Comparison of BRAF Mutation Screening Strategies in a Large Real-Life Series of Advanced Melanoma Patients
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Samples
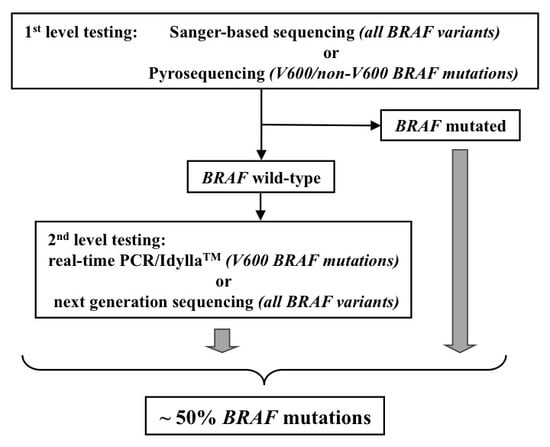
2.2. Molecular Testing
2.2.1. DNA Isolation and Screening
2.2.2. Sanger Sequencing (SS)
2.2.3. Pyrosequencing
2.2.4. Real-Time PCR (rtPCR) Test
2.2.5. Next-Generation Sequencing
2.3. Statistical Analysis
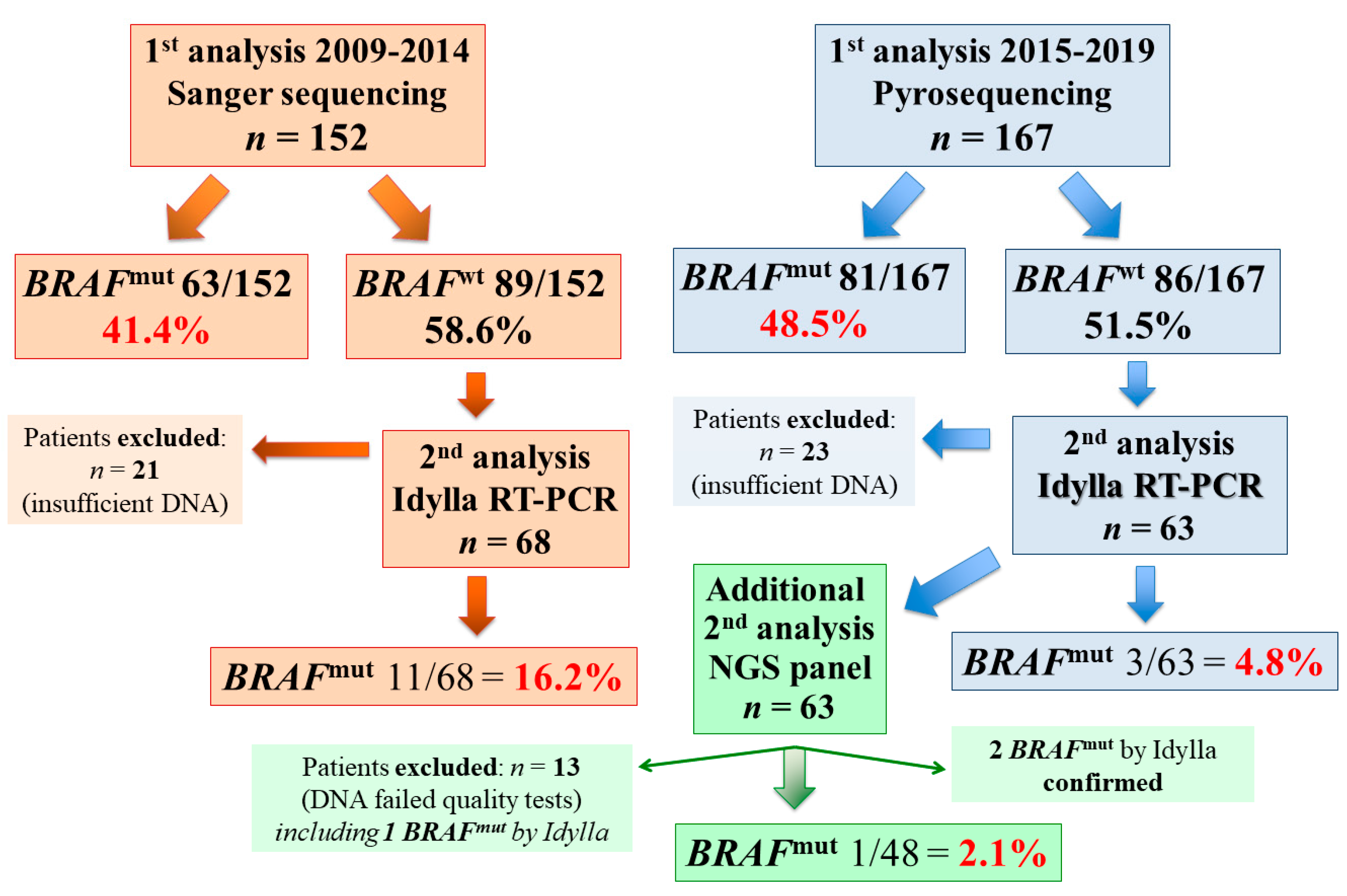
3. Results
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Cossu, A.; Casula, M.; Cesaraccio, R.; Lissia, A.; Colombino, M.; Sini, M.C.; Budroni, M.; Tanda, F.; Paliogiannis, P.; Palmieri, G. Epidemiology and genetic susceptibility of malignant melanoma in North Sardinia, Italy. Eur. J. Cancer Prev. 2017, 26, 263–267. [Google Scholar] [CrossRef] [PubMed]
- I Numeri del Cancro in Italia. Available online: https://www.aiom.it/wp-content/uploads/2019/09/2019_Numeri_Cancro-operatori-web.pdf (accessed on 11 May 2020).
- Parker, B.S.; Rautela, J.; Hertzog, P.J. Antitumor actions of interferons: Implications for cancer therapy. Nat. Rev. Cancer 2016, 16, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Luke, J.J.; Flaherty, K.T.; Ribas, A.; Long, G.V. Targeted agents and immunotherapies: Optimizing outcomes in melanoma. Nat. Rev. Clin. Oncol. 2017, 14, 463–482. [Google Scholar] [CrossRef]
- Su, M.Y.; Fisher, D.E. Immunotherapy in the precision medicine era: Melanoma and beyond. PLoS Med. 2016, 13, e1002196. [Google Scholar] [CrossRef]
- Sini, M.C.; Doneddu, V.; Paliogiannis, P.; Casula, M.; Colombino, M.; Manca, A.; Botti, G.; Ascierto, P.A.; Lissia, A.; Cossu, A.; et al. Genetic alterations in main candidate genes during melanoma progression. Oncotarget 2018, 9, 8531–8541. [Google Scholar] [CrossRef]
- Davies, H.; Bignell, G.R.; Cox, C.; Stephens, P.; Edkins, S.; Clegg, S.; Teague, J.; Woffendin, H.; Garnett, M.J.; Bottomley, W.; et al. Mutations of the BRAF gene in human cancer. Nature 2002, 417, 949–954. [Google Scholar] [CrossRef]
- Greaves, W.O.; Verma, S.; Patel, K.P.; Davies, M.A.; Barkoh, B.A.; Galbincea, J.M.; Yao, H.; Lazar, A.J.; Aldape, K.D.; Medeiros, L.J.; et al. Frequency and spectrum of BRAF mutations in a retrospective, single institution study of 1112 cases of melanoma. J. Mol. Diagn. 2013, 15, 220–226. [Google Scholar] [CrossRef]
- Wan, P.T.; Garnett, M.J.; Roe, S.M.; Lee, S.; Niculescu-Duvaz, D.; Good, V.M.; Project, C.G.; Jones, C.M.; Marshall, C.J.; Springer, C.J.; et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell 2004, 116, 855–867. [Google Scholar] [CrossRef]
- Chapman, P.B.; Hauschild, A.; Robert, C.; Haanen, J.B.; Ascierto, P.; Larkin, J.; Dummer, R.; Garbe, C.; Testori, A.; Maio, M.; et al. BRIM-3 Study Group. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 2011, 364, 2507–2516. [Google Scholar]
- Hauschild, A.; Grob, J.J.; Demidov, L.V.; Jouary, T.; Gutzmer, R.; Millward, M.; Rutkowski, P.; Blank, C.U.; Miller, W.H., Jr.; Kaempgen, E.; et al. Dabrafenib in BRAF-mutated metastatic melanoma: A multicentre, open-label, phase 3 randomised controlled trial. Lancet 2012, 380, 358–365. [Google Scholar] [CrossRef]
- Dummer, R.; Hauschild, A.; Lindenblatt, N.; Pentheroudakis, G.; Keilholz, U. Cutaneous melanoma: ESMO clinical practice guidelines for diagnosis, treatment, and follow-up. Ann. Oncol. 2016, 26 (Suppl. 5), 126–132. [Google Scholar] [CrossRef] [PubMed]
- Seth, R.; Messersmith, H.; Kaur, V.; Kirkwood, J.M.; Kudchadkar, R.; McQuade, J.L.; Provenzano, A.; Swami, U.; Weber, J.; Alluri, K.C.; et al. Systemic therapy for melanoma: ASCO Guideline. J. Clin. Oncol. 2020, 31, JCO2000198. [Google Scholar] [CrossRef] [PubMed]
- Ascierto, P.A.; Agarwala, S.S.; Botti, G.; Budillon, A.; Davies, M.A.; Dummer, R.; Ernstoff, M.; Ferrone, S.; Formenti, S.; Gajewski, T.F.; et al. Perspectives in melanoma: Meeting report from the Melanoma Bridge (November 29th-1 December 1st, 2018, Naples, Italy). J. Transl. Med. 2019, 17, 234. [Google Scholar] [CrossRef] [PubMed]
- National Comprehensive Cancer Network Guidelines. Available online: https://www.nccn.org/professionals/physician_gls/default.aspx (accessed on 11 May 2020).
- Michielin, O.; van Akkooi, A.C.J.; Ascierto, P.A.; Dummer, R.; Keilholz, U.; ESMO Guidelines Committee. Electronic address: Clinicalguidelines@esmo.org. Cutaneous melanoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2019, 30, 1884–1901. [Google Scholar] [CrossRef]
- Coit, D.G.; Thompson, J.A.; Albertini, M.R.; Barker, C.; Carson, W.E.; Contreras, C.; Daniels, G.A.; DiMaio, D.; Fields, R.C.; Fleming, M.D.; et al. Cutaneous Melanoma, Version 2.2019, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2019, 17, 367–402. [Google Scholar] [CrossRef]
- Garbe, C.; Amaral, T.; Peris, K.; Hauschild, A.; Arenberger, P.; Bastholt, L.; Bataille, V.; Del Marmol, V.; Dréno, B.; Fargnoli, M.C.; et al. European consensus-based interdisciplinary guideline for melanoma. Part 2: Treatment—Update 2019. Eur. J. Cancer 2020, 126, 159–177. [Google Scholar]
- Poklepovic, A.S.; Luke, J.J. Considering adjuvant therapy for stage II melanoma. Cancer 2020, 126, 1166–1174. [Google Scholar] [CrossRef]
- Long, G.V.; Hauschild, A.; Santinami, M.; Atkinson, V.; Mandalà, M.; Chiarion-Sileni, V.; Larkin, J.; Nyakas, M.; Dutriaux, C.; Haydon, A.; et al. Adjuvant Dabrafenib plus Trametinib in Stage III BRAF-Mutated Melanoma. N. Engl. J. Med. 2017, 377, 1813–1823. [Google Scholar] [CrossRef]
- Hauschild, A.; Dummer, R.; Schadendorf, D.; Santinami, M.; Atkinson, V.; Mandalà, M.; Chiarion-Sileni, V.; Larkin, J.; Nyakas, M.; Dutriaux, C.; et al. Longer Follow-Up Confirms Relapse-Free Survival Benefit with Adjuvant Dabrafenib Plus Trametinib in Patients with Resected BRAF V600-Mutant Stage III Melanoma. J. Clin. Oncol. 2018, 36, JCO1801219. [Google Scholar] [CrossRef]
- Hauschild, A.; Dummer, R.; Santinami, M.; Atkinson, V.; Mandalà, M.; Kirkwood, J.M.; Chiarion Sileni, V.; Larkin, J.M.G.; Nyakas, M.; Dutriaux, C.; et al. Long-term benefit of adjuvant dabrafenib + trametinib (D+T) in patients (pts) with resected stage III BRAF V600–mutant melanoma: Five-year analysis of COMBI-AD. J. Clin. Oncol. 2020, 38, 10001. [Google Scholar] [CrossRef]
- Dummer, R.; Ascierto, P.A.; Gogas, H.J.; Arance, A.; Mandala, M.; Liszkay, G.; Garbe, C.; Schadendorf, D.; Krajsova, I.; Gutzmer, R.; et al. Overall survival in patients with BRAF-mutant melanoma receiving encorafenib plus binimetinib versus vemurafenib or encorafenib (COLUMBUS): A multicentre, open-label, randomised, phase 3 trial. Lancet Oncol. 2018, 19, 1315–1327. [Google Scholar] [CrossRef]
- Ascierto, P.A.; Lewis, K.D.; Di Giacomo, A.M.; Demidov, L.; Mandalà, M.; Bondarenko, I.; Herbert, C.; Mackiewicz, A.; Rutkowski, P.; Guminski, A.; et al. Prognostic impact of baseline tumour immune infiltrate on disease-free survival in patients with completely resected, BRAFv600 mutation-positive melanoma receiving adjuvant vemurafenib. Ann. Oncol. 2020, 31, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Colombino, M.; Capone, M.; Lissia, A.; Cossu, A.; Rubino, C.; De Giorgi, V.; Massi, D.; Fonsatti, E.; Staibano, S.; Nappi, O.; et al. BRAF/NRAS mutation frequencies among primary tumors and metastases in patients with melanoma. J. Clin. Oncol. 2012, 30, 2522–2529. [Google Scholar] [CrossRef] [PubMed]
- Colombino, M.; Lissia, A.; Franco, R.; Botti, G.; Ascierto, P.A.; Manca, A.; Sini, M.C.; Pisano, M.; Paliogiannis, P.; Tanda, F.; et al. Unexpected distribution of cKIT and BRAF mutations among southern Italian patients with sinonasal melanoma. Dermatology 2013, 226, 279–284. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Network. Genomic classification of cutaneous melanoma. Cell 2015, 161, 1681–1696.
- Palmieri, G.; Ombra, M.; Colombino, M.; Casula, M.; Sini, M.; Manca, A.; Paliogiannis, P.; Ascierto, P.A.; Cossu, A. Multiple molecular pathways in melanomagenesis: Characterization of therapeutic targets. Front. Oncol. 2015, 5, 183. [Google Scholar] [CrossRef]
- Feller, J.K.; Yang, S.; Mahalingam, M. Immunohistochemistry with a mutation-specific monoclonal antibody as a screening tool for the BRAFV600E mutational status in primary cutaneous malignant melanoma. Mod. Pathol. 2013, 26, 414–420. [Google Scholar] [CrossRef]
- Sholl, L.M.; Andea, A.; Bridge, J.A.; Cheng, L.; Davies, M.A.; Ehteshami, M.; Gangadhar, T.C.; Kamel-Reid, S.; Lazar, A.; Raparia, K.; et al. Template for reporting results of biomarker testing of specimens from Patients with melanoma. Arch. Pathol. Lab. Med. 2016, 140, 355–357. [Google Scholar] [CrossRef]
- Vallée, A.; Denis-Musquer, M.; Herbreteau, G.; Théoleyre, S.; Bossard, C.; Denis, M.G. Prospective evaluation of two screening methods for molecular testing of metastatic melanoma: Diagnostic performance of BRAF V600E immunohistochemistry and of a NRAS-BRAF fully automated real-time PCR-based assay. PLoS ONE 2019, 14, e0221123. [Google Scholar] [CrossRef]
- Palomba, G.; Doneddu, V.; Cossu, A.; Paliogiannis, P.; Manca, A.; Casula, M.; Colombino, M.; Lanzillo, A.; Defraia, E.; Pazzola, A.; et al. Prognostic impact of KRAS, NRAS, BRAF, and PIK3CA mutations in primary colorectal carcinomas: A population-based study. J. Transl. Med. 2016, 14, 292. [Google Scholar] [CrossRef] [PubMed]
- Barel, F.; Guibourg, B.; Lambros, L.; Le Flahec, G.; Marcorelles, P.; Uguen, A. Evaluation of a Rapid, Fully Automated Platform for Detection of BRAF and NRAS Mutations in Melanoma. Acta Derm. Venereol. 2018, 98, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, G.; Colombino, M.; Casula, M.; Manca, A.; Mandalà, M.; Cossu, A.; Italian Melanoma Intergroup (IMI). Molecular pathways in melanomagenesis: What we learned from next-generation sequencing approaches. Curr. Oncol. Rep. 2018, 20, 86. [Google Scholar] [CrossRef]
- Ihle, M.A.; Fassunke, J.; König, K.; Grünewald, I.; Schlaak, M.; Kreuzberg, N.; Tietze, L.; Schildhaus, H.U.; Büttner, R.; Merkelbach-Bruse, S. Comparison of high resolution melting analysis, pyrosequencing, next gene-ration sequencing and immunohistochemistry to conventional Sanger sequencing for the detection of p.V600E and non-p.V600E BRAF mutations. BMC Cancer 2014, 14, 13. [Google Scholar] [CrossRef] [PubMed]
- Bruno, W.; Martinuzzi, C.; Andreotti, V.; Pastorino, L.; Spagnolo, F.; Dalmasso, B.; Cabiddu, F.; Gualco, M.; Ballestrero, A.; Bianchi-Scarrà, G.; et al. Heterogeneity and frequency of BRAF mutations in primary melanoma: Comparison between molecular methods and immunohistochemistry. Oncotarget 2017, 8, 8069–8082. [Google Scholar] [CrossRef] [PubMed]
- Mosko, M.J.; Nakorchevsky, A.A.; Flores, E.; Metzler, H.; Ehrich, M.; van den Boom, D.J.; Sherwood, J.L.; Nygren, A.O. Ultrasensitive detection of multiplexed somatic mutations using MALDI-TOF Mass Spectrometry. J. Mol. Diagn. 2016, 18, 23–31. [Google Scholar] [CrossRef]
- Casula, C.; Muggiano, A.; Cossu, A.; Budroni, M.; Caracò, C.; Ascierto, P.A.; Pagani, E.; Stanganelli, I.; Canzanella, S.; Sini, M.C.; et al. Role of key-regulator genes in melanoma susceptibility and pathogenesis among patients from South Italy. BMC Cancer 2009, 9, 352. [Google Scholar] [CrossRef]
- Palomba, G.; Loi, A.; Porcu, E.; Cossu, A.; Zara, I.; Budroni, M.; Dei, M.; Lai, S.; Mulas, A.; Olmeo, N.; et al. Genome-wide association study of susceptibility loci for breast cancer in Sardinian population. BMC Cancer 2015, 15, 383. [Google Scholar] [CrossRef][Green Version]
| Characteristics | Data |
|---|---|
| Age at diagnosis, median (range) | 65 (21–92) |
| Male gender, n (%) | 183 (57.4) |
| Primary melanoma localization (n = 319) | |
| Limbs, n (%) | 93 (29.2) |
| Head and neck, n (%) | 44 (13.8) |
| Trunk, n (%) | 152 (47.6) |
| Occult, n (%) | 30 (9.4) |
| Lymph node metastasis, n (%) | 22 (6.9) |
| Visceral metastasis, n (%) | 8 (2.5) |
| Histology (n = 289) | |
| SSM, n (%) | 146 (50.5) |
| Nodular, n (%) | 112 (38.7) |
| Acral, n (%) | 19 (6.6) |
| Lentigo maligna, n (%) | 8 (2.8) |
| Mucosal, n (%) | 4 (1.4) |
| Breslow class (n = 285) | |
| ≤1 mm, n (%) | 23 (8.1) |
| >1-≤2 mm, n (%) | 78 (27.4) |
| >2-≤4 mm, n (%) | 95 (33.3) |
| >4 mm, n (%) | 89 (31.2) |
| Ulceration (n = 248) | |
| Present, n (%) | 137 (55.2) |
| Absent, n (%) | 111 (44.8) |
| Mitosis number (n = 243) | |
| <1, n (%) | 47 (19.3) |
| ≥1, n (%) | 196 (80.7) |
| AJCC stage at diagnosis (n = 285) | |
| IA-IB, n (%) | 26 (9.2) |
| IIA-IIB, n (%) | 107 (37.5) |
| IIC, n (%) | 28 (9.8) |
| III, n (%) | 101 (35.4) |
| IV, n (%) | 23 (8.1) |
| Lymph node metastasis at diagnosis (n = 289) | |
| pN0, n (%) | 171 (59.2) |
| pN+, n (%) | 118 (40.8) |
| Characteristics | Mutated | Wild-Type | p |
|---|---|---|---|
| BRAF (n = 319) | |||
| Mutated, n (%) | 144 (45.1) | 175 (54.9) | |
| Gender, n (%) | |||
| Male, n (%) | 79 (54.9) | 104 (59.4) | 0.479 |
| Female, n (%) | 65 (45.1) | 71 (40.6) | |
| Age at diagnosis, n (%) | |||
| ≤55 years, n (%) | 58 (40.3) | 37 (21.1) | 0.003 |
| >55 years, n (%) | 86 (59.7) | 138 (78.9) | |
| NRAS (n = 272) | |||
| Mutated, n (%) | 40 (14.7) | 232 (85.3) | |
| Gender, n (%) | |||
| Male, n (%) | 25 (62.5) | 129 (55.6) | 0.522 |
| Female, n (%) | 15 (37.5) | 103 (44.4) | |
| Age at diagnosis, n (%) | |||
| ≤55 years, n (%) | 4 (10) | 73 (31.5) | 0.004 |
| >55 years, n (%) | 36 (90) | 159 (68.5) | |
| Exon | Mutation | Base Change | Amino Acid Change | Mutated Samples | % |
|---|---|---|---|---|---|
| BRAF | |||||
| 15 | V600D | 1799–1800 TG>AT | Val to Asp | 3 | 2.1 |
| 15 | V600E | 1799 T>A | Val to Glu | 117 | 81.2 |
| 15 | V600E | 1799_1800TG>AA | Val to Glu | 3 | 2.1 |
| 15 | V600K | 1798–99 GT>AA | Val to Lys | 19 | 13.2 |
| 15 | V600R | 1798–99 GT>AG | Val to Arg | 1 | 0.7 |
| 15 | K601E | 1790 T>G | Leu to Arg | 1 | 0.7 |
| NRAS | |||||
| 2 | G12A | 35 G>C | Gly to Ala | 1 | 2.5 |
| 2 | G13D | 38 G>A | Gly to Asp | 2 | 5.0 |
| 2 | G13R | 37 G>C | Gly to Arg | 2 | 5.0 |
| 3 | Q61H | 183 A>T | Gln to His | 1 | 2.5 |
| 3 | Q61K | 181 C>A | Gln to Lys | 12 | 30.0 |
| 3 | Q61L | 182 A>T | Gln to Leu | 3 | 7.5 |
| 3 | Q61R | 182 A>G | Gln to Arg | 19 | 47.5 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Colombino, M.; Rozzo, C.; Paliogiannis, P.; Casula, M.; Manca, A.; Doneddu, V.; Fedeli, M.A.; Sini, M.C.; Palomba, G.; Pisano, M.; et al. Comparison of BRAF Mutation Screening Strategies in a Large Real-Life Series of Advanced Melanoma Patients. J. Clin. Med. 2020, 9, 2430. https://doi.org/10.3390/jcm9082430
Colombino M, Rozzo C, Paliogiannis P, Casula M, Manca A, Doneddu V, Fedeli MA, Sini MC, Palomba G, Pisano M, et al. Comparison of BRAF Mutation Screening Strategies in a Large Real-Life Series of Advanced Melanoma Patients. Journal of Clinical Medicine. 2020; 9(8):2430. https://doi.org/10.3390/jcm9082430
Chicago/Turabian StyleColombino, Maria, Carla Rozzo, Panagiotis Paliogiannis, Milena Casula, Antonella Manca, Valentina Doneddu, Maria Antonietta Fedeli, Maria Cristina Sini, Grazia Palomba, Marina Pisano, and et al. 2020. "Comparison of BRAF Mutation Screening Strategies in a Large Real-Life Series of Advanced Melanoma Patients" Journal of Clinical Medicine 9, no. 8: 2430. https://doi.org/10.3390/jcm9082430
APA StyleColombino, M., Rozzo, C., Paliogiannis, P., Casula, M., Manca, A., Doneddu, V., Fedeli, M. A., Sini, M. C., Palomba, G., Pisano, M., Ascierto, P. A., Caracò, C., Lissia, A., Cossu, A., & Palmieri, G. (2020). Comparison of BRAF Mutation Screening Strategies in a Large Real-Life Series of Advanced Melanoma Patients. Journal of Clinical Medicine, 9(8), 2430. https://doi.org/10.3390/jcm9082430