft, A.S. Mechanisms Behind Resistance to PI3K Inhibitor Treatment Induced by the PIM Kinase. Mol. Cancer Ther. 2018, 17, 2710–2721. [Google Scholar] [CrossRef] [PubMed]
- Meja, K.; Stengel, C.; Sellar, R.; Huszar, D.; Davies, B.R.; Gale, R.E.; Linch, D.C.; Khwaja, A. PIM and AKT kinase inhibitors show synergistic cytotoxicity in acute myeloid leukaemia that is associated with convergence on mTOR and MCL1 pathways. Br. J. Haematol. 2014, 167, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Koblish, H.; Li, Y.-L.; Shin, N.; Hall, L.; Wang, Q.; Wang, K.; Covington, M.; Marando, C.; Bowman, K.; Boer, J.; et al. Preclinical characterization of INCB053914, a novel pan-PIM kinase inhibitor, alone and in combination with anticancer agents, in models of hematologic malignancies. PLoS ONE 2018, 13, e0199108. [Google Scholar] [CrossRef]
- Jain, N.; Curran, E.; Iyengar, N.M.; Diaz-Flores, E.; Kunnavakkam, R.; Popplewell, L.; Kirschbaum, M.H.; Karrison, T.; Erba, H.P.; Green, M.; et al. Phase II Study of the Oral Mek Inhibitor Selumetinib in Advanced Acute Myeloid Leukemia (AML): A University of Chicago Phase II Consortium Trial. Clin. Cancer Res. 2014, 20, 490–498. [Google Scholar] [CrossRef]
- Sandhöfer, N.; Metzeler, K.H.; Rothenberg, M.; Herold, T.; Tiedt, S.; Groiß, V.; Carlet, M.; Walter, G.; Hinrichsen, T.; Wachter, O.; et al. Dual PI3K/mTOR inhibition shows antileukemic activity in MLL-rearranged acute myeloid leukemia. Leukemia 2015, 29, 828–838. [Google Scholar] [CrossRef]
- Liu, H.; Diaz-Flores, E.; Poiré, X.; Odenike, O.; Koval, G.; Malnassy, G.; Ihonor, P.; Zhang, Y.; Le Beau, M.M.; Shannon, K.; et al. Combination of a MEK Inhibitor, AZD6244, and Dual PI3K/mTOR Inhibitor, NVP-BEZ235: An Effective Therapeutic Strategy for Acute Myeloid Leukemia. Blood 2010, 116, 3978. [Google Scholar] [CrossRef]
- Juric, D.; Soria, J.-C.; Sharma, S.; Banerji, U.; Azaro, A.; Desai, J.; Ringeisen, F.P.; Kaag, A.; Radhakrishnan, R.; Hourcade-Potelleret, F.; et al. A phase 1b dose-escalation study of BYL719 plus binimetinib (MEK162) in patients with selected advanced solid tumors. JCO 2014, 32, 9051. [Google Scholar] [CrossRef]
- Antar, A.I.; Otrock, Z.K.; Jabbour, E.; Mohty, M.; Bazarbachi, A. FLT3 inhibitors in acute myeloid leukemia: Ten frequently asked questions. Leukemia 2020, 34, 682–696. [Google Scholar] [CrossRef]
- Lam, S.S.Y.; Leung, A.Y.H. Overcoming Resistance to FLT3 Inhibitors in the Treatment of FLT3-Mutated AML. Int. J. Mol. Sci. 2020, 21, 1537. [Google Scholar] [CrossRef]
- Larrosa-Garcia, M.; Baer, M.R. FLT3 inhibitors in acute myeloid leukemia: Current status and future directions. Mol. Cancer Ther. 2017, 16, 991–1001. [Google Scholar] [CrossRef]
- Schlenk, R.F.; Weber, D.; Fiedler, W.; Salih, H.R.; Wulf, G.; Salwender, H.; Schroeder, T.; Kindler, T.; Lübbert, M.; Wolf, D.; et al. Midostaurin added to chemotherapy and continued single-agent maintenance therapy in acute myeloid leukemia with FLT3-ITD. Blood 2019, 133, 840–851. [Google Scholar] [CrossRef]
- Perl, A.E.; Martinelli, G.; Cortes, J.E.; Neubauer, A.; Berman, E.; Paolini, S.; Montesinos, P.; Baer, M.R.; Larson, R.A.; Ustun, C.; et al. Gilteritinib or Chemotherapy for Relapsed or Refractory FLT3-Mutated AML. N. Engl. J. Med. 2019, 381, 1728–1740. [Google Scholar] [CrossRef]
- Lindblad, O.; Cordero, E.; Puissant, A.; Macaulay, L.; Ramos, A.; Kabir, N.N.; Sun, J.; Vallon-Christersson, J.; Haraldsson, K.; Hemann, M.T.; et al. Aberrant activation of the PI3K/mTOR pathway promotes resistance to sorafenib in AML. Oncogene 2016, 35, 5119–5131. [Google Scholar] [CrossRef] [PubMed]
- Piloto, O.; Wright, M.; Brown, P.; Kim, K.-T.; Levis, M.; Small, D. Prolonged exposure to FLT3 inhibitors leads to resistance via activation of parallel signaling pathways. Blood 2007, 109, 1643–1652. [Google Scholar] [CrossRef] [PubMed]
- Seedhouse, C.H.; Hunter, H.M.; Lloyd-Lewis, B.; Massip, A.-M.; Pallis, M.; Carter, G.I.; Grundy, M.; Shang, S.; Russell, N.H. DNA repair contributes to the drug-resistant phenotype of primary acute myeloid leukaemia cells with FLT3 internal tandem duplications and is reversed by the FLT3 inhibitor PKC412. Leukemia 2006, 20, 2130–2136. [Google Scholar] [CrossRef] [PubMed]
- Gregory, M.A.; D’Alessandro, A.; Alvarez-Calderon, F.; Kim, J.; Nemkov, T.; Adane, B.; Rozhok, A.I.; Kumar, A.; Kumar, V.; Pollyea, D.A.; et al. ATM/G6PD-driven redox metabolism promotes FLT3 inhibitor resistance in acute myeloid leukemia. Proc. Natl. Acad. Sci. USA 2016, 113, E6669–E6678. [Google Scholar] [CrossRef] [PubMed]
- Menolfi, D.; Zha, S. ATM, ATR and DNA-PKcs kinases—The lessons from the mouse models: Inhibition ≠ deletion. Cell Biosci. 2020, 10, 8. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Li, L.; Small, D.; Rassool, F. Cells expressing FLT3/ITD mutations exhibit elevated repair errors generated through alternative NHEJ pathways: Implications for genomic instability and therapy. Blood 2010, 116, 5298–5305. [Google Scholar] [CrossRef]
- Scully, R.; Panday, A.; Elango, R.; Willis, N.A. DNA double strand break repair pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 2019, 20, 698–714. [Google Scholar] [CrossRef]
- Caron, M.-C.; Sharma, A.K.; O’Sullivan, J.; Myler, L.R.; Ferreira, M.T.; Rodrigue, A.; Coulombe, Y.; Ethier, C.; Gagné, J.-P.; Langelier, M.-F.; et al. Poly(ADP-ribose) polymerase-1 antagonizes DNA resection at double-strand breaks. Nat Commun. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Chen, A. PARP inhibitors: Its role in treatment of cancer. Chin. J. Cancer 2011, 30, 463–471. [Google Scholar] [CrossRef]
- Lord, C.J.; Tutt, A.N.J.; Ashworth, A. Synthetic lethality and cancer therapy: Lessons learned from the development of PARP inhibitors. Annu. Rev. Med. 2015, 66, 455–470. [Google Scholar] [CrossRef] [PubMed]
- Lord, C.J.; Ashworth, A. PARP inhibitors: Synthetic lethality in the clinic. Science 2017, 355, 1152–1158. [Google Scholar] [CrossRef] [PubMed]
- Audeh, M.W.; Carmichael, J.; Penson, R.T.; Friedlander, M.; Powell, B.; Bell-McGuinn, K.M.; Scott, C.; Weitzel, J.N.; Oaknin, A.; Loman, N.; et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: A proof-of-concept trial. Lancet 2010, 376, 245–251. [Google Scholar] [CrossRef]
- Tutt, A.; Robson, M.; Garber, J.E.; Domchek, S.M.; Audeh, M.W.; Weitzel, J.N.; Friedlander, M.; Arun, B.; Loman, N.; Schmutzler, R.K.; et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: A proof-of-concept trial. Lancet 2010, 376, 235–244. [Google Scholar] [CrossRef]
- Zhao, L.; So, C.W.E. PARP-inhibitor-induced synthetic lethality for acute myeloid leukemia treatment. Exp. Hematol. 2016, 44, 902–907. [Google Scholar] [CrossRef]
- Gaymes, T.J.; Shall, S.; MacPherson, L.J.; Twine, N.A.; Lea, N.C.; Farzaneh, F.; Mufti, G.J. Inhibitors of poly ADP-ribose polymerase (PARP) induce apoptosis of myeloid leukemic cells: Potential for therapy of myeloid leukemia and myelodysplastic syndromes. Haematologica 2009, 94, 638–646. [Google Scholar] [CrossRef][Green Version]
- Gafencu, G.A.; Tomuleasa, C.I.; Ghiaur, G. PARP inhibitors in acute myeloid leukaemia therapy: How a synthetic lethality approach can be a valid therapeutic alternative. Med. Hypotheses 2017, 104, 30–34. [Google Scholar] [CrossRef]
- Huang, T.-T.; Lampert, E.J.; Coots, C.; Lee, J.-M. Targeting the PI3K pathway and DNA damage response as a therapeutic strategy in ovarian cancer. Cancer Treat. Rev. 2020, 86, 102021. [Google Scholar] [CrossRef]
- Kumar, A.; Fernandez-Capetillo, O.; Fernadez-Capetillo, O.; Carrera, A.C. Nuclear phosphoinositide 3-kinase beta controls double-strand break DNA repair. Proc. Natl. Acad. Sci. USA 2010, 107, 7491–7496. [Google Scholar] [CrossRef] [PubMed]
- González-Billalabeitia, E.; Seitzer, N.; Song, S.J.; Song, M.S.; Patnaik, A.; Liu, X.-S.; Epping, M.T.; Papa, A.; Hobbs, R.M.; Chen, M.; et al. Vulnerabilities of PTEN-TP53-deficient prostate cancers to compound PARP-PI3K inhibition. Cancer Discov. 2014, 4, 896–904. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, Y.H.; García-García, C.; Serra, V.; He, L.; Torres-Lockhart, K.; Prat, A.; Anton, P.; Cozar, P.; Guzmán, M.; Grueso, J.; et al. PI3K inhibition impairs BRCA1/2 expression and sensitizes BRCA-proficient triple-negative breast cancer to PARP inhibition. Cancer Discov. 2012, 2, 1036–1047. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Li, C.; Zhang, Y.; Wang, M.; Jiang, N.; Xiang, L.; Li, T.; Roberts, T.M.; Zhao, J.J.; Cheng, H.; et al. Combined inhibition of PI3K and PARP is effective in the treatment of ovarian cancer cells with wild-type PIK3CA genes. Gynecol. Oncol. 2016, 142, 548–556. [Google Scholar] [CrossRef] [PubMed]
- De, P.; Sun, Y.; Carlson, J.H.; Friedman, L.S.; Leyland-Jones, B.R.; Dey, N. Doubling down on the PI3K-AKT-mTOR pathway enhances the antitumor efficacy of PARP inhibitor in triple negative breast cancer model beyond BRCA-ness. Neoplasia 2014, 16, 43–72. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).