Mast Cells, microRNAs and Others: The Role of Translational Research on Colorectal Cancer in the Forthcoming Era of Precision Medicine
Abstract
1. Introduction
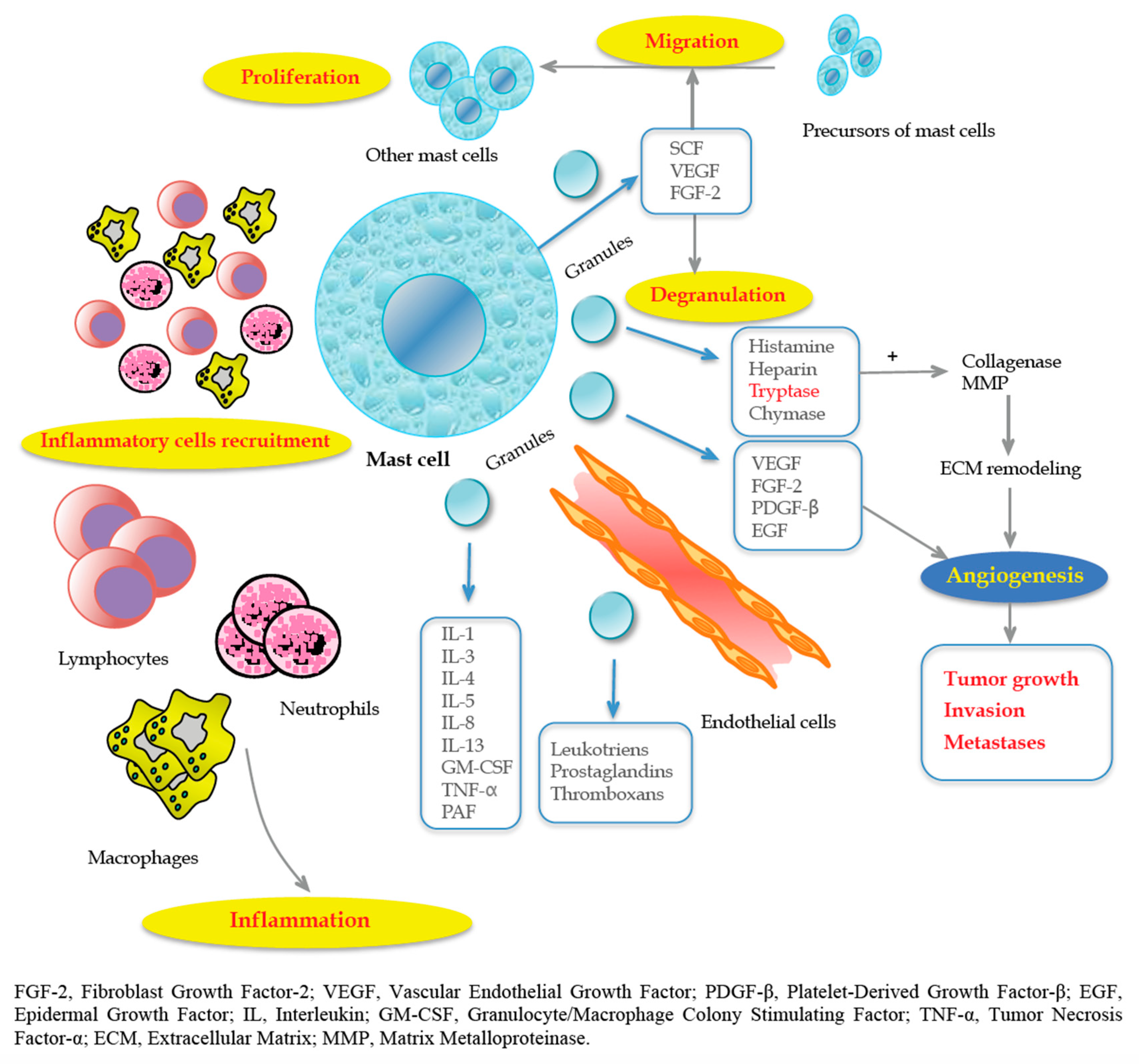
2. Mast Cells
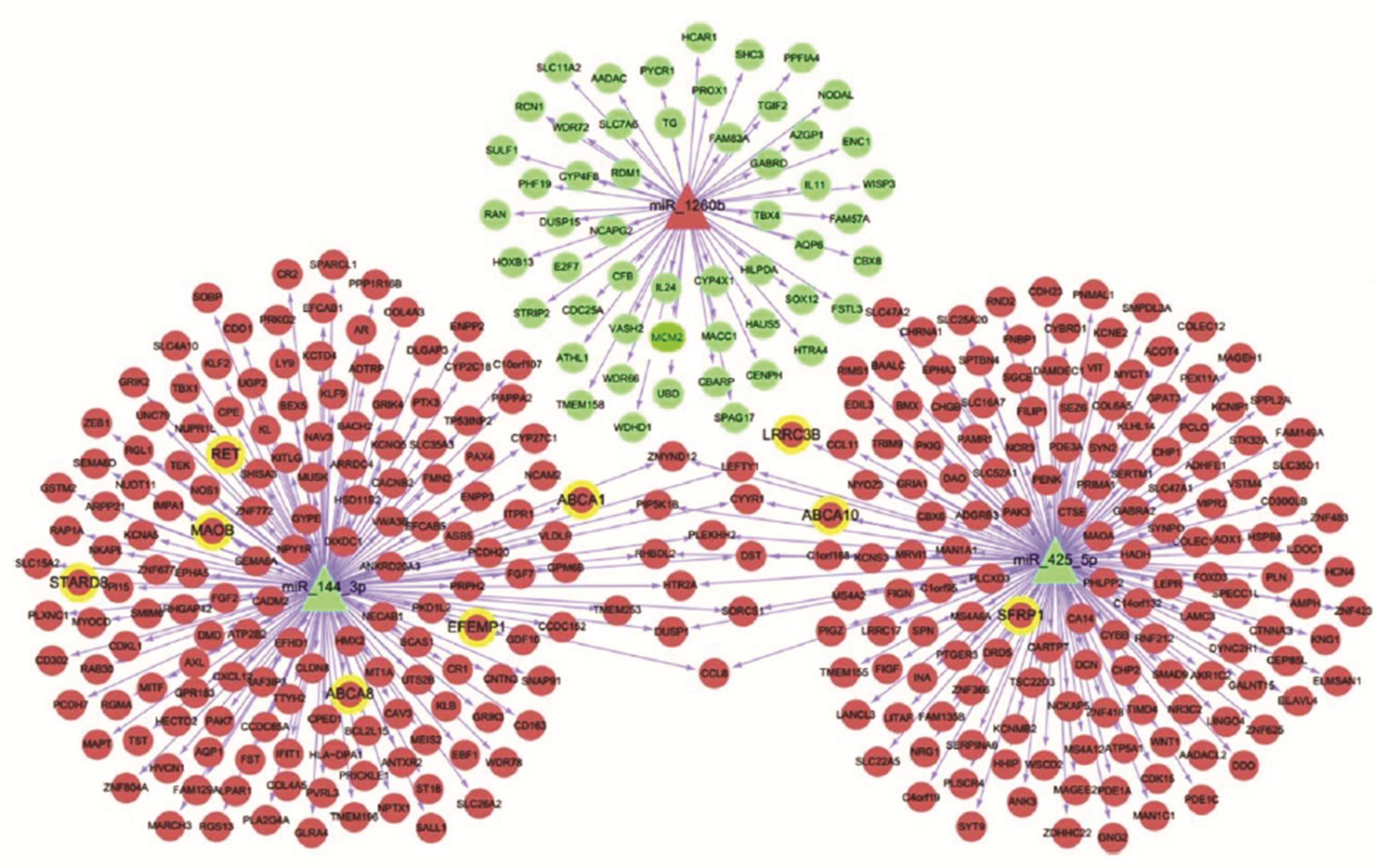
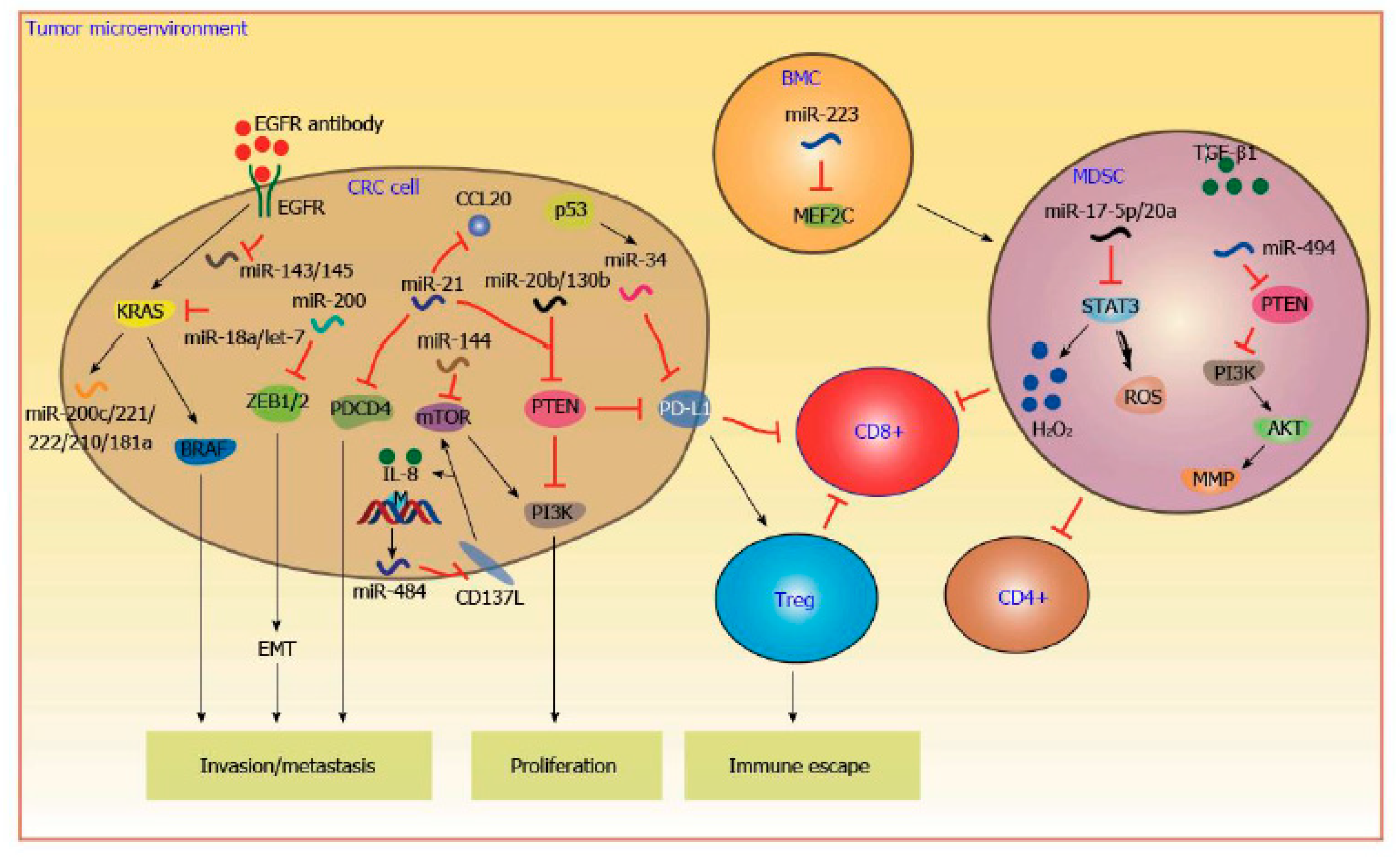
3. MicroRNA
4. KRAS
5. BRAF
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Molinari, C.; Marisi, G.; Passardi, A.; Matteucci, L.; De Maio, G.; Ulivi, P. Heterogeneity in Colorectal Cancer: A Challenge for Personalized Medicine? Int. J. Mol. Sci. 2018, 19, 3733. [Google Scholar] [CrossRef]
- Alvaro, E.; Cano, J.M.; Garcia, J.L.; Brandariz, L.; Olmedillas-Lopez, S.; Arriba, M.; Rueda, D.; Rodriguez, Y.; Canete, A.; Arribas, J.; et al. Clinical and Molecular Comparative Study of Colorectal Cancer Based on Age-of-Onset and Tumor Location: Two Main Criteria for Subclassifying Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 968. [Google Scholar] [CrossRef] [PubMed]
- Fearon, E.R.; Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 1990, 61, 759–767. [Google Scholar] [CrossRef]
- Arnold, M.; Sierra, M.S.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017, 66, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN Sources and Methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef] [PubMed]
- Gallo, G.; Sena, G.; Vescio, G.; Papandrea, M.; Sacco, R.; Trompetto, M.; Sammarco, G. The prognostic value of KRAS and BRAF in stage I-III colorectal cancer. A systematic review. Ann. Ital. Chir. 2019, 90, 127–137. [Google Scholar]
- Pellino, G.; Gallo, G.; Pallante, P.; Capasso, R.; De Stefano, A.; Maretto, I.; Malapelle, U.; Shengyang, Q.; Nikolaou, S.; Barina, A.; et al. Noninvasive Biomarkers of Colorectal Cancer: Role in Diagnosis and Personalised Treatment Perspectives. Gastroenterol. Res. Pract. 2018, 2018, 2397863. [Google Scholar] [CrossRef]
- Carethers, J.M.; Jung, B.H. Genetics and Genetic Biomarkers in Sporadic Colorectal Cancer. Gastroenterology 2015, 149, 1177–1190. [Google Scholar] [CrossRef]
- Hui, L.; Chen, Y. Tumor Microenvironment: Sanctuary of the Devil. Cancer Lett. 2015, 368, 7–13. [Google Scholar] [CrossRef]
- Gallo, G.; Kotze, P.G.; Spinelli, A. Surgery in ulcerative colitis: When? How? Best Pract. Res. Clin. Gastroenterol. 2018, 32, 71–78. [Google Scholar] [CrossRef]
- Pellino, G.; Marcellinaro, R.; Candilio, G.; De Fatico, G.S.; Guadagno, E.; Campione, S.; Santangelo, G.; Reginelli, A.; Sciaudone, G.; Riegler, G.; et al. The Experience of a Referral Centre and Literature Overview of GIST and Carcinoid Tumours in Inflammatory Bowel Diseases. Int. J. Surg. 2016, 28, S133–S141. [Google Scholar] [CrossRef] [PubMed]
- Wisselink, D.D.; Braakhuis, L.L.F.; Gallo, G.; van Grevenstein, W.M.U.; van Dieren, S.; Kok, N.F.M.; de Reuver, P.R.; Tanis, P.J.; de Hingh, I.H.J.T. Systematic review of published literature on Oxaliplatin and Mitomycin C as chemotherapeutic agents for Hyperthermic Intraperitoneal Chemotherapy in patients with peritoneal metastases from colorectal cancer. Crit. Rev. Oncol. Hematol. 2019, 142, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Rawson, J.B.; Bapat, B. Epigenetic Biomarkers in Colorectal Cancer Diagnostics. Expert Rev. Mol. Diagn. 2012, 12, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Greegor, D.H. Diagnosis of Large-Bowel Cancer in the Asymptomatic Patient. JAMA 1967, 201, 943–945. [Google Scholar] [CrossRef] [PubMed]
- Sidransky, D.; Tokino, T.; Hamilton, S.R.; Kinzler, K.W.; Levin, B.; Frost, P.; Vogelstein, B. Identification of Ras oncogene mutations in the stool of patients with curable colorectal tumors. Science 1992, 256, 102–105. [Google Scholar] [CrossRef]
- Von Karsa, L.; Patnick, J.; Segnan, N.; Atkin, W.; Halloran, S.; Lansdorp-Vogelaar, I.; Malila, N.; Minozzi, S.; Moss, S.; Quirke, P.; et al. European Guidelines for Quality Assurance in Colorectal Cancer Screening and Diagnosis: Overview and Introduction to the Full Supplement Publication. Endoscopy 2013, 45, 51–59. [Google Scholar]
- Rosman, A.S.; Korsten, M.A. Meta-analysis comparing CT colonography, air contrast barium enema, and colonoscopy. Am. J. Med. 2007, 120, 203–210. [Google Scholar] [CrossRef]
- Rockey, D.C.; Paulson, E.; Niedzwiecki, D.; Davis, W.; Bosworth, H.B.; Sanders, L.; Yee, J.; Henderson, J.; Hatten, P.; Burdick, S.; et al. Analysis of air contrast barium enema, computed tomographic colonography, and colonoscopy: Prospective comparison. Lancet 2005, 365, 305–311. [Google Scholar] [CrossRef]
- Graser, A.; Stieber, P.; Nagel, D.; Schäfer, C.; Horst, D.; Becker, C.R.; Nikolaou, K.; Lottes, A.; Geisbüsch, S.; Kramer, H.; et al. Comparison of CT colonography, colonoscopy, sigmoidoscopy and faecal occult blood tests for the detection of advanced adenoma in an average risk population. Gut 2009, 58, 241–248. [Google Scholar] [CrossRef]
- Gallo, G. Preoperative Colorectal-Cancer Detection: Do We Need Anything Else? An Invited Brief Commentary on Is CT Scan More Accurate than Endoscopy in Identifying Distance from the Anal Verge for Left-sided Colon Cancer? A Comparative Cohort Analysis. J. Investig. Surg. 2020, 33, 281–282. [Google Scholar] [CrossRef]
- Ehrlich, P. Beiträge zur Theorie und Praxis der Histologischen Färbung. In The Collected Papers of Paul Ehrlich; Elsevier: Pergamon, Turkey, 2013; pp. 29–64. [Google Scholar]
- Crivellato, E.; Beltrami, C.A.; Mallardi, F.; Ribatti, D. Paul Ehrlich’s doctoral thesis: A milestone in the study of mast cells. Br. J. Haematol. 2003, 123, 19–21. [Google Scholar] [PubMed]
- Blank, U. The mechanism of exocytosis in Mast Cells. Adv. Exp. Med. Biol. 2011, 716, 107–122. [Google Scholar] [PubMed]
- Wernersson, S.; Pejler, G. Mast cell secretory granules: Armed for battle. Nat. Rev. Immunol. 2014, 14, 478–494. [Google Scholar] [CrossRef] [PubMed]
- Silver, R.; Silverman, A.J.; Vitkovic, L.; Lederhendler, I.I. Mast cells in the brain: Evidence and functional significance. Trends. Neurosci. 1996, 19, 25–31. [Google Scholar] [CrossRef]
- Okayama, Y.; Kawakami, T. Development, Migration, and Survival of Mast Cells. Immunol. Res. 2006, 34, 97–115. [Google Scholar] [CrossRef]
- Westphal, E. Uber Mastzellen. In Farbenanalytische Untersuchungen; Ehrlich, E., Ed.; Hirschwald: Berlin, Germany, 1891; pp. 17–41. [Google Scholar]
- Conti, P.; Castellani, M.L.; Kempuraj, D.; Salini, V.; Vecchiet, J.; Tetè, S.; Mastrangelo, F.; Perrella, A.; De Lutiis, M.A.; Tagen, M.; et al. Role of mast cells in tumor growth. Ann. Clin. Lab. Sci. 2007, 37, 315–322. [Google Scholar] [PubMed]
- Acikalin, M.F.; Oner, U.; Topcu, I.; Yasar, B.; Kiper, H.; Colak, E. Tumour angiogenesis and mast cell density in the prognostic assessment of colorectal carcinomas. Dig. Liver Dis. 2005, 37, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Melillo, R.M.; Guarino, V.; Avilla, E.; Galdiero, M.R.; Liotti, F.; Prevete, N.; Rossi, F.W.; Basolo, F.; Ugolini, C.; de Paulis, A.; et al. Mast cells have a protumorigenic role in human thyroid cancer. Oncogene 2010, 29, 6203–6215. [Google Scholar] [CrossRef] [PubMed]
- Ammendola, M.; Sacco, R.; Donato, G.; Zuccala, V.; Russo, E.; Luposella, M.; Vescio, G.; Rizzuto, A.; Patruno, R.; De Sarro, G.; et al. Mast cell positivity to tryptase correlates with metastatic lymph nodes in gastrointestinal cancer patients treated surgically. Oncology 2013, 85, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Malfettone, A.; Silvestris, N.; Saponaro, C.; Ranieri, G.; Russo, A.; Caruso, S.; Popescu, O.; Simone, G.; Paradiso, A.; Mangia, A. High density of tryptase-positive mast cells in human colorectal cancer: A poor prognostic factor related to protease-activated receptor 2 expression. J. Cell. Mol. Med. 2013, 17, 1025–1037. [Google Scholar] [CrossRef] [PubMed]
- Ammendola, M.; Rosario, S.; Giuseppe, S.; Giuseppe, D.; Montemurro, S.; Ruggieri, E.; Patruno, R.; Marech, I.; Cariello, M.; Vacca, A.; et al. Correlation between serum tryptase, mast cells positive to tryptase and microvascular density in colo-rectal cancer patients: Possible biological-clinical significance. PLoS ONE 2014, 9, e99512. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Yang, S.; Shen, P.; Wu, B.; Sun, T.; Sun, H.; Ji, F.; Zhou, D. SCF/c-KIT signaling promotes mucus secretion of colonic goblet cells and development of mucinous colorectal adenocarcinoma. Am. J. Cancer Res. 2018, 8, 1064–1073. [Google Scholar] [PubMed]
- Marech, I.; Ammendola, M.; Gadaleta, C.; Zizzo, N.; Oakley, C.; Gadaleta, C.D.; Ranieri, G. Possible biological and translational significance of mast cells density in colorectal cancer. World J. Gastroenterol. 2014, 20, 8910–8920. [Google Scholar] [PubMed]
- Yu, Y.; Blokhuis, B.; Derks, Y.; Kumari, S.; Garssen, J.; Redegeld, F. Human mast cells promote colon cancer growth via bidirectional crosstalk: Studies in 2D and 3D coculture models. Oncoimmunology 2018, 7, e1504729. [Google Scholar] [CrossRef]
- Coussens, L.M.; Raymond, W.W.; Bergers, G.; Laig-Webster, M.; Behrendtsen, O.; Werb, Z.; Caughey, G.H.; Hanahan, D. Inflammatory mast cells up-regulate angiogenesis during squamous epithelial carcinogenesis. Genes Dev. 1999, 13, 1382–1397. [Google Scholar] [CrossRef] [PubMed]
- Cimpean, A.M.; Tamma, R.; Ruggieri, S.; Nico, B.; Toma, A.; Ribatti, D. Mast cells in breast cancer angiogenesis. Crit. Rev. Oncol. Hematol. 2017, 115, 23–26. [Google Scholar] [CrossRef]
- Folkman, J. What is the evidence that tumors are angiogenesis dependent? J. Natl. Cancer Inst. 1990, 82, 4–6. [Google Scholar] [CrossRef]
- Engel, C.J.; Bennett, S.T.; Chambers, A.F.; Doig, G.S.; Kerkvliet, N.; O’Malley, F.P. Tumor angiogenesis predicts recurrence in invasive colorectal cancer when controlled for Dukes staging. Am. J. Surg. Pathol. 1996, 20, 1260–1265. [Google Scholar] [CrossRef]
- Frank, R.E.; Saclarides, T.J.; Leurgans, S.; Speziale, N.J.; Drab, E.A.; Rubin, D.B. Tumor angiogenesis as a predictor of recurrence and survival in patients with node-negative colon cancer. Ann. Surg. 1995, 222, 695–699. [Google Scholar] [CrossRef]
- Detoraki, A.; Staiano, R.I.; Granata, F.; Giannattasio, G.; Prevete, N.; de Paulis, A.; Ribatti, D.; Genovese, A.; Triggiani, M.; Marone, G. Vascular endothelial growth factors synthesized by human lung mast cells exert angiogenic effects. J. Allergy Clin. Immunol. 2009, 123, 1142–1149. [Google Scholar] [CrossRef]
- Sammarco, G.; Varricchi, G.; Ferraro, V.; Ammendola, M.; De Fazio, M.; Altomare, D.F.; Luposella, M.; Maltese, L.; Currò, G.; Marone, G.; et al. Mast Cells, Angiogenesis and Lymphangiogenesis in Human Gastric Cancer. Int. J. Mol. Sci. 2019, 20, 2106. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Zhai, L.; Xue, R.; Shi, J.; Zeng, Q.; Gao, C. Mast cell tryptase contributes to pancreatic cancer growth through promoting angiogenesis via activation of angiopoietin-1. Int. J. Mol. Sci. 2016, 17, 834. [Google Scholar] [CrossRef]
- Payne, V.; Kam, P.C.A. Mast cell tryptase: A review of its physiology and clinical significance. Anaesthesia 2004, 59, 695–703. [Google Scholar] [CrossRef] [PubMed]
- Marichal, T.; Tsai, M.; Galli, S.J. Mast cells: Potential positive and negative roles in tumor biology. Cancer Immunol. Res. 2013, 1, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Adams, W.J.; Morris, D.L. Short-course cimetidine and survival with colorectal cancer. Lancet 1994, 344, 1768–1769. [Google Scholar] [CrossRef]
- Matsumoto, S. Cimetidine and survival with colorectal cancer. Lancet 1995, 346, 115. [Google Scholar] [CrossRef]
- Gulubova, M.; Vlaykova, T. Prognostic significance of mast cell number and microvascular density for the survival of patients with primary colorectal cancer. J. Gastroenterol. Hepatol. 2009, 24, 1265–1275. [Google Scholar] [CrossRef]
- Ammendola, M.; Zuccalà, V.; Patruno, R.; Russo, E.; Luposella, M.; Amorosi, A.; Vescio, G.; Sammarco, G.; Montemurro, S.; De Sarro, G.; et al. Tryptase-positive mast cells and angiogenesis in keloids: A new possible post-surgical target for prevention. Updates Surg. 2013, 65, 53–57. [Google Scholar] [CrossRef]
- Patruno, R.; Marech, I.; Zizzo, N.; Ammendola, M.; Nardulli, P.; Gadaleta, C.; Introna, M.; Capriuolo, G.; Rubini, R.A.; Ribatti, D.; et al. c-Kit expression, angiogenesis, and grading in canine mast cell tumour: A unique model to study c-Kit driven human malignancies. BioMed. Res. Int. 2014, 2014, 730246. [Google Scholar] [CrossRef]
- Ammendola, M.; Sacco, R.; Sammarco, G.; Luposella, M.; Patruno, R.; Gadaleta, C.D.; De Sarro, G.; Ranieri, G. Mast Cell-Targeted Strategies in Cancer Therapy. Transfus. Med. Hemother. 2016, 43, 109–113. [Google Scholar] [CrossRef]
- Ferrero, G.; Cordero, F.; Tarallo, S.; Arigoni, M.; Riccardo, F.; Gallo, G.; Ronco, G.; Allasia, M.; Kulkarni, N.; Matullo, G.; et al. Small non-coding RNA profiling in human biofluids and surrogate tissues from healthy individuals: Description of the diverse and most represented species. Oncotarget 2017, 9, 3097–3111. [Google Scholar] [CrossRef] [PubMed]
- Schetter, A.J.; Leung, S.Y.; Sohn, J.J.; Zanetti, K.A.; Bowman, E.D.; Yanaihara, N.; Yuen, A.T.; Chan, T.L.; Kwong, D.L.W.; Au, G.K.H.; et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA 2008, 299, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, M.; Calin, G.A. Epigenetics and miRNAs in human cancer. Adv. Genet. 2010, 70, 87–99. [Google Scholar]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. Elegans Heterochronic Gene lin-4 Encodes Small RNAs with Antisense Complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional Regulation of the Heterochronic Gene lin-14 by lin-4 Mediates Temporal Pattern Formation in C. Elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs-microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef]
- Ambros, V.; Bartel, B.; Bartel, D.P.; Burge, C.B.; Carrington, J.C.; Chen, X.; Dreyfuss, G.; Eddy, S.R.; Griffiths-Jones, S.; Mashall, M.; et al. A uniform system for microRNA annotation. RNA 2003, 9, 277–279. [Google Scholar] [CrossRef]
- Mendell, J.T.; Olson, E.N. MicroRNAs in stress signaling and human disease. Cell 2012, 148, 1172–1187. [Google Scholar] [CrossRef]
- Tan, Y.; Lin, J.J.; Yang, X.; Gou, D.M.; Fu, L.; Li, F.R.; Yu, X.F. A panel of three plasma microRNAs for colorectal cancer diagnosis. Cancer Epidemiol. 2019, 60, 67–76. [Google Scholar] [CrossRef]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef]
- Poursheikhani, A.; Abbaszadegan, M.R.; Nokhandani, N.; Kerachian, M.A. Integration analysis of long non-coding RNA (lncRNA) role in tumorigenesis of colon adenocarcinoma. BMC Med. Genom. 2020, 13, 108. [Google Scholar] [CrossRef] [PubMed]
- Sipos, F.; Galamb, O. Epithelial-to-mesenchymal and mesenchymal-toepithelial transitions in the colon. World J. Gastroenterol. 2012, 18, 601–608. [Google Scholar] [CrossRef] [PubMed]
- Fish, J.E.; Santoro, M.M.; Morton, S.U.; Yu, S.; Yeh, R.F.; Wythe, J.D.; Ivey, K.N.; Bruneau, B.G.; Stainer, D.Y.; Srivastava, D. miR-126 regulates angiogenic signaling and vascular integrity. Dev. Cell 2008, 105, 1516–1521. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Croce, C.M. MicroRNA-cancer connection: The beginning of a new tale. Cancer Res. 2006, 66, 7390–7394. [Google Scholar] [CrossRef]
- Chauhan, N.; Dasmana, A.; Jaggi, M.; Chauhan, S.C.; Yallapu, M.M. miR-205: A Potential Biomedicine for Cancer Therapy. Cells 2020, 9, 1957. [Google Scholar] [CrossRef]
- Li, X.; Nie, J.; Mei, Q.; Han, W.D. MicroRNAs: Novel immunotherapeutic targets in colorectal carcinoma. World J. Gastroenterol. 2016, 22, 5317–5331. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef]
- Hollis, M.; Nair, K.; Vyas, A.; Chaturvedi, L.S.; Gambhir, S.; Vyas, D. MicroRNAs potential utility in colon cancer: Early detection, prognosis, and chemosensitivity. World J. Gastroenterol. 2015, 21, 8284–8292. [Google Scholar] [CrossRef]
- Ghareib, A.F.; Mohamed, R.H.; Abd El-Fatah, A.R.; Saadawy, S.F. Assessment of Serum MicroRNA-21 Gene Expression for Diagnosis and Prognosis of Colorectal Cancer. J. Gastrointest. Cancer 2020, 51, 818–823. [Google Scholar] [CrossRef]
- Thomas, A.M.; Manghi, P.; Asnicar, F.; Pasolli, E.; Armanini, F.; Moreno, Z.; Beghini, F.; Manara, S.; Karcher, N.; Pozzi, C.; et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat. Med. 2019, 25, 667–678. [Google Scholar] [CrossRef]
- Nagel, R.; le Sage, C.; Diosdado, B.; van der Waal, M.; Oude Vrielink, J.A.F.; Brolijn, A.; Meijer, G.A.; Agami, R. Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res. 2008, 68, 5795–5802. [Google Scholar] [CrossRef]
- Fang, Z.; Tang, J.; Bai, Y.; Lin, H.; You, H.; Jin, H.; Lin, L.; You, P.; Li, J.; Dai, Z.; et al. Plasma levels of microRNA-24, microRNA-320a, and microRNA-423-5p are potential biomarkers for colorectal carcinoma. J. Exp. Clin. Cancer Res. 2015, 34, 86. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Weiner, H.L. Control of the gut microbiome by fecal microRNA. Microb. Cell 2016, 3, 176–177. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, F.E.; Ahmed, N.C.; Vos, P.W.; Bonnerup, C.; Atkins, J.N.; Casey, M.; Nuovo, G.J.; Naziri, W.; Wiley, J.E.; Mota, H.; et al. Diagnostic microRNA markers to screen for sporadic human colon cancer in stool: I. Proof of principle. Cancer Genom. Proteom. 2013, 10, 93–113. [Google Scholar]
- Ahlquist, D.A. Molecular detection of colorectal neoplasia. Gastroenterology 2010, 138, 2127–2139. [Google Scholar] [CrossRef] [PubMed]
- Tarallo, S.; Ferrero, G.; Gallo, G.; Francavilla, A.; Clerico, G.; Realis Luc, A.; Manghi, P.; Thomas, A.M.; Vineis, P.; Segata, N.; et al. Altered Fecal Small RNA Profiles in Colorectal Cancer Reflect Gut Microbiome Composition in Stool Samples. Msystems 2019, 4, e00289–e00319. [Google Scholar] [CrossRef] [PubMed]
- Koga, Y.; Yasunaga, M.; Takahashi, A.; Kuroda, J.; Moriya, Y.; Akasu, T.; Fujita, S.; Yamamoto, S.; Baba, H.; Matsumura, Y. MicroRNA expression profiling of exfoliated colonocytes isolated from feces for colorectal cancer screening. Cancer Prev. Res. 2010, 3, 1435–1442. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.W.; Ng, S.S.; Dong, Y.J.; Ng, S.C.; Leung, W.W.; Lee, C.W.; Wong, Y.N.; Chan, F.K.L.; Sung, J.J.Y. Detection of miR-92a and miR-21 in stool samples as potential screening biomarkers for colorectal cancer and polyps. Gut 2012, 61, 739–745. [Google Scholar] [CrossRef]
- Link, A.; Balaguer, F.; Shen, Y.; Nagasaka, T.; Lozano, J.J.; Boland, C.R.; Goel, A. Fecal MicroRNAs as novel biomarkers for colon cancer screening. Cancer Epidemiol. Biomark. Prev. 2010, 19, 1766–1774. [Google Scholar] [CrossRef]
- Sarshar, M.; Scribano, D.; Ambrosi, C.; Palamara, A.T.; Masotti, A. Fecal microRNAs as Innovative Biomarkers of Intestinal Diseases and Effective Players in Host-Microbiome Interactions. Cancers 2020, 12, 2174. [Google Scholar] [CrossRef] [PubMed]
- Michael, M.Z.; O’Connor, S.M.; van Holst Pellekaan, N.G.; Young, G.P.; James, R.J. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol. Cancer Res. 2003, 1, 882–891. [Google Scholar] [PubMed]
- Ng, E.K.; Tsang, W.P.; Ng, S.S.M.; Jin, H.C.; Yu, J.; Li, J.J.; Rocken, C.; Ebert, M.P.A.; Kwok, T.T.; Sung, J.J.Y. MicroRNA-143 targets DNA methyltransferases 3A in colorectal cancer. Br. J. Cancer 2009, 101, 699–706. [Google Scholar] [CrossRef] [PubMed]
- Bandrés, E.; Cubedo, E.; Agirre, X.; Malumbres, R.; Zarate, R.; Ramirez, N.; Abajo, A.; Navarro, A.; Moreno, I.; Monzo, M.; et al. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol. Cancer 2006, 5, 29. [Google Scholar] [CrossRef]
- Fabbri, M.; Paone, A.; Calore, F.; Galli, R.; Gaudio, E.; Santhanam, R.; Lovat, F.; Fadda, P.; Mao, C.; Nuovo, G.J.; et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. USA 2012, 109, E2110–E2116. [Google Scholar] [CrossRef]
- Schepeler, T.; Reinert, J.T.; Ostenfeld, M.S.; Christensen, L.L.; Silahtaroglu, A.N.; Dyrskjot, L.; Wiuf, C.; Sorensen, F.J.; Kruhoffer, M.; Laurberg, S.; et al. Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res. 2008, 68, 6416–6424. [Google Scholar] [CrossRef]
- Milanesi, E.; Dobre, M.; Bucuroiu, A.I.; Herlea, V.; Manuc, T.E.; Salvi, E.; De Petro, G.; Manuc, M.; Becheanu, G. miRNAs-Based Molecular Signature for KRAS Mutated and Wild Type Colorectal Cancer: An Explorative Study. J. Immunol. Res. 2020, 2020, 4927120. [Google Scholar] [CrossRef]
- Gasparello, J.; Papi, C.; Allegretti, M.; Giordani, E.; Carboni, F.; Zazza, S.; Pescarmona, E.; Romania, P.; Giacomini, P.; Scapoli, C.; et al. A distinctive microRNA (miRNA) SIgature in the Blood of Colorectal Cancer (CRC) Patients at Surgery. Cancers 2020, 12, 2410. [Google Scholar] [CrossRef]
- Caramés, C.; Cristóbal, I.; Moreno, V.; del Puerto, L.; Moreno, I.; Rodriguez, M.; Marín, J.P.; Correa, A.V.; Hernández, R.; Zenzola, V.; et al. MicroRNA-21 predicts response to preoperative chemoradiotherapy in locally advanced rectal cancer. Int. J. Colorectal Dis. 2015, 30, 899–906. [Google Scholar] [CrossRef] [PubMed]
- Kjersem, J.B.; Ikdahl, T.; Lingjaerde, O.C.; Guren, T.; Tveit, K.M.; Kure, E.H. Plasma microRNAs predicting clinical outcome in metastatic colorectal cancer patients receiving first-line oxaliplatin-based treatment. Mol. Oncol. 2014, 8, 59–67. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, S.J.; Choo, J.; Heo, J.; Yoo, J.W.; Jung, Y.; Ree, S.H.; Im, E. miR-23a-3p is a Key Regulator of IL-17C-Induced Tumor Angiogenesis in Colorectal Cancer. Cells 2020, 9, 1363. [Google Scholar] [CrossRef] [PubMed]
- Amerizadeh, F.; Khazaei, M.; Maftouh, M.; Mardani, R.; Bahrami, A. miRNA Targeting Angiogenesis as a Potential Therapeutic Approach in the Treatment of Colorectal Cancers. Curr. Pharm. Des. 2018, 24, 4668–4674. [Google Scholar] [CrossRef] [PubMed]
- Ruzzo, A.; Graziano, F.; Vincenzi, B.; Canestrari, E.; Perrone, G.; Galluccio, N.; Catalano, V.; Loupakis, F.; Rabitti, C.; Santini, D.; et al. High let-7° microRNA levels in KRAS-mutated colorectal carcinomas may rescue anti-EGFR therapy effects in patients with chemotherapyrefractory metastatic disease. Oncologist 2012, 17, 823–829. [Google Scholar] [CrossRef] [PubMed]
- Valeri, N.; Gasparini, P.; Braconi, C.; Paone, A.; Lovat, F.; Fabbri, M.; Sumani, K.M.; Alder, H.; Amadori, D.; Patel, T.; et al. Croce CM. MicroRNA-21 induces resistance to 5-fluorouracil by down-regulating human DNA MutS homolog 2 (hMSH2). Proc. Natl. Acad. Sci. USA 2010, 107, 21098–21103. [Google Scholar] [CrossRef]
- Wagenaar, T.R.; Zabludoff, S.; Ahn, S.M.; Allerson, C.; Arlt, H.; Baffa, R.; Cao, H.; Davis, S.; Garcia-Echeverria, C.; Gaur, R.; et al. Anti-miR-21 Suppresses Hepatocellular Carcinoma Growth via Broad Transcriptional Network Deregulation. Mol. Cancer Res. 2015, 13, 1009–1021. [Google Scholar] [CrossRef]
- Nishida, N.; Yamashita, S.; Mimori, K.; Sudo, T.; Tanaka, F.; Shibata, K.; Yamamoto, H.; Ishii, H.; Doki, Y.; Mori, M. MicroRNA-10b is a prognostic indicator in colorectal cancer and confers resistance to the chemotherapeutic agent 5-fluorouracil in colorectal cancer cells. Ann. Surg. Oncol. 2012, 19, 3065–3071. [Google Scholar] [CrossRef] [PubMed]
- Poorebrahim, M.; Sadeghi, S.; Ghanbarian, M.; Kalhor, H.; Mehrtash, A.; Teimoori-Toolabi, L. Identification of candidate genes and miRNAs for sensitizing resistant colorectal cancer cells to oxaliplatin and irinotecan. Cancer Chemother. Pharmacol. 2020, 85, 153–171. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Medarde, A.; Santos, E. Ras in cancer and developmental diseases. Genes Cancer 2011, 2, 344–358. [Google Scholar] [CrossRef] [PubMed]
- Ciombor, K.K.; Bekaii-Saab, T. A Comprehensive Review of Sequencing and Combination Strategies of Targeted Agents in Metastatic Colorectal Cancer. Oncologist 2018, 23, 25–34. [Google Scholar] [CrossRef]
- Cox, A.D.; Fesik, S.W.; Kimmelman, A.C.; Luo, J.; Der, C.J. Drugging the undruggable RAS: Mission possible? Nat. Rev. Drug. Discov. 2014, 13, 828–851. [Google Scholar] [CrossRef]
- Irahara, N.; Baba, Y.; Nosho, K.; Shima, K.; Yan, L.; Dias-Santagata, D.; Iafrate, A.J.; Fuchs, C.S.; Haigis, K.M.; Ogino, S. NRAS mutations are rare in colorectal cancer. Diagn. Mol. Pathol. 2010, 19, 157–163. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Thorrez, L.; Siegfried, G.; Meulemans, S.; Evrard, S.; Tejpar, S.; Khatib, A.-M.; Creemers, J.W.M. The proprotein convertase furin is a pro-oncogenic driver in KRAS and BRAF driven colorectal cancer. Oncogene 2020, 39, 3571–3587. [Google Scholar] [CrossRef] [PubMed]
- Yokota, T.; Shibata, N.; Ura, T.; Takahari, D.; Shitara, K.; Muro, K.; Yatabe, Y. Cycleave polymerase chain reaction method is practically applicable for V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS)/V-raf murine sarcoma viral oncogene homolog B1 (BRAF) genotyping in colorectal cancer. Transl. Res. 2010, 156, 98–105. [Google Scholar] [PubMed]
- Smakman, N.; Borel Rinkes, I.H.M.; Voest, E.E.; Kranenburg, O. Control of colorectal metastasis formation by K-Ras. Biochim. Biophys. Acta 2005, 1756, 103–114. [Google Scholar] [CrossRef]
- Allgayer, H.; Wang, H.; Shirasawa, S.; Sasazuki, T.; Boyd, D. Targeted disruption of the K-ras oncogene in an invasive colon cancer cell line down-regulates urokinase receptor expression and plasminogen-dependent proteolysis. Br. J. Cancer 1999, 80, 1884–1891. [Google Scholar] [CrossRef][Green Version]
- Di Mauro, C.; Pesapane, A.; Formisano, L.; Rosa, R.; D’amato, V.; Ciciola, P.; Servetto, A.; Marciano, R.; Orsini, R.C.; Monteleone, F.; et al. Urokinase-type plasminogen acrivator receptor (uPAR) expression enhances invasion and metastasis in RAS mutated tumors. Sci. Rep. 2017, 7, 9388. [Google Scholar] [CrossRef]
- Yamamoto, H.; Itoh, F.; Senota, A.; Adachi, Y.; Yoshimoto, M.; Endoh, T.; Hinoda, Y.; Yachi, A.; Imai, K. Expression of matrix metalloproteinase matrilysin (MMP-7) was induced by activated Ki-ras via AP-1 activation in SW1417 colon cancer cells. J. Clin. Lab. Anal. 1995, 9, 297–301. [Google Scholar] [CrossRef]
- Samowitz, W.S.; Curtin, K.; Schaffer, D.; Robertson, M.; Leppert, M.; Slattery, M.L. Relationship of Ki-ras mutations in colon cancers to tumor location, stage, and survival: A population-based study. Cancer Epidemiol. Biomark. Prev. 2000, 9, 1193–1197. [Google Scholar]
- Bazan, V.; Migliavacca, M.; Zanna, I.; Tubiolo, C.; Grassi, N.; Latteri, M.A.; La Farina, M.; Albanese, I.; Dardanoni, G.; Salerno, S.; et al. Specific codon 13 K-ras mutations are predictive of clinical outcome in colorectal cancer patients, whereas codon 12 K-ras mutations are associated with mucinous histotype. Ann. Oncol. 2002, 13, 1438–1446. [Google Scholar] [CrossRef]
- Jiang, Y.; Kimchi, E.T.; Staveley-O’Carroll, K.F.; Cheng, H.; Ajani, J.A. Assessment of K-ras mutation: A step toward personalized medicine for patients with colorectal cancer. Cancer 2009, 115, 3609–3617. [Google Scholar] [CrossRef]
- Binefa, G.; Rodriguez-Moranta, F.; Teule, A.; Medina-Hayas, M. Colorectal cancer: From prevention to personalized medicine. World J. Gastroenterol. 2014, 20, 6786–6808. [Google Scholar] [CrossRef]
- Rose, J.S.; Serna, D.S.; Martin, L.K.; Li, X.; Weatherby, L.M.; Abdel-Misih, S.; Zhao, W.; Bekaii-Saab, T. Influence of KRAS mutation status in metachronous and synchronous metastatic colorectal adenocarcinoma. Cancer 2012, 118, 6243–6252. [Google Scholar] [CrossRef]
- Charlton, M.E.; Kahl, A.R.; Greenbaum, A.A.; Karlitz, J.J.; Lin, C.; Lynch, C.F.; Chen, V.W. KRAS Testing, Tumor Location, and Survival in Patients with Stage IV Colorectal Cancer: SEER 2010–2013. J. Natl. Compr. Cancer Netw. 2017, 15, 1484–1493. [Google Scholar] [CrossRef]
- Tsilimigras, D.I.; Ntanasis-Stathopoulos, I.; Bagante, F.; Moris, D.; Cloyd, J.; Spartalis, E.; Pawlik, T.M. Clinical significance and prognostic relevance of KRAS, BRAF, PI3K and TP53 genetic mutation analysis for resectable and unresectable colorectal liver metastases: A systematic review of the current evidence. Surg. Oncol. 2018, 27, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Denbo, J.W.; Yamashita, S.; Passot, G.; Egger, M.; Chun, Y.S.; Kopetz, S.E.; Maru, D.; Brudvik, K.W.; Wei, S.H.; Conrad, C.; et al. RAS Mutation Is Associated with Decreased Survival in Patients Undergoing Repeat Hepatectomy for Colorectal Liver Metastases. J. Gastrointest. Surg. 2017, 21, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Lo Nigro, C.; Ricci, V.; Vivenza, D.; Granetto, C.; Fabozzi, T.; Miraglio, E.; Merlano, M.C. Prognostic and predictive biomarkers in metastatic colorectal cancer anti-EGFR therapy. World J. Gastroenterol. 2016, 22, 6944–6954. [Google Scholar] [CrossRef] [PubMed]
- Ulz, P.; Heitzer, E.; Geigl, J.B.; Speicher, M.R. Patient monitoring through liquid biopsies using circulating tumor DNA. Int. J. Cancer 2017, 141, 887–896. [Google Scholar] [CrossRef]
- Oellerich, M.; Schutz, E.; Beck, J.; Walson, P.D. Circulating Cell-Free DNA-Diagnostic and Prognostic Applications in Personalized Cancer Therapy. Ther. Drug. Monit. 2019, 41, 115–120. [Google Scholar] [CrossRef]
- Furuki, H.; Yamada, T.; Takahashi, G.; Iwai, T.; Koizumi, M.; Shinji, S.; Yokoyama, Y.; Takeda, K.; Taniai, N.; Uchida, E. Evaluation of liquid biopsies for detection of emerging mutated genes in metastatic colorectal cancer. Eur. J. Surg. Oncol. 2018, 44, 975–982. [Google Scholar] [CrossRef]
- Fearon, E.R. Molecular genetics of colorectal cancer. Annu. Rev. Pathol. 2011, 6, 479–507. [Google Scholar] [CrossRef]
- Sepulveda, A.R.; Hamilton, S.R.; Allegra, C.J.; Grody, W.; Cushman-Vokoun, A.M.; Funkhouser, W.K.; Kopetz, S.E.; Lieu, C.; Lindor, N.M.; Minsky, B.D.; et al. Molecular Biomarkers for the Evaluation of Colorectal Cancer: Guideline Summary From the American Society for Clinical Pathology, College of American Pathologists, Association for Molecular Pathology, and American Society of Clinical Oncology. J. Oncol. Pract. 2017, 13, 333–337. [Google Scholar] [CrossRef] [PubMed]
- Sanz-Garcia, E.; Argiles, G.; Elez, E.; Tabernero, J. BRAF mutant colorectal cancer: Prognosis, treatment, and new perspectives. Ann. Oncol. 2017, 28, 2648–2657. [Google Scholar] [CrossRef] [PubMed]
- Barras, D. BRAF Mutation in Colorectal Cancer: An Update. Biomark. Cancer 2015, 7, 9–12. [Google Scholar] [CrossRef] [PubMed]
- Matallanas, D.; Birtwistle, M.; Romano, D.; Zebisch, A.; Rauch, J.; von Kriegsheim, A.; Kolch, W. Raf family kinases: Old dogs have learned new tricks. Genes Cancer 2011, 2, 232–260. [Google Scholar] [CrossRef]
- Wan, P.T.C.; Garnett, M.J.; Roe, S.M.; Lee, S.; Niculescu-Duvaz, D.; Good, V.M.; Jones, C.M.; Marshall, C.J.; Springer, C.J.; Barford, D.; et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell 2004, 116, 855–867. [Google Scholar] [CrossRef]
- Davies, H.; Bignell, G.R.; Cox, C.; Stephens, P.; Edkins, S.; Clegg, S.; Teague, J.; Woffendin, H.; Garnett, M.J.; Bottomley, W.; et al. Mutations of the BRAF gene in human cancer. Nature 2002, 417, 949–954. [Google Scholar] [CrossRef]
- Cohen, R.; Pudlarz, T.; Delattre, J.F.; Colle, R.; André, T. Molecular Targets for the Treatement of Metastatic Colorectal Cancer. Cancers 2020, 19, 2350. [Google Scholar] [CrossRef]
- Chen, D.; Huang, J.F.; Liu, K.; Zhang, L.Q.; Yang, Z.; Chuai, Z.R.; Wang, Y.X.; Shi, D.C.; Huang, Q.; Fu, W.L. BRAFV600E mutation and its association with clinicopathological features of colorectal cancer: A systematic review and meta-analysis. PLoS ONE 2014, 9, e90607. [Google Scholar] [CrossRef]
- Clarke, C.N.; Kopetz, E.S. BRAF mutant colorectal cancer as a distinct subset of colorectal cancer: Clinical characteristics, clinical behavior, and response to targeted therapies. J. Gastrointest. Oncol. 2015, 6, 660–667. [Google Scholar]
- Ahronian, L.G.; Sennott, E.M.; Van Allen, E.M.; Wagle, N.; Kwak, E.L.; Faris, J.E.; Godfrey, J.T.; Nishimura, K.; Lynch, K.D.; Mermel, C.H.; et al. Clinical Acquired Resistance to RAF Inhibitor Combinations in BRAF-Mutant Colorectal Cancer through MAPK Pathway Alterations. Cancer Discov. 2015, 5, 358–367. [Google Scholar] [CrossRef]
- Tie, J.; Desai, J. Targeting BRAF mutant metastatic colorectal cancer: Clinical implications and emerging therapeutic strategies. Target Oncol. 2015, 10, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Clancy, C.; Burke, J.P.; Kalady, M.F.; Coffey, J.C. BRAF mutation is associated with distinct clinicopathological characteristics in colorectal cancer: A systematic review and meta-analysis. Colorectal Dis. 2013, 15, e711–e718. [Google Scholar] [CrossRef] [PubMed]
- Missiaglia, E.; Jacobs, B.; D’Ario, G.; Di Narzo, A.F.; Soneson, C.; Budinska, E.; Popovici, V.; Vecchione, L.; Gerster, S.; Yan, P.; et al. Distal and proximal colon cancers differ in terms of molecular, pathological, and clinical features. Ann. Oncol. 2014, 25, 1995–2001. [Google Scholar] [CrossRef] [PubMed]
- Weisenberger, D.J.; Siegmund, K.D.; Campan, M.; Young, J.; Long, T.I.; Faasse, M.A.; Kang, G.H.; Widschwendter, M.; Weener, D.; Buchanan, D.; et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat. Genet. 2006, 38, 787–793. [Google Scholar] [CrossRef] [PubMed]
- Tie, J.; Gibbs, P.; Lipton, L.; Christie, M.; Jorissen, R.N.; Burgess, A.W.; Croxford, M.; Jones, I.; Langland, R.; Kosmider, S.; et al. Optimizing targeted therapeutic development: Analysis of a colorectal cancer patient population with the BRAF(V600E) mutation. Int. J. Cancer 2011, 128, 2075–2084. [Google Scholar] [CrossRef]
- Lochhead, P.; Kuchiba, A.; Imamura, Y.; Liao, X.; Yamauchi, M.; Nishihara, R.; Qian, Z.R.; Morikawa, T.; Shen, J.; Meyerhardt, J.A.; et al. Microsatellite instability and BRAF mutation testing in colorectal cancer prognostication. J. Natl. Cancer Inst. 2013, 105, 1151–1156. [Google Scholar] [CrossRef]
- Landau, M.S.; Kuan, S.F.; Chiosea, S.; Pai, R.K. BRAF-mutated microsatellite stable colorectal carcinoma: An aggressive adenocarcinoma with reduced CDX2 and increased cytokeratin 7 immunohistochemical expression. Hum. Pathol. 2014, 45, 1704–1712. [Google Scholar] [CrossRef]
- Dankner, M.; Rose, A.A.N.; Rajkumar, S.; Siegel, P.M.; Watson, I.R. Classifying BRAF alterations in cancer: New rational therapeutic strategies for actionable mutations. Oncogene 2018, 37, 3183–3199. [Google Scholar] [CrossRef]
- De Roock, W.; Claes, B.; Bernasconi, D.; De Schutter, J.; Biesmans, B.; Fountzilas, G.; Kalogeras, K.T.; Kotoula, V.; Papamichael, D.; Laurent-Puig, P.; et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: A retrospective consortium analysis. Lancet Oncol. 2010, 11, 753–762. [Google Scholar] [CrossRef]
- Fanelli, G.N.; Dal Pozzo, C.A.; Depetris, I.; Schirripa, M.; Brignola, S.; Biason, P.; Balistreri, M.; Dal Santo, L.; Lonardi, S.; Munari, G.; et al. The heterogeneous clinical and pathological landscapes of metastatic Braf-mutated colorectal cancer. Cancer Cell Int. 2020, 20, 30. [Google Scholar] [CrossRef]
- Ogino, S.; Shima, K.; Meyerhardt, J.A.; McCleary, J.; Ng, K.; Hollis, D.; Saltz, L.B.; Mayer, R.J.; Schaefer, P.; Whittom, R.; et al. Predictive and prognostic roles of BRAF mutation in stage III colon cancer: Results from intergroup trial CALGB 89803. Clin. Cancer Res. 2012, 18, 890–900. [Google Scholar] [CrossRef] [PubMed]
- Loupakis, F.; Intini, R.; Cremolini, C.; Orlandi, A.; Sartore-Bianchi, A.; Pietrantonio, F.; Pella, N.; Spallanzani, A.; Dell’Aquila, E.; Scartozzi, M.; et al. A validated prognostic classifier for (V600E)BRAF-mutated metastatic colorectal cancer: The ‘BRAF BeCool’ study. Eur. J. Cancer 2019, 118, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Di Nicolantonio, F.; Martini, M.; Molinari, F.; Sartore-Bianchi, A.; Arena, S.; Saletti, P.; De Dosso, S.; Mazzucchelli, L.; Frattini, M.; Siena, S.; et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J. Clin. Oncol. 2008, 26, 5705–5712. [Google Scholar] [CrossRef] [PubMed]
- Modest, D.P.; Martens, U.M.; Riera-Knorrenschild, J.; Greeve, J.; Florschutz, A.; Wessendorf, S.; Ettrich, T.; Kanzler, S.; Norenberg, D.; Ricke, J. FOLFOXIRI Plus Panitumumab As First-Line Treatment of RAS Wild-Type Metastatic Colorectal Cancer: The Randomized, Open-Label, Phase II VOLFI Study (AIO KRK0109). J. Clin. Oncol. 2019, 37, 3401–3411. [Google Scholar] [CrossRef]
- Flaherty, K.T.; Puzanov, I.; Kim, K.B.; Ribas, A.; McArthur, G.A.; Sosman, J.A.; O’Dwye, P.J.; Lee, R.J.; Grippo, J.F.; Nolop, K.; et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N. Engl. J. Med. 2010, 363, 809–819. [Google Scholar] [CrossRef]
- Prahallad, A.; Sun, C.; Huang, S.; Di Nicolantonio, F.; Salazar, R.; Zecchin, D.; Beijersbergen, R.L.; Bardelli, A.; Bernards, R. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 2012, 483, 100–103. [Google Scholar] [CrossRef]
- Kopetz, S.; Grothey, A.; Yaeger, R.; Van Cutsem, E.; Desai, J.; Yoshino, T.; Wasan, H.; Ciardiello, F.; Loupakis, F.; Sang Hong, Y.; et al. Encorafenib, Binimetinib, and Cetuximab in BRAF V600E-Mutated Colorectal Cancer. N. Engl. J. Med. 2019, 381, 1632–1643. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sammarco, G.; Gallo, G.; Vescio, G.; Picciariello, A.; De Paola, G.; Trompetto, M.; Currò, G.; Ammendola, M. Mast Cells, microRNAs and Others: The Role of Translational Research on Colorectal Cancer in the Forthcoming Era of Precision Medicine. J. Clin. Med. 2020, 9, 2852. https://doi.org/10.3390/jcm9092852
Sammarco G, Gallo G, Vescio G, Picciariello A, De Paola G, Trompetto M, Currò G, Ammendola M. Mast Cells, microRNAs and Others: The Role of Translational Research on Colorectal Cancer in the Forthcoming Era of Precision Medicine. Journal of Clinical Medicine. 2020; 9(9):2852. https://doi.org/10.3390/jcm9092852
Chicago/Turabian StyleSammarco, Giuseppe, Gaetano Gallo, Giuseppina Vescio, Arcangelo Picciariello, Gilda De Paola, Mario Trompetto, Giuseppe Currò, and Michele Ammendola. 2020. "Mast Cells, microRNAs and Others: The Role of Translational Research on Colorectal Cancer in the Forthcoming Era of Precision Medicine" Journal of Clinical Medicine 9, no. 9: 2852. https://doi.org/10.3390/jcm9092852
APA StyleSammarco, G., Gallo, G., Vescio, G., Picciariello, A., De Paola, G., Trompetto, M., Currò, G., & Ammendola, M. (2020). Mast Cells, microRNAs and Others: The Role of Translational Research on Colorectal Cancer in the Forthcoming Era of Precision Medicine. Journal of Clinical Medicine, 9(9), 2852. https://doi.org/10.3390/jcm9092852





