Identification of microRNAs Targeting the Transporter Associated with Antigen Processing TAP1 in Melanoma
Abstract
:1. Introduction
2. Experimental Section
2.1. Cell Lines and Cell Culture Conditions
2.2. Human Melanoma Tissues
2.3. Plasmids and Cloning
2.4. MiRNA Trapping by RNA In Vitro Affinity Purification (miTRAP)
2.5. Isolation of Plasmid DNA, Cellular RNA, miRNA and qPCR Analysis
2.6. Protein Extraction and Western Blot Analysis
2.7. Luciferase Reporter Assay
2.8. Transfection of miR
2.9. Flow Cytometry
2.10. CD107a Degranulation Assay
2.11. Immunohistochemical Staining of the Paraffin-Embedded Tissue Sections of Melanoma Patients
2.12. Next-Generation Sequencing Analysis
2.13. Functional and Pathway Enrichment Analyses
2.14. Bioinformatics—Survival Analysis
2.15. Statistical Analysis
3. Results
3.1. Clinical Relevance of TAP1 and HLA Class I Molecules Regarding Survival of Tumor Patients
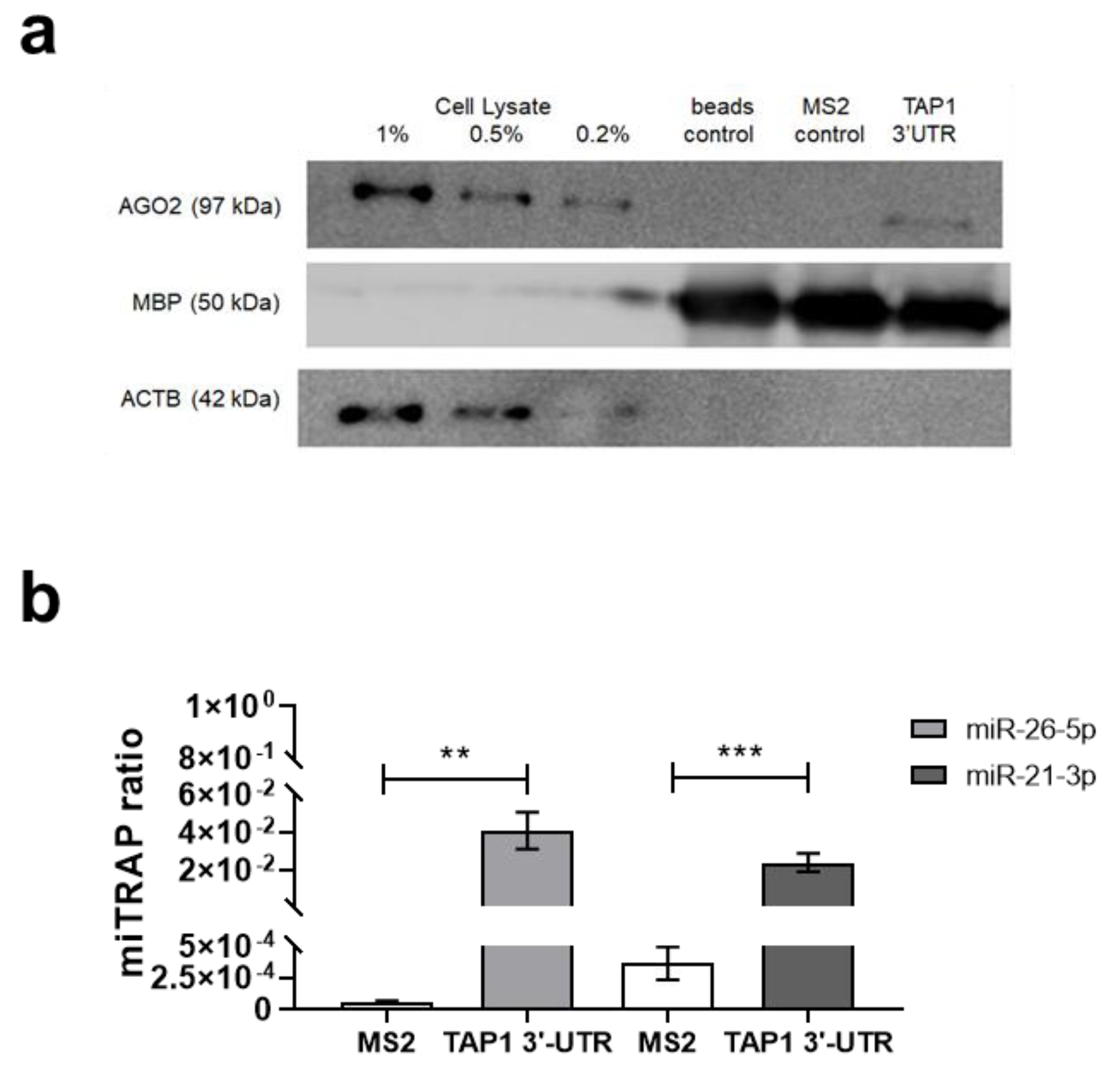
3.2. Identification of New Candidate miR Targeting TAP1 Using the miTRAP Assay
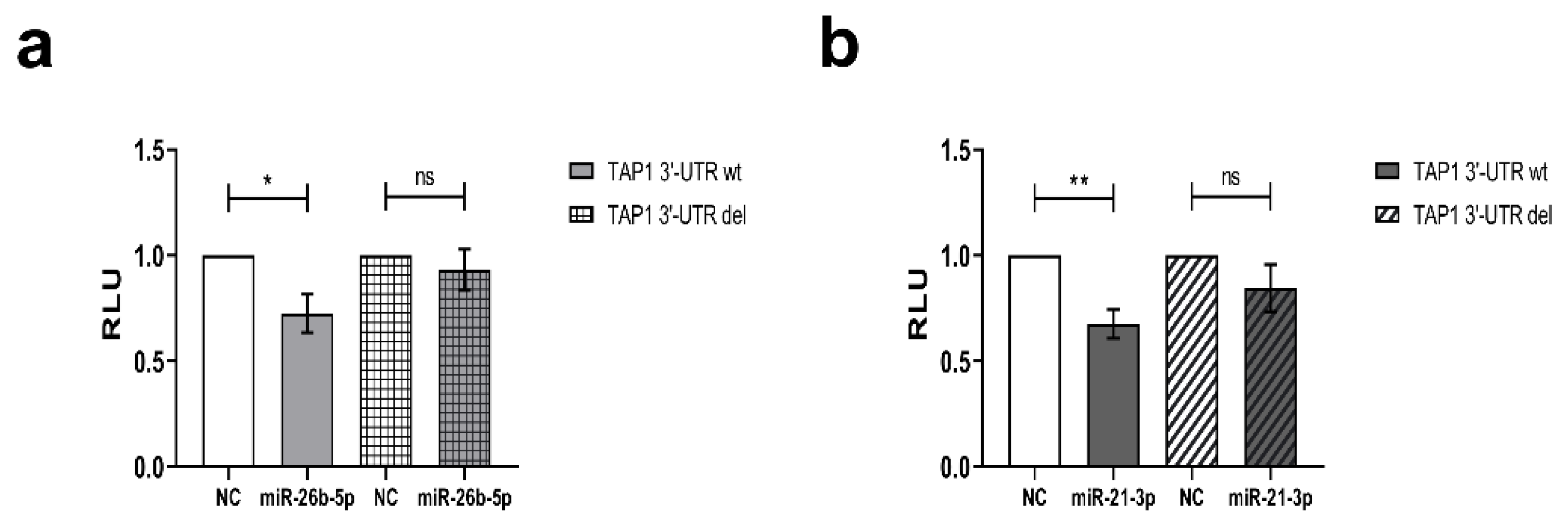
3.3. Direct Interaction of miR-26b-5p and miR-21-3p with TAP1 3′ UTR
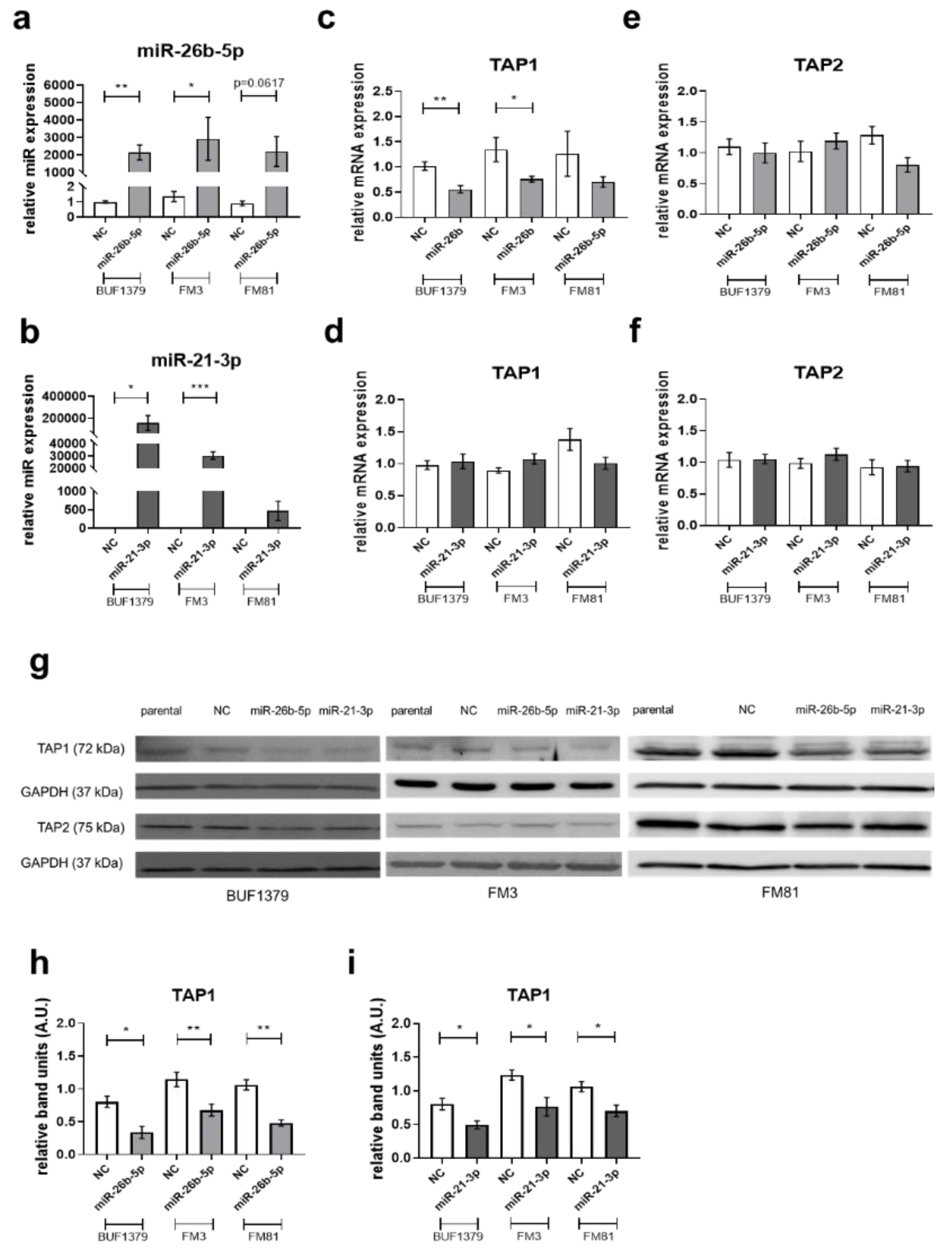
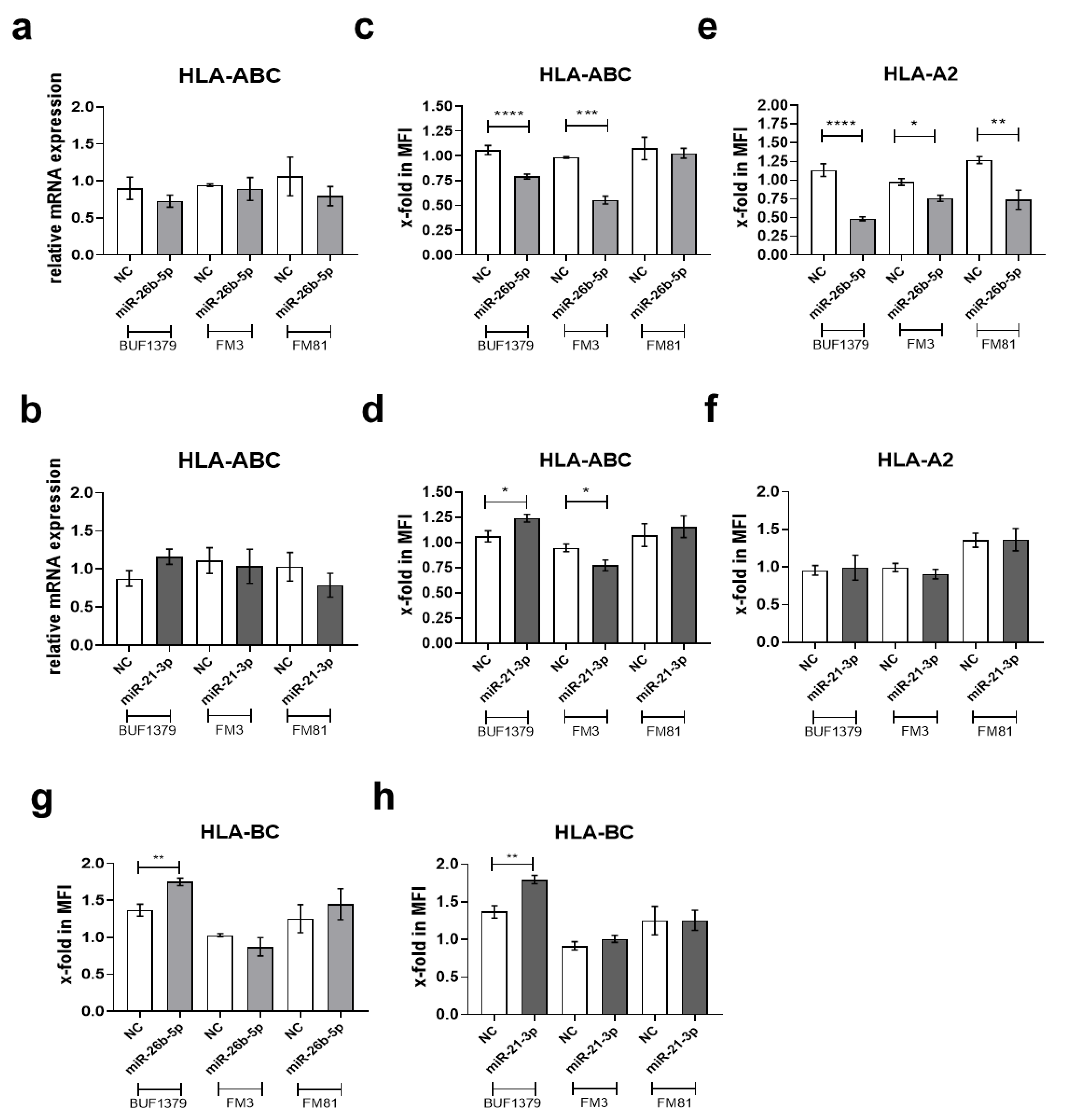
3.4. Downregulation of TAP1 Expression by miR-26b-5p and miR-21-3p
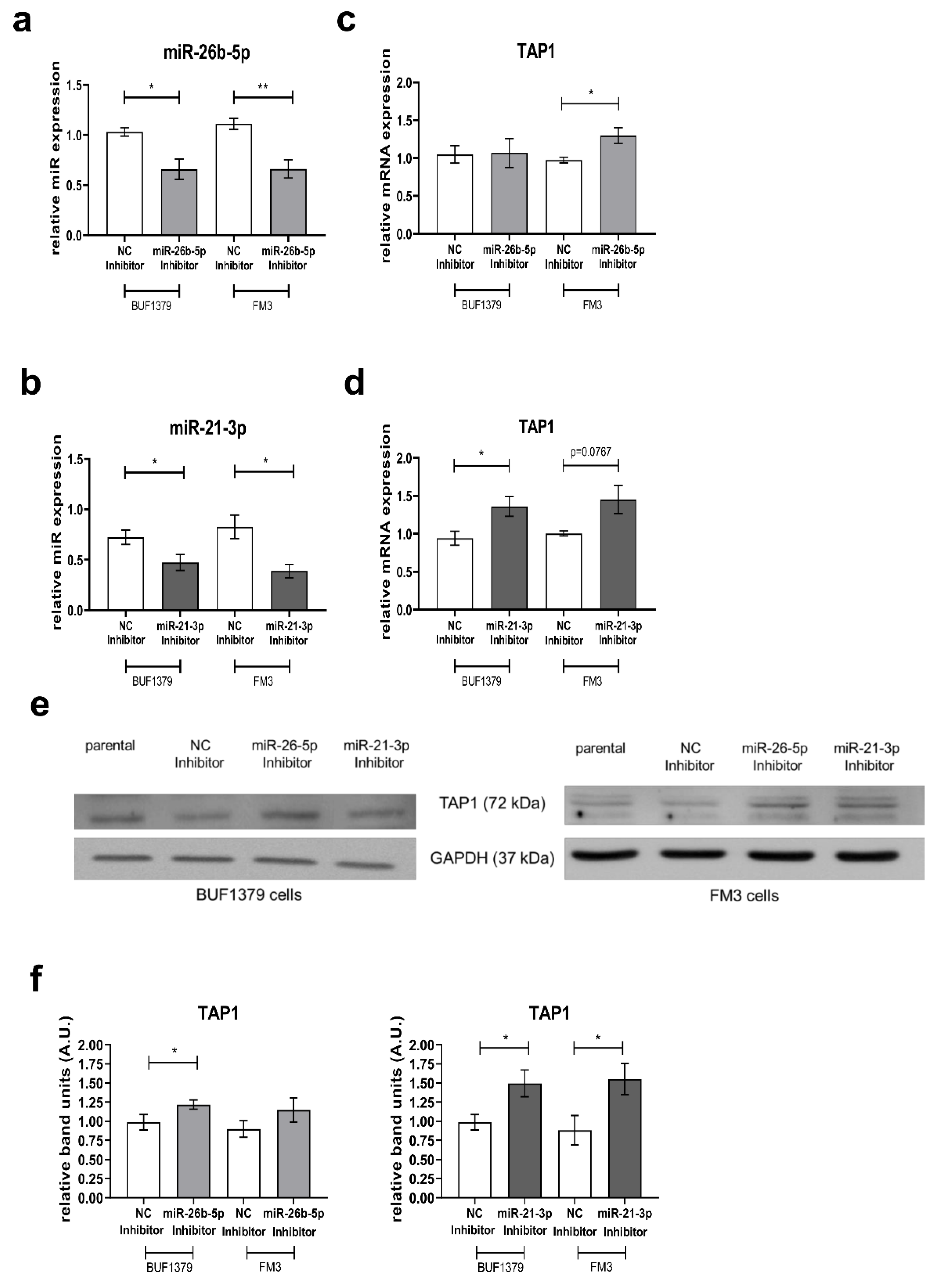
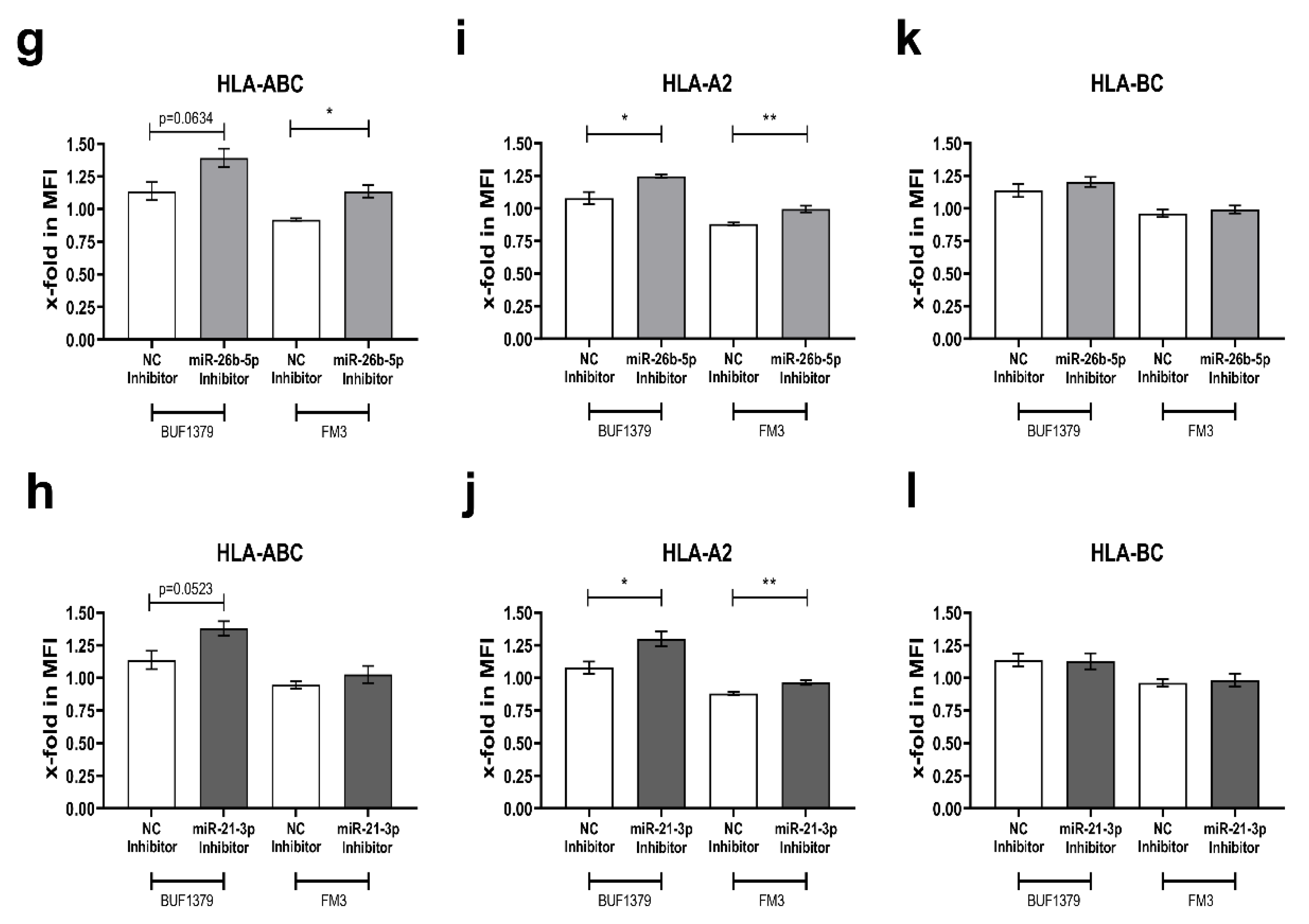
3.5. Reversion of the miR Effect by Inhibition of miR-26b-5p and miR-21-3p
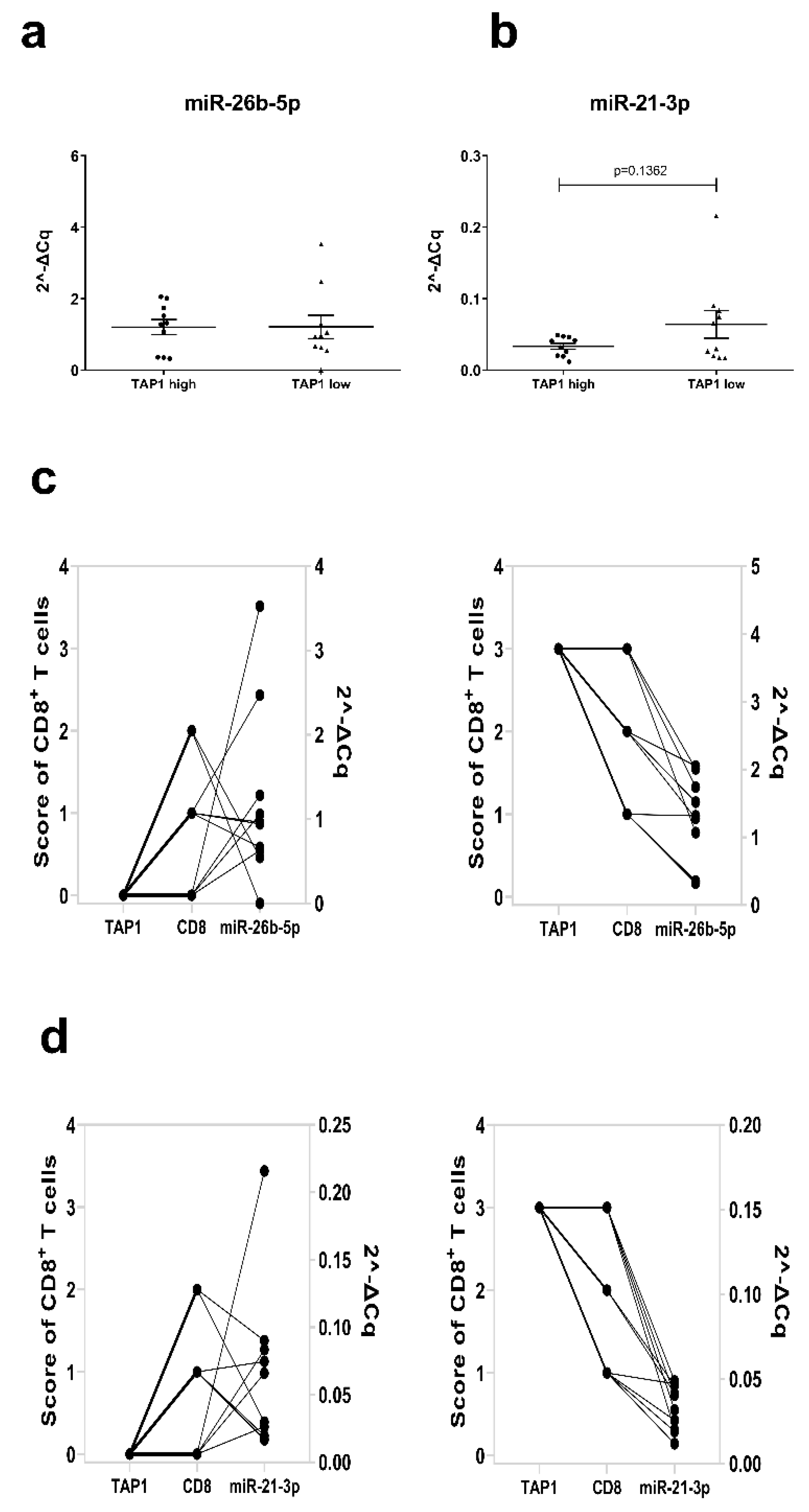
3.6. Correlation of the miR26b-5p-Mediated Downregulation of TAP1 with Decreased T Cell Recognition
3.7. Correlation between miR-26b-5p or miR-21-3p Expression with TAP1 and Immune Cell Infiltration in Melanoma Lesions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Cai, L.; Michelakos, T.; Yamada, T.; Fan, S.; Wang, X.; Schwab, J.H.; Ferrone, C.R.; Ferrone, S. Defective HLA class I antigen processing machinery in cancer. Cancer Immunol. Immunother. 2018, 67, 999–1009. [Google Scholar] [CrossRef] [PubMed]
- Seliger, B.; Ferrone, S. HLA Class I Antigen Processing Machinery Defects in Cancer Cells-Frequency, Functional Significance, and Clinical Relevance with Special Emphasis on Their Role in T Cell-Based Immunotherapy of Malignant Disease. Methods Mol. Biol. 2020, 2055, 325–350. [Google Scholar] [CrossRef] [PubMed]
- Seliger, B. Molecular mechanisms of HLA class I-mediated immune evasion of human tumors and their role in resistance to immunotherapies. HLA 2016, 88, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Andersson, E.; Villabona, L.; Bergfeldt, K.; Carlson, J.W.; Ferrone, S.; Kiessling, R.; Seliger, B.; Masucci, G.V. Correlation of HLA-A02* genotype and HLA class I antigen down-regulation with the prognosis of epithelial ovarian cancer. Cancer Immunol. Immunother. 2012, 61, 1243–1253. [Google Scholar] [CrossRef] [PubMed]
- Kalbasi, A.; Ribas, A. Tumour-intrinsic resistance to immune checkpoint blockade. Nat. Rev. Immunol. 2020, 20, 25–39. [Google Scholar] [CrossRef]
- Mittal, D.; Gubin, M.M.; Schreiber, R.D.; Smyth, M.J. New insights into cancer immunoediting and its three component phases—Elimination, equilibrium and escape. Curr. Opin. Immunol. 2014, 27, 16–25. [Google Scholar] [CrossRef] [Green Version]
- Jabrane-Ferrat, N.; Faille, A.; Loiseau, P.; Poirier, O.; Charron, D.; Calvo, F. Effect of gamma interferon on HLA class-I and -II transcription and protein expression in human breast adenocarcinoma cell lines. Int. J. Cancer 1990, 45, 1169–1176. [Google Scholar] [CrossRef]
- Seliger, B. Novel insights into the molecular mechanisms of HLA class I abnormalities. Cancer Immunol. Immunother. 2012, 61, 249–254. [Google Scholar] [CrossRef]
- Snyder, S.R.; Waring, J.F.; Zhu, S.Z.; Kaplan, S.; Schultz, J.; Ginder, G.D. A 3′-transcribed region of the HLA-A2 gene mediates posttranscriptional stimulation by IFN-gamma. J. Immunol. 2001, 166, 3966–3974. [Google Scholar] [CrossRef] [Green Version]
- Campoli, M.; Ferrone, S. HLA antigen changes in malignant cells: Epigenetic mechanisms and biologic significance. Oncogene 2008, 27, 5869–5885. [Google Scholar] [CrossRef] [Green Version]
- Rene, C.; Lozano, C.; Eliaou, J.F. Expression of classical HLA class I molecules: Regulation and clinical impacts: Julia Bodmer Award Review 2015. HLA 2016, 87, 338–349. [Google Scholar] [CrossRef]
- Dovhey, S.E.; Ghosh, N.S.; Wright, K.L. Loss of interferon-gamma inducibility of TAP1 and LMP2 in a renal cell carcinoma cell line. Cancer Res. 2000, 60, 5789–5796. [Google Scholar] [PubMed]
- Reches, A.; Nachmani, D.; Berhani, O.; Duev-Cohen, A.; Shreibman, D.; Ophir, Y.; Seliger, B.; Mandelboim, O. HNRNPR Regulates the Expression of Classical and Nonclassical MHC Class I Proteins. J. Immunol. 2016, 196, 4967–4976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eichmuller, S.B.; Osen, W.; Mandelboim, O.; Seliger, B. Immune Modulatory microRNAs Involved in Tumor Attack and Tumor Immune Escape. J. Natl. Cancer Inst. 2017, 109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Q.; Cao, W.; Wang, Z.; Zhang, B.; Liu, J. Regulation of cancer immune escape: The roles of miRNAs in immune checkpoint proteins. Cancer Lett. 2018, 431, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Cano, F.; Rapiteanu, R.; Winkler, G.S.; Lehner, P.J. A non-proteolytic role for ubiquitin in deadenylation of MHC-I mRNA by the RNA-binding E3-ligase MEX-3C. Nat. Commun. 2015, 6, 8670. [Google Scholar] [CrossRef] [Green Version]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [Green Version]
- Jiang, C.; Chen, X.; Alattar, M.; Wei, J.; Liu, H. MicroRNAs in tumorigenesis, metastasis, diagnosis and prognosis of gastric cancer. Cancer Gene Ther. 2015, 22, 291–301. [Google Scholar] [CrossRef] [Green Version]
- Nana-Sinkam, S.P.; Croce, C.M. MicroRNA regulation of tumorigenesis, cancer progression and interpatient heterogeneity: Towards clinical use. Genome Biol. 2014, 15, 445. [Google Scholar] [CrossRef] [Green Version]
- Mari, L.; Hoefnagel, S.J.M.; Zito, D.; van de Meent, M.; van Endert, P.; Calpe, S.; Serra, M.D.C.S.; Heemskerk, M.H.M.; van Laarhoven, H.W.M.; Hulshof, M.; et al. microRNA 125a Regulates MHC-I Expression on Esophageal Adenocarcinoma Cells, Associated With Suppression of Antitumor Immune Response and Poor Outcomes of Patients. Gastroenterology 2018, 155, 784–798. [Google Scholar] [CrossRef] [PubMed]
- Jasinski-Bergner, S.; Stoehr, C.; Bukur, J.; Massa, C.; Braun, J.; Huttelmaier, S.; Spath, V.; Wartenberg, R.; Legal, W.; Taubert, H.; et al. Clinical relevance of miR-mediated HLA-G regulation and the associated immune cell infiltration in renal cell carcinoma. Oncoimmunology 2015, 4, e1008805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, F.; Zhao, Z.L.; Zhao, W.T.; Fan, Q.R.; Wang, S.C.; Li, J.; Zhang, Y.Q.; Shi, J.W.; Lin, X.L.; Yang, S.; et al. miR-9 modulates the expression of interferon-regulated genes and MHC class I molecules in human nasopharyngeal carcinoma cells. Biochem. Biophys. Res. Commun. 2013, 431, 610–616. [Google Scholar] [CrossRef] [PubMed]
- Jasinski-Bergner, S.; Reches, A.; Stoehr, C.; Massa, C.; Gonschorek, E.; Huettelmaier, S.; Braun, J.; Wach, S.; Wullich, B.; Spath, V.; et al. Identification of novel microRNAs regulating HLA-G expression and investigating their clinical relevance in renal cell carcinoma. Oncotarget 2016, 7, 26866–26878. [Google Scholar] [CrossRef] [PubMed]
- Manaster, I.; Goldman-Wohl, D.; Greenfield, C.; Nachmani, D.; Tsukerman, P.; Hamani, Y.; Yagel, S.; Mandelboim, O. MiRNA-mediated control of HLA-G expression and function. PLoS ONE 2012, 7, e33395. [Google Scholar] [CrossRef]
- Albanese, M.; Tagawa, T.; Bouvet, M.; Maliqi, L.; Lutter, D.; Hoser, J.; Hastreiter, M.; Hayes, M.; Sugden, B.; Martin, L.; et al. Epstein-Barr virus microRNAs reduce immune surveillance by virus-specific CD8+ T cells. Proc. Natl. Acad. Sci. USA 2016, 113, E6467–E6475. [Google Scholar] [CrossRef] [Green Version]
- Knox, B.; Wang, Y.; Rogers, L.J.; Xuan, J.; Yu, D.; Guan, H.; Chen, J.; Shi, T.; Ning, B.; Kadlubar, S.A. A functional SNP in the 3′-UTR of TAP2 gene interacts with microRNA hsa-miR-1270 to suppress the gene expression. Environ. Mol. Mutagen. 2018, 59, 134–143. [Google Scholar] [CrossRef] [Green Version]
- Bartoszewski, R.; Brewer, J.W.; Rab, A.; Crossman, D.K.; Bartoszewska, S.; Kapoor, N.; Fuller, C.; Collawn, J.F.; Bebok, Z. The unfolded protein response (UPR)-activated transcription factor X-box-binding protein 1 (XBP1) induces microRNA-346 expression that targets the human antigen peptide transporter 1 (TAP1) mRNA and governs immune regulatory genes. J. Biol. Chem. 2011, 286, 41862–41870. [Google Scholar] [CrossRef] [Green Version]
- Marzagalli, M.; Ebelt, N.D.; Manuel, E.R. Unraveling the crosstalk between melanoma and immune cells in the tumor microenvironment. Semin. Cancer Biol. 2019, 59, 236–250. [Google Scholar] [CrossRef]
- Gettinger, S.; Choi, J.; Hastings, K.; Truini, A.; Datar, I.; Sowell, R.; Wurtz, A.; Dong, W.; Cai, G.; Melnick, M.A.; et al. Impaired HLA Class I Antigen Processing and Presentation as a Mechanism of Acquired Resistance to Immune Checkpoint Inhibitors in Lung Cancer. Cancer Discov. 2017, 7, 1420–1435. [Google Scholar] [CrossRef] [Green Version]
- Kalbasi, A.; Ribas, A. Antigen Presentation Keeps Trending in Immunotherapy Resistance. Clin. Cancer Res. 2018, 24, 3239–3241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cortez, M.A.; Anfossi, S.; Ramapriyan, R.; Menon, H.; Atalar, S.C.; Aliru, M.; Welsh, J.; Calin, G.A. Role of miRNAs in immune responses and immunotherapy in cancer. Genes Chromosomes Cancer 2019, 58, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Hu-Lieskovan, S.; Wargo, J.A.; Ribas, A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell 2017, 168, 707–723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gide, T.N.; Wilmott, J.S.; Scolyer, R.A.; Long, G.V. Primary and Acquired Resistance to Immune Checkpoint Inhibitors in Metastatic Melanoma. Clin. Cancer Res. 2018, 24, 1260–1270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tretbar, U.S.; Friedrich, M.; Lazaridou, M.F.; Seliger, B. Identification of Immune Modulatory miRNAs by miRNA Enrichment via RNA Affinity Purification. Methods Mol. Biol. 2019, 1913, 81–101. [Google Scholar] [CrossRef] [PubMed]
- Braun, J.; Misiak, D.; Busch, B.; Krohn, K.; Huttelmaier, S. Rapid identification of regulatory microRNAs by miTRAP (miRNA trapping by RNA in vitro affinity purification). Nucleic Acids Res. 2014, 42, e66. [Google Scholar] [CrossRef] [Green Version]
- Friedrich, M.; Jasinski-Bergner, S.; Lazaridou, M.F.; Subbarayan, K.; Massa, C.; Tretbar, S.; Mueller, A.; Handke, D.; Biehl, K.; Bukur, J.; et al. Tumor-induced escape mechanisms and their association with resistance to checkpoint inhibitor therapy. Cancer Immunol. Immunother. 2019, 68, 1689–1700. [Google Scholar] [CrossRef]
- Pullen, A.M.; Kappler, J.W.; Marrack, P. Tolerance to self antigens shapes the T-cell repertoire. Immunol. Rev. 1989, 107, 125–139. [Google Scholar] [CrossRef]
- Weiss, S.A.; Wolchok, J.D.; Sznol, M. Immunotherapy of Melanoma: Facts and Hopes. Clin. Cancer Res. 2019, 25, 5191–5201. [Google Scholar] [CrossRef] [Green Version]
- Curtin, J.A.; Fridlyand, J.; Kageshita, T.; Patel, H.N.; Busam, K.J.; Kutzner, H.; Cho, K.H.; Aiba, S.; Brocker, E.B.; LeBoit, P.E.; et al. Distinct sets of genetic alterations in melanoma. N. Engl. J. Med. 2005, 353, 2135–2147. [Google Scholar] [CrossRef]
- Shankaran, V.; Ikeda, H.; Bruce, A.T.; White, J.M.; Swanson, P.E.; Old, L.J.; Schreiber, R.D. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature 2001, 410, 1107–1111. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, T.N.; Schreiber, R.D. Neoantigens in cancer immunotherapy. Science 2015, 348, 69–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Borresen-Dale, A.L.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef] [Green Version]
- Gajewski, T.F.; Schreiber, H.; Fu, Y.X. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 2013, 14, 1014–1022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maeurer, M.J.; Gollin, S.M.; Martin, D.; Swaney, W.; Bryant, J.; Castelli, C.; Robbins, P.; Parmiani, G.; Storkus, W.J.; Lotze, M.T. Tumor escape from immune recognition: Lethal recurrent melanoma in a patient associated with downregulation of the peptide transporter protein TAP-1 and loss of expression of the immunodominant MART-1/Melan-A antigen. J. Clin. Investig. 1996, 98, 1633–1641. [Google Scholar] [CrossRef]
- Passarelli, A.; Mannavola, F.; Stucci, L.S.; Tucci, M.; Silvestris, F. Immune system and melanoma biology: A balance between immunosurveillance and immune escape. Oncotarget 2017, 8, 106132–106142. [Google Scholar] [CrossRef]
- Cao, W.; Cheng, W.; Wu, W. MicroRNAs Reprogram Tumor Immune Response. Methods Mol. Biol. 2018, 1699, 67–74. [Google Scholar] [CrossRef]
- Jasinski-Bergner, S.; Vaxevanis, C.; Heimer, N.; Lazaridou, M.F.; Friedrich, M.; Seliger, B. An altered miTRAP method for miRNA affinity purification with its pros and cons. Methods Enzymol. 2020, 636, 323–337. [Google Scholar] [CrossRef]
- Bignell, G.R.; Greenman, C.D.; Davies, H.; Butler, A.P.; Edkins, S.; Andrews, J.M.; Buck, G.; Chen, L.; Beare, D.; Latimer, C.; et al. Signatures of mutation and selection in the cancer genome. Nature 2010, 463, 893–898. [Google Scholar] [CrossRef]
- Pawelec, G.; Marsh, S.G. ESTDAB: A collection of immunologically characterised melanoma cell lines and searchable databank. Cancer Immunol. Immunother. 2006, 55, 623–627. [Google Scholar] [CrossRef]
- Wulfanger, J.; Biehl, K.; Tetzner, A.; Wild, P.; Ikenberg, K.; Meyer, S.; Seliger, B. Heterogeneous expression and functional relevance of the ubiquitin carboxyl-terminal hydrolase L1 in melanoma. Int. J. Cancer 2013, 133, 2522–2532. [Google Scholar] [CrossRef] [PubMed]
- Salter, R.D.; Cresswell, P. Impaired assembly and transport of HLA-A and -B antigens in a mutant TxB cell hybrid. EMBO J. 1986, 5, 943–949. [Google Scholar] [CrossRef] [PubMed]
- Koelblinger, P.; Emberger, M.; Drach, M.; Cheng, P.F.; Lang, R.; Levesque, M.P.; Bauer, J.W.; Dummer, R. Increased tumour cell PD-L1 expression, macrophage and dendritic cell infiltration characterise the tumour microenvironment of ulcerated primary melanomas. J. Eur. Acad. Dermatol. Venereol. JEADV 2019, 33, 667–675. [Google Scholar] [CrossRef] [PubMed]
- Fander, J.; Kielstein, H.; Buttner, M.; Koelblinger, P.; Dummer, R.; Bauer, M.; Handke, D.; Wickenhauser, C.; Seliger, B.; Jasinski-Bergner, S. Characterizing CD44 regulatory microRNAs as putative therapeutic agents in human melanoma. Oncotarget 2019, 10, 6509–6525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, J.H.; Srikantan, S.; Gorospe, M. MS2-TRAP (MS2-tagged RNA affinity purification): Tagging RNA to identify associated miRNAs. Methods 2012, 58, 81–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lazaridou, M.-F.; Gonschorek, E.; Massa, C.; Friedrich, M.; Handke, D.; Mueller, A.; Jasinski-Bergner, S.; Dummer, R.; Koelblinger, P.; Seliger, B. Identification of miR-200a-5p targeting the peptide transporter TAP1 and its association with the clinical outcome of melanoma patients. Oncoimmunology 2020, 9, 1774323. [Google Scholar] [CrossRef]
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005, 33, e179. [Google Scholar] [CrossRef]
- Varkonyi-Gasic, E.; Wu, R.; Wood, M.; Walton, E.F.; Hellens, R.P. Protocol: A highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 2007, 3, 12. [Google Scholar] [CrossRef] [Green Version]
- Kramer, M.F. Stem-loop RT-qPCR for miRNAs. Curr. Protoc. Mol. Biol. 2011. [Google Scholar] [CrossRef]
- Bukur, J.; Herrmann, F.; Handke, D.; Recktenwald, C.; Seliger, B. Identification of E2F1 as an important transcription factor for the regulation of tapasin expression. J. Biol. Chem. 2010, 285, 30419–30426. [Google Scholar] [CrossRef] [Green Version]
- Alter, G.; Malenfant, J.M.; Altfeld, M. CD107a as a functional marker for the identification of natural killer cell activity. J. Immunol. Methods 2004, 294, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo-Herrero, S.; Sordo-Bahamonde, C.; Gonzalez, S.; Lopez-Soto, A. CD107a Degranulation Assay to Evaluate Immune Cell Antitumor Activity. Methods Mol. Biol. 2019, 1884, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, J.; Lai, J.; Lehman, M.L.; Nelson, C.C. miRDeep*: An integrated application tool for miRNA identification from RNA sequencing data. Nucleic Acids Res. 2013, 41, 727–737. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [Green Version]
- Wei, L.; He, F.; Zhang, W.; Chen, W.; Yu, B. Bioinformatics analysis of microarray data to reveal the pathogenesis of diffuse intrinsic pontine glioma. Biol. Res. 2018, 51, 26. [Google Scholar] [CrossRef] [Green Version]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.H.; Yang, J.H. StarBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.H.; Li, J.H.; Shao, P.; Zhou, H.; Chen, Y.Q.; Qu, L.H. Starbase: A database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res. 2011, 39, D202–D209. [Google Scholar] [CrossRef] [Green Version]
- Guan, J.; Gupta, R.; Filipp, F.V. Cancer systems biology of TCGA SKCM: Efficient detection of genomic drivers in melanoma. Sci. Rep. 2015, 5, 7857. [Google Scholar] [CrossRef] [Green Version]
- Ellrott, K.; Bailey, M.H.; Saksena, G.; Covington, K.R.; Kandoth, C.; Stewart, C.; Hess, J.; Ma, S.; Chiotti, K.E.; McLellan, M.; et al. Scalable Open Science Approach for Mutation Calling of Tumor Exomes Using Multiple Genomic Pipelines. Cell Syst. 2018, 6, 271–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Q.; Liang, W.W.; Foltz, S.M.; Mutharasu, G.; Jayasinghe, R.G.; Cao, S.; Liao, W.W.; Reynolds, S.M.; Wyczalkowski, M.A.; Yao, L.; et al. Driver Fusions and Their Implications in the Development and Treatment of Human Cancers. Cell Rep. 2018, 23, 227–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoadley, K.A.; Yau, C.; Hinoue, T.; Wolf, D.M.; Lazar, A.J.; Drill, E.; Shen, R.; Taylor, A.M.; Cherniack, A.D.; Thorsson, V.; et al. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell 2018, 173, 291–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Lichtenberg, T.; Hoadley, K.A.; Poisson, L.M.; Lazar, A.J.; Cherniack, A.D.; Kovatich, A.J.; Benz, C.C.; Levine, D.A.; Lee, A.V.; et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018, 173, 400–416. [Google Scholar] [CrossRef] [Green Version]
- Sanchez-Vega, F.; Mina, M.; Armenia, J.; Chatila, W.K.; Luna, A.; La, K.C.; Dimitriadoy, S.; Liu, D.L.; Kantheti, H.S.; Saghafinia, S.; et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018, 173, 321–337. [Google Scholar] [CrossRef] [Green Version]
- Taylor, A.M.; Shih, J.; Ha, G.; Gao, G.F.; Zhang, X.; Berger, A.C.; Schumacher, S.E.; Wang, C.; Hu, H.; Liu, J.; et al. Genomic and Functional Approaches to Understanding Cancer Aneuploidy. Cancer Cell 2018, 33, 676–689. [Google Scholar] [CrossRef] [Green Version]
- Sinn, B.V.; Weber, K.E.; Schmitt, W.D.; Fasching, P.A.; Symmans, W.F.; Blohmer, J.U.; Karn, T.; Taube, E.T.; Klauschen, F.; Marme, F.; et al. Human leucocyte antigen class I in hormone receptor-positive, HER2-negative breast cancer: Association with response and survival after neoadjuvant chemotherapy. Breast Cancer Res. 2019, 21, 142. [Google Scholar] [CrossRef] [Green Version]
- Helgadottir, H.; Andersson, E.; Villabona, L.; Kanter, L.; van der Zanden, H.; Haasnoot, G.W.; Seliger, B.; Bergfeldt, K.; Hansson, J.; Ragnarsson-Olding, B.; et al. The common Scandinavian human leucocyte antigen ancestral haplotype 62.1 as prognostic factor in patients with advanced malignant melanoma. Cancer Immunol. Immunother. 2009, 58, 1599–1608. [Google Scholar] [CrossRef]
- Seliger, B.; Ritz, U.; Abele, R.; Bock, M.; Tampe, R.; Sutter, G.; Drexler, I.; Huber, C.; Ferrone, S. Immune escape of melanoma: First evidence of structural alterations in two distinct components of the MHC class I antigen processing pathway. Cancer Res. 2001, 61, 8647–8650. [Google Scholar]
- Garrido, F.; Aptsiauri, N. Cancer immune escape: MHC expression in primary tumours versus metastases. Immunology 2019, 158, 255–266. [Google Scholar] [CrossRef] [Green Version]
- Luebker, S.A.; Zhang, W.; Koepsell, S.A. Comparing the genomes of cutaneous melanoma tumors to commercially available cell lines. Oncotarget 2017, 8, 114877–114893. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Chen, J.; Li, Z.; Li, X.; Hu, X.; Huang, Y.; Zhao, X.; Liang, C.; Wang, Y.; Sun, L.; et al. Integrated profiling of microRNAs and mRNAs: microRNAs located on Xq27.3 associate with clear cell renal cell carcinoma. PLoS ONE 2010, 5, e15224. [Google Scholar] [CrossRef] [PubMed]
- Dweep, H.; Gretz, N. miRWalk2.0: A comprehensive atlas of microRNA-target interactions. Nat. Methods 2015, 12, 697. [Google Scholar] [CrossRef] [PubMed]
- Dweep, H.; Sticht, C.; Pandey, P.; Gretz, N. miRWalk—Database: Prediction of possible miRNA binding sites by “walking” the genes of three genomes. J. Biomed. Inform. 2011, 44, 839–847. [Google Scholar] [CrossRef] [Green Version]
- Betel, D.; Wilson, M.; Gabow, A.; Marks, D.S.; Sander, C. The microRNA.org resource: Targets and expression. Nucleic Acids Res. 2008, 36, D149–D153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, N.; Wang, X. miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015, 43, D146–D152. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4. [Google Scholar] [CrossRef]
- Miranda, K.C.; Huynh, T.; Tay, Y.; Ang, Y.S.; Tam, W.L.; Thomson, A.M.; Lim, B.; Rigoutsos, I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 2006, 126, 1203–1217. [Google Scholar] [CrossRef] [Green Version]
- Rehmsmeier, M.; Steffen, P.; Hochsmann, M.; Giegerich, R. Fast and effective prediction of microRNA/target duplexes. RNA 2004, 10, 1507–1517. [Google Scholar] [CrossRef] [Green Version]
- Ribas, A.; Wolchok, J.D. Cancer immunotherapy using checkpoint blockade. Science 2018, 359, 1350–1355. [Google Scholar] [CrossRef] [Green Version]
- O’Donnell, J.S.; Teng, M.W.L.; Smyth, M.J. Cancer immunoediting and resistance to T cell-based immunotherapy. Nat. Rev. Clin. Oncol. 2019, 16, 151–167. [Google Scholar] [CrossRef] [PubMed]
- Andersen, R.; Westergaard, M.C.W.; Kjeldsen, J.W.; Muller, A.; Pedersen, N.W.; Hadrup, S.R.; Met, O.; Seliger, B.; Kromann-Andersen, B.; Hasselager, T.; et al. T-cell Responses in the Microenvironment of Primary Renal Cell Carcinoma-Implications for Adoptive Cell Therapy. Cancer Immunol. Res. 2018, 6, 222–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, L.; Malu, S.; McKenzie, J.A.; Andrews, M.C.; Talukder, A.H.; Tieu, T.; Karpinets, T.; Haymaker, C.; Forget, M.A.; Williams, L.J.; et al. The RNA-binding Protein MEX3B Mediates Resistance to Cancer Immunotherapy by Downregulating HLA-A Expression. Clin. Cancer Res. 2018, 24, 3366–3376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grasso, C.S.; Giannakis, M.; Wells, D.K.; Hamada, T.; Mu, X.J.; Quist, M.; Nowak, J.A.; Nishihara, R.; Qian, Z.R.; Inamura, K.; et al. Genetic Mechanisms of Immune Evasion in Colorectal Cancer. Cancer Discov. 2018, 8, 730–749. [Google Scholar] [CrossRef] [Green Version]
- Donia, M.; Harbst, K.; van Buuren, M.; Kvistborg, P.; Lindberg, M.F.; Andersen, R.; Idorn, M.; Ahmad, S.M.; Ellebaek, E.; Mueller, A.; et al. Acquired Immune Resistance Follows Complete Tumor Regression without Loss of Target Antigens or IFNgamma Signaling. Cancer Res. 2017, 77, 4562–4566. [Google Scholar] [CrossRef] [Green Version]
- Gilles, M.E.; Slack, F.J. Let-7 microRNA as a potential therapeutic target with implications for immunotherapy. Expert Opin. Ther. Targets 2018, 22, 929–939. [Google Scholar] [CrossRef]
- Paladini, L.; Fabris, L.; Bottai, G.; Raschioni, C.; Calin, G.A.; Santarpia, L. Targeting microRNAs as key modulators of tumor immune response. J. Exp. Clin. Cancer Res. 2016, 35, 103. [Google Scholar] [CrossRef] [Green Version]
- Seliger, B. Immune modulatory microRNAs as a novel mechanism to revert immune escape of tumors. Cytokine Growth Factor Rev. 2017, 36, 49–56. [Google Scholar] [CrossRef]
- Colangelo, T.; Polcaro, G.; Ziccardi, P.; Pucci, B.; Muccillo, L.; Galgani, M.; Fucci, A.; Milone, M.R.; Budillon, A.; Santopaolo, M.; et al. Proteomic screening identifies calreticulin as a miR-27a direct target repressing MHC class I cell surface exposure in colorectal cancer. Cell Death Dis. 2016, 7, e2120. [Google Scholar] [CrossRef]
- Neefjes, J.; Gottfried, E.; Roelse, J.; Gromme, M.; Obst, R.; Hammerling, G.J.; Momburg, F. Analysis of the fine specificity of rat, mouse and human TAP peptide transporters. Eur. J. Immunol. 1995, 25, 1133–1136. [Google Scholar] [CrossRef]
- Cresswell, P.; Bangia, N.; Dick, T.; Diedrich, G. The nature of the MHC class I peptide loading complex. Immunol. Rev. 1999, 172, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Heemels, M.T.; Ploegh, H. Generation, translocation, and presentation of MHC class I-restricted peptides. Annu. Rev. Biochem. 1995, 64, 463–491. [Google Scholar] [CrossRef] [PubMed]
- Lankat-Buttgereit, B.; Tampe, R. The transporter associated with antigen processing: Function and implications in human diseases. Physiol. Rev. 2002, 82, 187–204. [Google Scholar] [CrossRef] [Green Version]
- Pfeffer, S.R.; Yang, C.H.; Pfeffer, L.M. The Role of miR-21 in Cancer. Drug Dev. Res. 2015, 76, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Bautista-Sanchez, D.; Arriaga-Canon, C.; Pedroza-Torres, A.; De La Rosa-Velazquez, I.A.; Gonzalez-Barrios, R.; Contreras-Espinosa, L.; Montiel-Manriquez, R.; Castro-Hernandez, C.; Fragoso-Ontiveros, V.; Alvarez-Gomez, R.M.; et al. The Promising Role of miR-21 as a Cancer Biomarker and Its Importance in RNA-Based Therapeutics. Mol. Ther. Nucleic Acids 2020, 20, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Babapoor, S.; Wu, R.; Kozubek, J.; Auidi, D.; Grant-Kels, J.M.; Dadras, S.S. Identification of microRNAs associated with invasive and aggressive phenotype in cutaneous melanoma by next-generation sequencing. Lab. Investig. A J. Tech. Methods Pathol. 2017, 97, 636–648. [Google Scholar] [CrossRef]
- Neagu, M.; Constantin, C.; Cretoiu, S.M.; Zurac, S. miRNAs in the Diagnosis and Prognosis of Skin Cancer. Front. Cell Dev. Biol. 2020, 8, 71. [Google Scholar] [CrossRef] [Green Version]
- Saldanha, G.; Potter, L.; Lee, Y.S.; Watson, S.; Shendge, P.; Pringle, J.H. MicroRNA-21 expression and its pathogenetic significance in cutaneous melanoma. Melanoma Res. 2016, 26, 21–28. [Google Scholar] [CrossRef]
- Melnik, B.C. MiR-21: An environmental driver of malignant melanoma? J. Transl. Med. 2015, 13, 202. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Liu, Q.G. The role of miR-26 in tumors and normal tissues (Review). Oncol. Lett. 2011, 2, 1019–1023. [Google Scholar] [CrossRef] [Green Version]
- Jin, F.; Wang, Y.; Li, M.; Zhu, Y.; Liang, H.; Wang, C.; Wang, F.; Zhang, C.Y.; Zen, K.; Li, L. MiR-26 enhances chemosensitivity and promotes apoptosis of hepatocellular carcinoma cells through inhibiting autophagy. Cell Death Dis. 2017, 8, e2540. [Google Scholar] [CrossRef] [PubMed]
- Cochetti, G.; Poli, G.; Guelfi, G.; Boni, A.; Egidi, M.G.; Mearini, E. Different levels of serum microRNAs in prostate cancer and benign prostatic hyperplasia: Evaluation of potential diagnostic and prognostic role. Onco Targets Ther. 2016, 9, 7545–7553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Sun, B.; Sun, H.; Zhao, X.; Wang, X.; Zhao, N.; Zhang, Y.; Li, Y.; Gu, Q.; Liu, F.; et al. Regulation of proliferation, angiogenesis and apoptosis in hepatocellular carcinoma by miR-26b-5p. Tumour. Biol. 2016, 37, 10965–10979. [Google Scholar] [CrossRef]
- Shaikh, I.; Ansari, A.; Ayachit, G.; Gandhi, M.; Sharma, P.; Bhairappanavar, S.; Joshi, C.G.; Das, J. Differential gene expression analysis of HNSCC tumors deciphered tobacco dependent and independent molecular signatures. Oncotarget 2019, 10, 6168–6183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lekchnov, E.A.; Amelina, E.V.; Bryzgunova, O.E.; Zaporozhchenko, I.A.; Konoshenko, M.Y.; Yarmoschuk, S.V.; Murashov, I.S.; Pashkovskaya, O.A.; Gorizkii, A.M.; Zheravin, A.A.; et al. Searching for the Novel Specific Predictors of Prostate Cancer in Urine: The Analysis of 84 miRNA Expression. Int. J. Mol. Sci. 2018, 19, 88. [Google Scholar] [CrossRef] [Green Version]
- Shen, G.; Lin, Y.; Yang, X.; Zhang, J.; Xu, Z.; Jia, H. MicroRNA-26b inhibits epithelial-mesenchymal transition in hepatocellular carcinoma by targeting USP9X. BMC Cancer 2014, 14, 393. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, K.; Seki, N.; Matsushita, R.; Yonemori, M.; Yoshino, H.; Nakagawa, M.; Enokida, H. Tumour-suppressive miRNA-26a-5p and miR-26b-5p inhibit cell aggressiveness by regulating PLOD2 in bladder cancer. Br. J. Cancer 2016, 115, 354–363. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; He, M.L.; Wang, L.; Chen, Y.; Liu, X.; Dong, Q.; Chen, Y.C.; Peng, Y.; Yao, K.T.; Kung, H.F.; et al. MiR-26a inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma through repression of EZH2. Cancer Res. 2011, 71, 225–233. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Liu, X.X.; He, J.R.; Zhou, C.X.; Guo, M.; He, M.; Li, M.F.; Chen, G.Q.; Zhao, Q. Pathologically decreased miR-26a antagonizes apoptosis and facilitates carcinogenesis by targeting MTDH and EZH2 in breast cancer. Carcinogenesis 2011, 32, 2–9. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Long, C.; Yang, G.; Luo, Y.; Du, H. MiR-26b inhibits melanoma cell proliferation and enhances apoptosis by suppressing TRAF5-mediated MAPK activation. Biochem. Biophys. Res. Commun. 2016, 471, 361–367. [Google Scholar] [CrossRef]
- Wei, C.Y.; Wang, L.; Zhu, M.X.; Deng, X.Y.; Wang, D.H.; Zhang, S.M.; Ying, J.H.; Yuan, X.; Wang, Q.; Xuan, T.F.; et al. TRIM44 activates the AKT/mTOR signal pathway to induce melanoma progression by stabilizing TLR4. J. Exp. Clin. Cancer Res. 2019, 38, 137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedrich, M.; Heimer, N.; Stoehr, C.; Steven, A.; Wach, S.; Taubert, H.; Hartmann, A.; Seliger, B. CREB1 is affected by the microRNAs miR-22-3p, miR-26a-5p, miR-27a-3p, and miR-221-3p and correlates with adverse clinicopathological features in renal cell carcinoma. Sci. Rep. 2020, 10, 6499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Wang, Y.; Song, Y. MicroRNA-26b inhibits the immune response to Mycobacterium tuberculosis (M.tb) infection in THP-1 cells via targeting TGFbeta-activated kinase-1 (TAK1), a promoter of the NF-kappaB pathway. Int. J. Clin. Exp. Pathol. 2018, 11, 1218–1227. [Google Scholar] [PubMed]
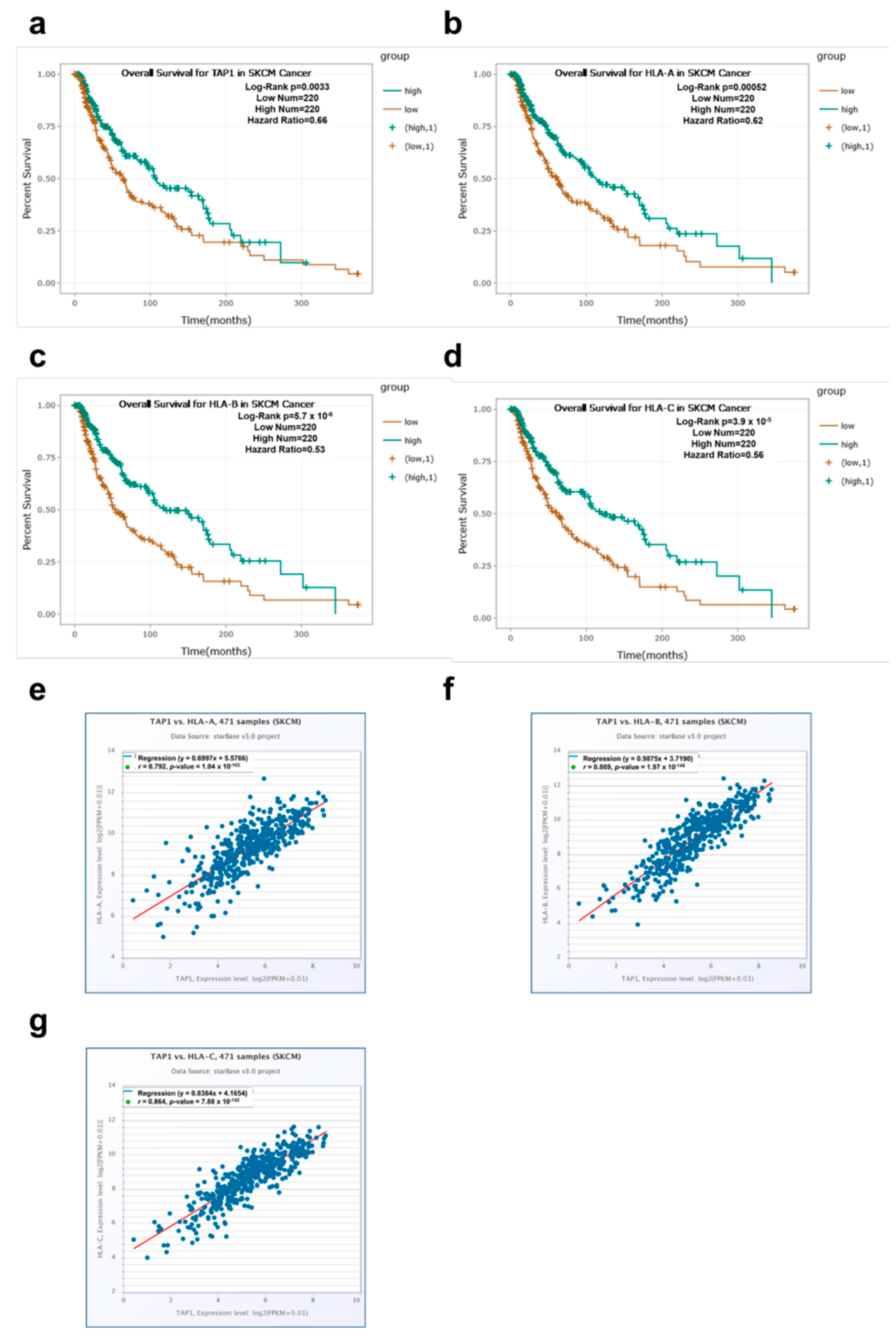

| a: TAP1 | |||||
|---|---|---|---|---|---|
| Abbreviation | Type of Cancer | Number of Cases | Median Gene Expression | Correlation | p-Value |
| OV | ovarian serous adenocarcinoma | 374 | 18.68 | positive | 0.00053 |
| LGG | brain lower grade glioma | 523 | 11.19 | negative | 0.0032 |
| SKCM | skin cutaneous melanoma | 440 | 41.39 | positive | 0.0033 |
| UVM | uveal melanoma | 80 | 9.02 | negative | 0.0069 |
| COAD | colon adenocarcinoma | 447 | 44.99 | positive | 0.044 |
| THYM | thymoma | 118 | 24.35 | negative | 0.1 |
| UCEC | uterine corpus endometrial carcinoma | 537 | 21.75 | positive | 0.56 |
| THCA | thyroid carcinoma | 509 | 12.61 | negative | 0.8 |
| b: HLA-A | |||||
| LGG | brain lower grade glioma | 523 | 133.35 | negative | 0.00012 |
| SKCM | skin cutaneous melanoma | 440 | 671.89 | positive | 0.00052 |
| UCEC | uterine corpus endometrial carcinoma | 537 | 523.84 | positive | 0.0074 |
| UVM | uveal melanoma | 80 | 502.23 | negative | 0.0081 |
| THCA | thyroid carcinoma | 509 | 468.03 | positive | 0.043 |
| OV | ovarian serous adenocarcinoma | 374 | 219.47 | positive | 0.059 |
| THYM | thymoma | 118 | 325.13 | negative | 0.063 |
| COAD | colon adenocarcinoma | 447 | 516.21 | positive | 0.26 |
| c: HLA-B | |||||
| SKCM | skin cutaneous melanoma | 440 | 620.14 | positive | 5.7 × 10−6 |
| LGG | brain lower grade glioma | 523 | 129.28 | negative | 0.00027 |
| UVM | uveal melanoma | 80 | 238.66 | negative | 0.0011 |
| THYM | thymoma | 118 | 390.69 | negative | 0.0026 |
| OV | ovarian serous cystadenocarcinoma | 374 | 300.77 | positive | 0.0069 |
| UCEC | uterine corpus endometrial carcinoma | 537 | 545.19 | positive | 0.053 |
| COAD | colon adenocarcinoma | 447 | 629.70 | positive | 0.18 |
| THCA | thyroid carcinoma | 509 | 503.74 | positive | 0.22 |
| d: HLA-C | |||||
| LGG | brain lower grade glioma | 523 | 117.10 | negative | 2.0 × 10−7 |
| SKCM | skin cutaneous melanoma | 440 | 458.38 | positive | 3.9 × 10−5 |
| THYM | thymoma | 118 | 260.27 | negative | 0.0081 |
| UVM | uveal melanoma | 80 | 247.30 | negative | 0.011 |
| UCEC | uterine corpus endometrial carcinoma | 537 | 406.42 | positive | 0.2 |
| OV | ovarian serous cystadenocarcinoma | 374 | 270.08 | positive | 0.4 |
| THCA | thyroid carcinoma | 509 | 334.74 | positive | 0.36 |
| COAD | colon adenocarcinoma | 447 | 461.01 | positive | 0.85 |
| miRBase Accession Number | miR | RNA-Seq Enrichment Ratio | miRWalk | Microrna.org | miRDB | TargetScan | RNA22 | RNAhybrid | In Silico Prediction Tools | Binding Energy (kcal/mol) |
|---|---|---|---|---|---|---|---|---|---|---|
| MIMAT0020601 | hsa-miR-1273f | 0.9 | yes | Yes | no | yes | no | yes | 4 | −30.1 |
| MIMAT0005797 | hsa-miR-1301-3p | 9.2 | yes | Yes | no | yes | yes | yes | 5 | −29.0 |
| MIMAT0004597 | hsa-miR-140-3p | 1.3 | yes | Yes | no | yes | yes | yes | 5 | −24.0 |
| MIMAT0004697 | hsa-miR-151a-5p | 19.1 | yes | Yes | no | yes | yes | yes | 5 | −26.1 |
| MIMAT0010214 | hsa-miR-151b | 6.3 | yes | Yes | no | yes | no | yes | 4 | −24.8 |
| MIMAT0004494 | hsa-miR-21-3p | 1036.0 | yes | Yes | yes | yes | no | yes | 5 | −20.3 |
| MIMAT0000077 | hsa-miR-22-3p | 102.9 | yes | Yes | no | yes | yes | yes | 5 | −21.6 |
| MIMAT0000079 | hsa-miR-24-1-5p | 5.0 | yes | Yes | no | yes | no | yes | 4 | −25.8 |
| MIMAT0004497 | hsa-miR-24-2-5p | 17.2 | yes | Yes | no | yes | no | yes | 4 | −25.9 |
| MIMAT0000082 | hsa-miR-26a-5p | 74.1 | yes | Yes | no | yes | no | yes | 4 | −25.1 |
| MIMAT0000083 | hsa-miR-26b-5p | 92.1 | yes | Yes | no | yes | no | yes | 4 | −25.4 |
| MIMAT0000085 | hsa-miR-28-5p | 16.1 | yes | Yes | no | yes | no | yes | 4 | −20.9 |
| MIMAT0000751 | hsa-miR-330-3p | 21.7 | yes | Yes | no | yes | no | yes | 4 | −24.8 |
| MIMAT0026612 | hsa-miR-504-3p | 16.2 | yes | Yes | no | yes | no | yes | 4 | −27.2 |
| MIMAT0004778 | hsa-miR-508-5p | 3.4 | yes | Yes | no | yes | no | yes | 4 | −25.5 |
| MIMAT0002823 | hsa-miR-512-3p | 4.0 | yes | Yes | no | yes | no | yes | 4 | −27.0 |
| MIMAT0002888 | hsa-miR-532-5p | 65.5 | yes | Yes | no | yes | no | yes | 4 | −20.5 |
| MIMAT0003254 | hsa-miR-548b-3p | 1.3 | yes | Yes | yes | yes | no | yes | 5 | −22.4 |
| MIMAT0004801 | hsa-miR-590-3p | 289.2 | yes | Yes | yes | yes | no | yes | 5 | −12.8 |
| MIMAT0026619 | hsa-miR-597-3p | 2.7 | yes | Yes | no | yes | no | yes | 4 | −23.5 |
| MIMAT0004926 | hsa-miR-708-5p | 13.8 | yes | Yes | no | yes | no | yes | 4 | −24.3 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lazaridou, M.-F.; Massa, C.; Handke, D.; Mueller, A.; Friedrich, M.; Subbarayan, K.; Tretbar, S.; Dummer, R.; Koelblinger, P.; Seliger, B. Identification of microRNAs Targeting the Transporter Associated with Antigen Processing TAP1 in Melanoma. J. Clin. Med. 2020, 9, 2690. https://doi.org/10.3390/jcm9092690
Lazaridou M-F, Massa C, Handke D, Mueller A, Friedrich M, Subbarayan K, Tretbar S, Dummer R, Koelblinger P, Seliger B. Identification of microRNAs Targeting the Transporter Associated with Antigen Processing TAP1 in Melanoma. Journal of Clinical Medicine. 2020; 9(9):2690. https://doi.org/10.3390/jcm9092690
Chicago/Turabian StyleLazaridou, Maria-Filothei, Chiara Massa, Diana Handke, Anja Mueller, Michael Friedrich, Karthikeyan Subbarayan, Sandy Tretbar, Reinhard Dummer, Peter Koelblinger, and Barbara Seliger. 2020. "Identification of microRNAs Targeting the Transporter Associated with Antigen Processing TAP1 in Melanoma" Journal of Clinical Medicine 9, no. 9: 2690. https://doi.org/10.3390/jcm9092690




