The Clinical Significance of RAS, PIK3CA, and PTEN Mutations in Non-Small Cell Lung Cancer Using Cell-Free DNA
Abstract
1. Introduction
2. Material and Methods
2.1. Patients
2.2. Detection of EGFR Mutations in Tumor Tissues
2.3. cfDNA Extraction
2.4. Next-Generation Sequencing (NGS) and Sequencing Data Analysis
2.5. Droplet Digital PCR
2.6. Statistical Analysis
3. Results
3.1. Detection of Somatic Mutations from cfDNA in 124 Patients with NSCLC
3.1.1. Detection of Activating EGFR Mutations and EGFR T790M Mutation by cfDNA ICP Analysis
3.1.2. Detection of RAS, PIK3CA and PTEN Mutations Using cfDNA ICP Analysis
3.1.3. Validation with ddPCR: EGFR Mutations and KRAS Mutation
3.2. The Clinical Characteristics of the 124 Patients with NSCLC
Patients with RAS/PIK3CA/PTEN Mutations
3.3. The Clinical Characteristics of Patients with NSCLC with Comprehensive EGFR Activating Mutations
Patients with RAS/PIK3CA/PTEN Mutations
3.4. EGFR-TKI Treatment Response in Patients Treated with EGFR-TKIs
Patients with RAS/PIK3CA/PTEN Mutations
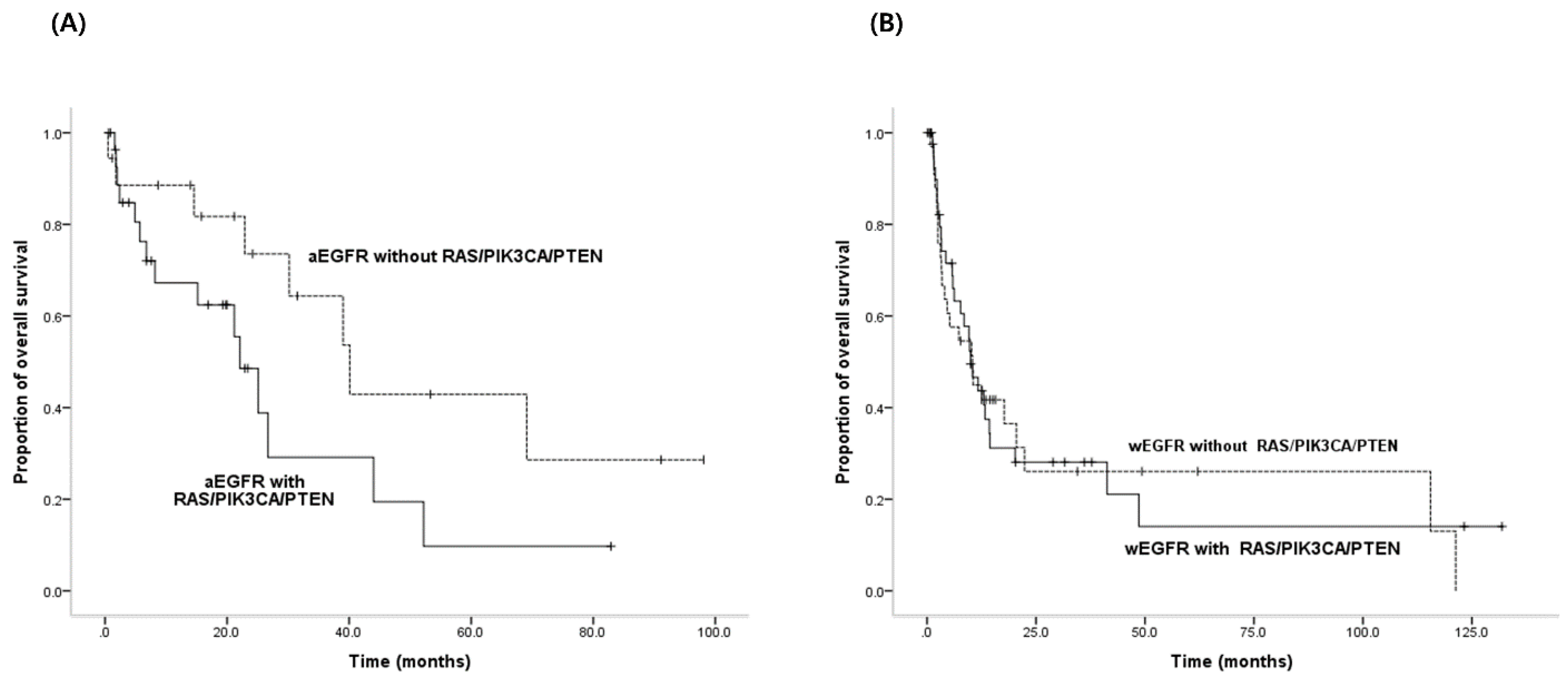
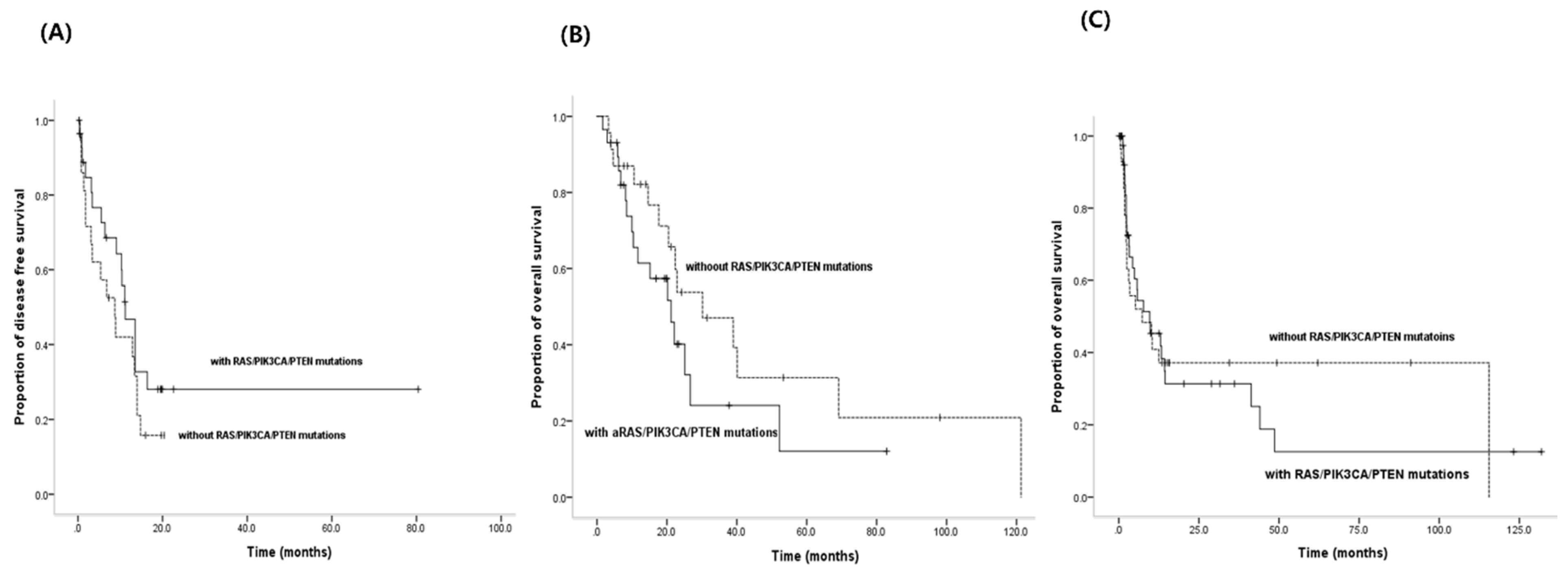
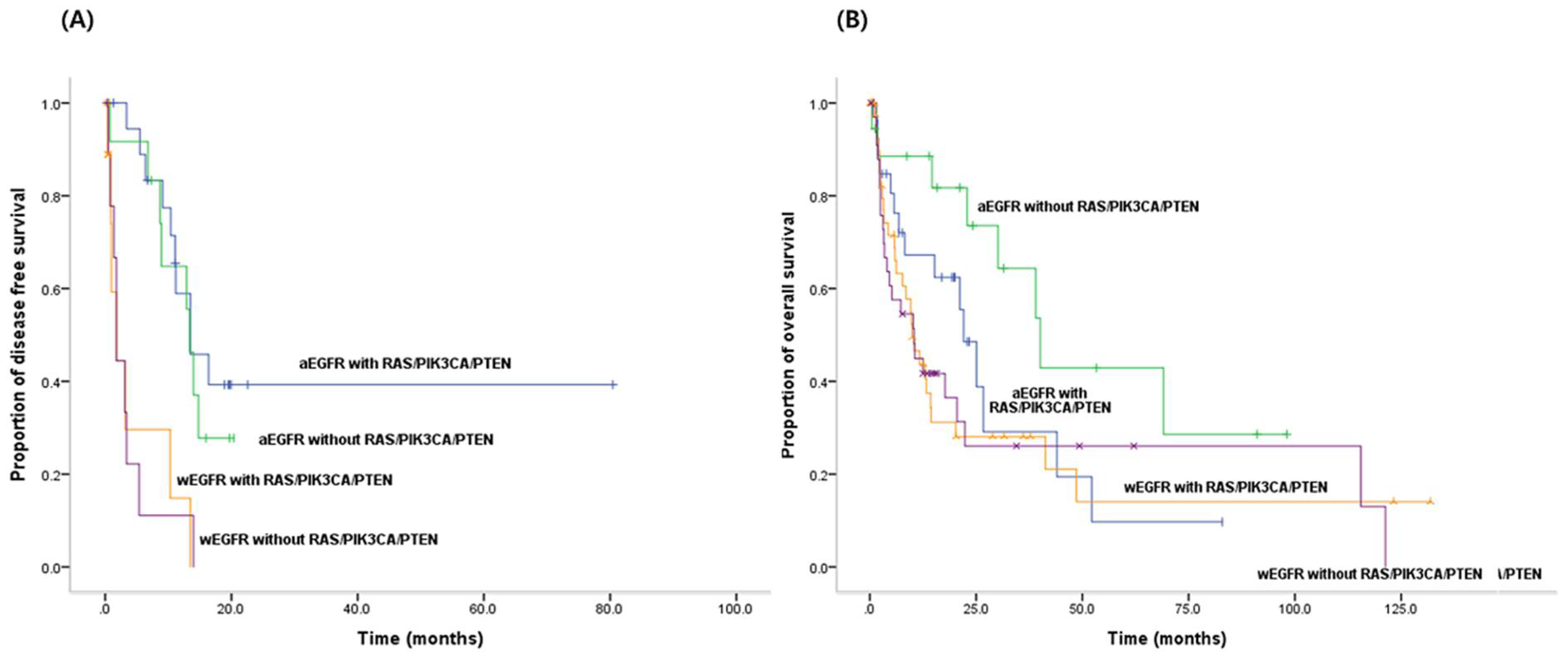
3.5. PFS for EGFR-TKIs and OS According to both Activating EGFR Mutation Status and Status of RAS/PIK3CA/PTEN Mutations
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sequist, L.V.; Yang, J.C.; Yamamoto, N.; O’Byrne, K.; Hirsh, V.; Mok, T.; Geater, S.L.; Orlov, S.; Tsai, C.M.; Boyer, M.; et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J. Clin. Oncol. 2013, 31, 3327–3334. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, S.; Boggon, T.J.; Dayaram, T.; Jänne, P.A.; Kocher, O.; Meyerson, M.; Johnson, B.E.; Eck, M.J.; Tenen, D.G.; Halmos, B. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2005, 352, 786–792. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Y.; Cheng, Y.N.; Han, L.; Wei, F.; Yu, W.W.; Zhang, X.W.; Cao, S.; Yu, J.P. Predictive value of K-ras and PIK3CA in non-small cell lung cancer patients treated with EGFR-TKIs: A systemic review and meta-analysis. Cancer Biol. Med. 2015, 12, 126–139. [Google Scholar] [PubMed]
- Shepherd, F.A.; Domerg, C.; Hainaut, P.; Jänne, P.A.; Pignon, J.P.; Graziano, S.; Douillard, J.Y.; Brambilla, E.; Le Chevalier, T.; Seymour, L.; et al. Pooled analysis of the prognostic and predictive effects of KRAS mutation status and KRAS mutation subtype in early-stage resected non-small-cell lung cancer in four trials of adjuvant chemotherapy. J. Clin. Oncol. 2013, 31, 2173–2181. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, L.; Zhu, Y.; Huang, C.; Qin, Y.; Liu, H.; Ren-Heidenreich, L.; Shi, B.; Ren, H.; Chu, X.; et al. Coexistence of EGFR with KRAS, or BRAF, or PIK3CA somatic mutations in lung cancer: A comprehensive mutation profiling from 5125 Chinese cohorts. Br. J. Cancer 2014, 110, 2812–2820. [Google Scholar] [CrossRef]
- Jin, G.; Kim, M.J.; Jeon, H.S.; Choi, J.E.; Kim, D.S.; Lee, E.B.; Cha, S.I.; Yoon, G.S.; Kim, C.H.; Jung, T.H.; et al. PTEN mutations and relationship to EGFR, ERBB2, KRAS, and TP53 mutations in non-small cell lung cancers. Lung Cancer 2010, 69, 279–283. [Google Scholar] [CrossRef]
- Rachiglio, A.M.; Abate, R.E.; Sacco, A.; Pasquale, R.; Fenizia, F.; Lambiase, M.; Morabito, A.; Montanino, A.; Rocco, G.; Romano, C.; et al. Limits and potential of targeted sequencing analysis of liquid biopsy in patients with lung and colon carcinoma. Oncotarget 2016, 7, 66595–66605. [Google Scholar] [CrossRef]
- McGranahan, N.; Favero, F.; de Bruin, E.C.; Birkbak, N.J.; Szallasi, Z.; Swanton, C. Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci. Transl. Med. 2015, 7, 283ra254. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Hoon, D.S.; Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef]
- Fenizia, F.; De Luca, A.; Pasquale, R.; Sacco, A.; Forgione, L.; Lambiase, M.; Iannaccone, A.; Chicchinelli, N.; Franco, R.; Rossi, A.; et al. EGFR mutations in lung cancer: From tissue testing to liquid biopsy. Future Oncol. 2015, 11, 1611–1623. [Google Scholar] [CrossRef]
- Sung, J.S.; Chong, H.Y.; Kwon, N.J.; Kim, H.M.; Lee, J.W.; Kim, B.; Lee, S.B.; Park, C.W.; Choi, J.Y.; Chang, W.J.; et al. Detection of somatic variants and EGFR mutations in cell-free DNA from non-small cell lung cancer patients by ultra-deep sequencing using the ion ampliseq cancer hotspot panel and droplet digital polymerase chain reaction. Oncotarget 2017, 8, 106901–106912. [Google Scholar] [CrossRef] [PubMed]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef] [PubMed]
- Normanno, N.; Denis, M.G.; Thress, K.S.; Ratcliffe, M.; Reck, M. Guide to detecting epidermal growth factor receptor (EGFR) mutations in ctDNA of patients with advanced non-small-cell lung cancer. Oncotarget 2017, 8, 12501–12516. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014, 511, 543–550. [Google Scholar] [CrossRef] [PubMed]
- Zill, O.A.; Banks, K.C.; Fairclough, S.R.; Mortimer, S.A.; Vowles, J.V.; Mokhtari, R.; Gandara, D.R.; Mack, P.C.; Odegaard, J.I.; Nagy, R.J.; et al. The landscape of actionable genomic alterations in cell-free circulating tumor DNA from 21,807 advanced cancer patients. Clin. Cancer Res. 2018, 24, 3528–3538. [Google Scholar] [CrossRef]
- Gormally, E.; Caboux, E.; Vineis, P.; Hainaut, P. Circulating free DNA in plasma or serum as biomarker of carcinogenesis: Practical aspects and biological significance. Mutat. Res. Rev. Mutat. Res. 2007, 635, 105–117. [Google Scholar] [CrossRef]
- Kim, S.T.; Lee, W.S.; Lanman, R.B.; Mortimer, S.; Zill, O.A.; Kim, K.M.; Jang, K.T.; Kim, S.H.; Park, S.H.; Park, J.O.; et al. Prospective blinded study of somatic mutation detection in cell-free DNA utilizing a targeted 54-gene next generation sequencing panel in metastatic solid tumor patients. Oncotarget 2015, 6, 40360–40369. [Google Scholar] [CrossRef][Green Version]
- Tie, J.; Kinde, I.; Wang, Y.; Wong, H.L.; Roebert, J.; Christie, M.; Tacey, M.; Wong, R.; Singh, M.; Karapetis, C.S.; et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann. Oncol. 2015, 26, 1715–1722. [Google Scholar] [CrossRef]
- Arriola, E.; Paredes-Lario, A.; Garcia-Gomez, R.; Diz-Tain, P.; Constenla, M.; García-Girón, C.; Márquez, G.; Reck, M.; López-Vivanco, G. Comparison of plasma ctDNA and tissue/cytology-based techniques for the detection of EGFR mutation status in advanced NSCLC: Spanish data subset from ASSESS. Clin. Transl. Oncol. 2018, 20, 1261–1267. [Google Scholar] [CrossRef]
- VanderLaan, P.A.; Rangachari, D.; Mockus, S.M.; Spotlow, V.; Reddi, H.V.; Malcolm, J.; Huberman, M.S.; Joseph, L.J.; Kobayashi, S.S.; Costa, D.B. Mutations in TP53, PIK3CA, PTEN and other genes in EGFR mutated lung cancers: Correlation with clinical outcomes. Lung Cancer 2017, 106, 17–21. [Google Scholar] [CrossRef]
- Hollander, M.C.; Blumenthal, G.M.; Dennis, P.A. PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nat. Rev. Cancer 2011, 11, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Hu, C.P.; He, B.X.; Chen, X.; Lu, X.X.; Xie, M.X.; Li, W.; He, S.Y.; You, S.J.; Chen, Q. PTEN expression is a prognostic marker for patients with non-small cell lung cancer: A systematic review and meta-analysis of the literature. Oncotarget 2016, 7, 57832–57840. [Google Scholar] [CrossRef] [PubMed]
- Shugg, R.P.; Thomson, A.; Tanabe, N.; Kashishian, A.; Steiner, B.H.; Puri, K.D.; Pereverzev, A.; Lannutti, B.J.; Jirik, F.R.; Dixon, S.J.; et al. Effects of isoform-selective phosphatidylinositol 3-kinase inhibitors on osteoclasts: Actions on cytoskeletal organization, survival, and resorption. J. Biol. Chem. 2013, 288, 35346–35357. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liu, J.; Wang, Y.; Tan, X.; Zhao, W.; Xing, X.; Qiu, Y.; Wang, R.; Jin, M.; Fan, G.; et al. Phosphatidylinositol 3-kinase beta and delta isoforms play key roles in metastasis of prostate cancer DU145 cells. FASEB J. 2018, 32, 5967–5975. [Google Scholar] [CrossRef] [PubMed]
- Confavreux, C.B.; Girard, N.; Pialat, J.B.; Bringuier, P.P.; Devouassoux-Shisheboran, M.; Rousseau, J.C.; Isaac, S.; Thivolet-Bejui, F.; Clezardin, P.; Brevet, M. Mutational profiling of bone metastases from lung adenocarcinoma: Results of a prospective study (POUMOS-TEC). BoneKEy Rep. 2014, 3, 580. [Google Scholar] [CrossRef]
- Mao, C.; Qiu, L.X.; Liao, R.Y.; Du, F.B.; Ding, H.; Yang, W.C.; Li, J.; Chen, Q. KRAS mutations and resistance to EGFR-TKIs treatment in patients with non-small cell lung cancer: A meta-analysis of 22 studies. Lung Cancer 2010, 69, 272–278. [Google Scholar] [CrossRef]
| Patient No. | Status of EGFR Mutations | ||
|---|---|---|---|
| TTG | ICP (%) | ddPCR (%) | |
| 1 | E19 | W | 0.04 |
| 2 | E19 | W | 0.02 |
| 3 | E19 | W | 0.00 |
| 4 | E19 | W | 0.00 |
| 5 | E19 | W | 0.12 |
| 6 | E19 | W | 0.00 |
| 7 | E19 | W | 0.00 |
| 8 | E19 | W | 0.08 |
| 9 | E19 | W | 0.00 |
| 10 | E19 | W | 39.00 |
| 11 | E19 | W | Failed |
| 12 | E21 | W | 0.01 |
| 13 | E21 | W | 0.00 |
| 14 | E21 | W | 0.00 |
| 15 | W | E19 (0.32) | 0.00 |
| 16 | W | E19 (0.70) | 0.27 |
| 17 | W | E19 (7.40) | 8.00 |
| 18 | W | E19 (0.69) | 5.60 |
| 19 | W | E19 (0.67) | 0.00 |
| 20 | W | E19 (0.73) | 0.00 |
| 21 | W | E19 (1.04) | NA |
| 22 | W | E19 (0.25), E21 (0.23) | 0.03, 0 |
| 23 | W | E19 (3.34), E21 (0.20) | 0.80, 0.12 |
| 24 | W | E21 (0.32) | 0.45 |
| 25 | W | E21 (0.19) | 0.00 |
| 26 | W | E21 (2.10) | 9.20 |
| 27 | W | E21 (0.68), E20 (0.87) | 0.03, 0.05 |
| 28 | W | E21 (0.45), E20 (0.68) | 0.05, 0.06 |
| 29 | W | E21 (0.65), E20 (2.06) | 0, 0.08 |
| 30 | W | E21 (1.08), E20 (1.24) | NA, 0.29 |
| 31 | W | E21 (0.58), E20 (0.61) | 0.01, 0.03 |
| 32 | W | E21 (0.25), E20 (0.41) | 0, 0.15 |
| 33 | W | E21 (0.21) | 0.01 |
| 34 | W | E21 (0.63), E20 (0.23) | 0, 0 |
| 35 | W | E20 (0.30) | 0.00 |
| 36 | W | E20 (0.37) | 0.11 |
| 37 | W | E20 (0.50) | 0.07 |
| 38 | W | E20 (0.24) | 0.02 |
| 39 | W | E20 (0.30) | 0.03 |
| 40 | N | E19 (3.11) | 4.00 |
| 41 | N | E19 (1.64) | 0.01 |
| 42 | N | E21 (2.65), E20 (2.41) | 0, 0 |
| 43 | N | E21 (0.36), E20 (0.62) | 0, 0.03 |
| 44 | N | E21 (0.31), E20 (0.57) | 0, 0.01 |
| 45 | N | E21 (1.36), E20 (1.28) | 0, 0.20 |
| 46 | N | E21 (0.25), E20 (0.23) | 0, 0.05 |
| 47 | N | E20 (2.90) | 0.03 |
| 48 | N | E20 (0.22) | 0.09 |
| 49 | N | E21 (0.16), E20 (10.31) | 0, 0.53 |
| 50 | E19 | E19 (0.18), E21 (0.16), E20 (0.42) | 0.80, 0, 0.23 |
| 51 | E19 | E19 (7.36), E20 (0.24) | 17.2, NA |
| 52 | E19 | E19 (44.82), E21 (3.05), E20 (1.5) | 58.00, NA, 0 |
| 53 | E19 | E19 (2.72), E20 (0.97) | 4.48, 1.27 |
| 54 | E21 | E21 (0.27), E20 (1.11) | 0.07, 0 |
| 55 | E21 | E21 (0.21), E20 (9.56) | 0.06, 0.06 |
| 56 | E21 | E21 (0.52), E20 (3.19) | 0, 0.04 |
| 57 | E21 | E21 (2.34) | 1.69 |
| 58 | E21 | E19 (3.17), E21 (2.32) | 0.50, NA |
| 59 | E21 | E19 (1.08), E20 (1.56) | NA, 0 |
| 60 | E21 | E19 (0.39), E20 (0.19) | 0, 0 |
| 61 | E21 | E19 (0.54), E21 (0.21) | 0, NA |
| Patient No. | Status of KRAS Mutation | |
|---|---|---|
| ICP (%) | ddPCR (%) | |
| 1 | G13G (0.15) | 0.03 |
| 2 | G12D (1.90) | 0.01 |
| 3 | Q61H (6.72) | 28.90 |
| 4 | G12S (0.46), G13D (0.19), G13G (0.24) | 0.05, 0.02, 0 |
| 5 | Q61K (0.14) | 0.00 |
| 6 | G12S (0.48), G13G (0.19) | NA, NA |
| 7 | G12S (0.50), G12C (1.51), G12D (0.15), G13D (0.17), G13G (0.25), Q61H (0.50) | 1.07, 1.30, 0.05, 0.01, 0.02, 0.01 |
| 8 | G12S (0.20) | 0.05 |
| 9 | G12C (0.72) | 4.02 |
| 10 | G13G (0.15) | 0.06 |
| 11 | G12S (0.16) | 0.02 |
| 12 | G13D (0.18) | 0.03 |
| 13 | G12V (0.69) | 1.30 |
| 14 | G12C (0.36) | 1.30 |
| 15 | G13G (0.20) | 0.00 |
| 16 | G12C (0.19) | 0.37 |
| 17 | G12S (0.28) | Failed |
| 18 | G13G (0.20) | Failed |
| 19 | A59T (0.10) | NA |
| RAS/PIK3CA/PTEN (+) (n = 72, %) | RAS/PIK3CA/PTEN (−) (n = 52, %) | p | |
|---|---|---|---|
| Age (years), median | 65 (27–87) | 64 (42–84) | 0.728 |
| Gender | |||
| Male | 44 (63.8) | 38 (73.1) | 0.249 |
| Female | 25 (36.2) | 14 (26.9) | |
| Smoking status | |||
| Never smoker | 34 (49.3) | 28 (53.8) | 0.671 |
| Ex-smoker | 18 (26.1) | 11 (21.2) | |
| Current smoker | 17 (24.6) | 13 (25.0) | |
| Histology | |||
| Adenocarcinoma | 52 (75.4) | 34 (65.4) | 0.121 |
| Squamous carcinoma | 7 (10.1) | 29 (34.6) | |
| Others | 10 (14.5) | ||
| Stage at sample acquisition | |||
| II | 0 | 1 (1.9) | 0.023 |
| III | 6 (8.3) | 4 (7.7) | |
| IV | 64 (88.9) | 38 (73.1) | |
| Relapsed | 2 (2.8) | 9 (17.3) | |
| No. of metastatic organ | |||
| ≤2 | 36 (50) | 31 (59.6) | 0.362 |
| >2 | 36 (50) | 21 (40.4) | |
| Bone metastasis | |||
| Present | 33 (47.8) | 20 (38.5) | 0.273 |
| Absent | 36 (52.2) | 32 (61.5) | |
| Brain metastasis | |||
| Present | 20 (29) | 16 (30.8) | 0.841 |
| Absent | 49 (71) | 36 (69.2) | |
| Activating EGFR mutation | |||
| Present | 29 (40.3) | 18 (34.6) | 0.576 |
| Absent | 43 (59.7) | 34 (65.4) |
| Number | Median OS (Months) | p (Univariate) | p (Multivariate) | ||
|---|---|---|---|---|---|
| Age, years | ≤65 | 67 | 20.5 | 0.016 | 0.003 |
| >65 | 57 | 10.4 | |||
| Sex | Male | 83 | 11.7 | 0.012 | 0.235 |
| Female | 41 | 40.1 | |||
| Smoking | Yes | 59 | 10.2 | 0.013 | 0.235 |
| No | 65 | 22.9 | |||
| EGFR mutations | Wild-type | 77 | 10.4 | 0.001 | <0.001 |
| Activated | 47 | 30.2 | |||
| RAS/PIK3CA/PTEN mutations in activating EGFR mutations | Yes | 29 | 22.1 | 0.098 | 0.019 |
| No | 18 | 40.1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, W.J.; Sung, J.S.; Lee, S.Y.; Kang, E.J.; Kwon, N.-J.; Kim, H.M.; Shin, S.W.; Choi, J.Y.; Choi, Y.J.; Kim, J.W.; et al. The Clinical Significance of RAS, PIK3CA, and PTEN Mutations in Non-Small Cell Lung Cancer Using Cell-Free DNA. J. Clin. Med. 2020, 9, 2642. https://doi.org/10.3390/jcm9082642
Chang WJ, Sung JS, Lee SY, Kang EJ, Kwon N-J, Kim HM, Shin SW, Choi JY, Choi YJ, Kim JW, et al. The Clinical Significance of RAS, PIK3CA, and PTEN Mutations in Non-Small Cell Lung Cancer Using Cell-Free DNA. Journal of Clinical Medicine. 2020; 9(8):2642. https://doi.org/10.3390/jcm9082642
Chicago/Turabian StyleChang, Won Jin, Jae Sook Sung, Sung Yong Lee, Eun Joo Kang, Nak-Jung Kwon, Hae Mi Kim, Sang Won Shin, Jung Yoon Choi, Yoon Ji Choi, Ju Won Kim, and et al. 2020. "The Clinical Significance of RAS, PIK3CA, and PTEN Mutations in Non-Small Cell Lung Cancer Using Cell-Free DNA" Journal of Clinical Medicine 9, no. 8: 2642. https://doi.org/10.3390/jcm9082642
APA StyleChang, W. J., Sung, J. S., Lee, S. Y., Kang, E. J., Kwon, N.-J., Kim, H. M., Shin, S. W., Choi, J. Y., Choi, Y. J., Kim, J. W., Park, K. H., & Kim, Y. H. (2020). The Clinical Significance of RAS, PIK3CA, and PTEN Mutations in Non-Small Cell Lung Cancer Using Cell-Free DNA. Journal of Clinical Medicine, 9(8), 2642. https://doi.org/10.3390/jcm9082642






