Deciphering the Molecular Landscape of Cutaneous Squamous Cell Carcinoma for Better Diagnosis and Treatment
Abstract
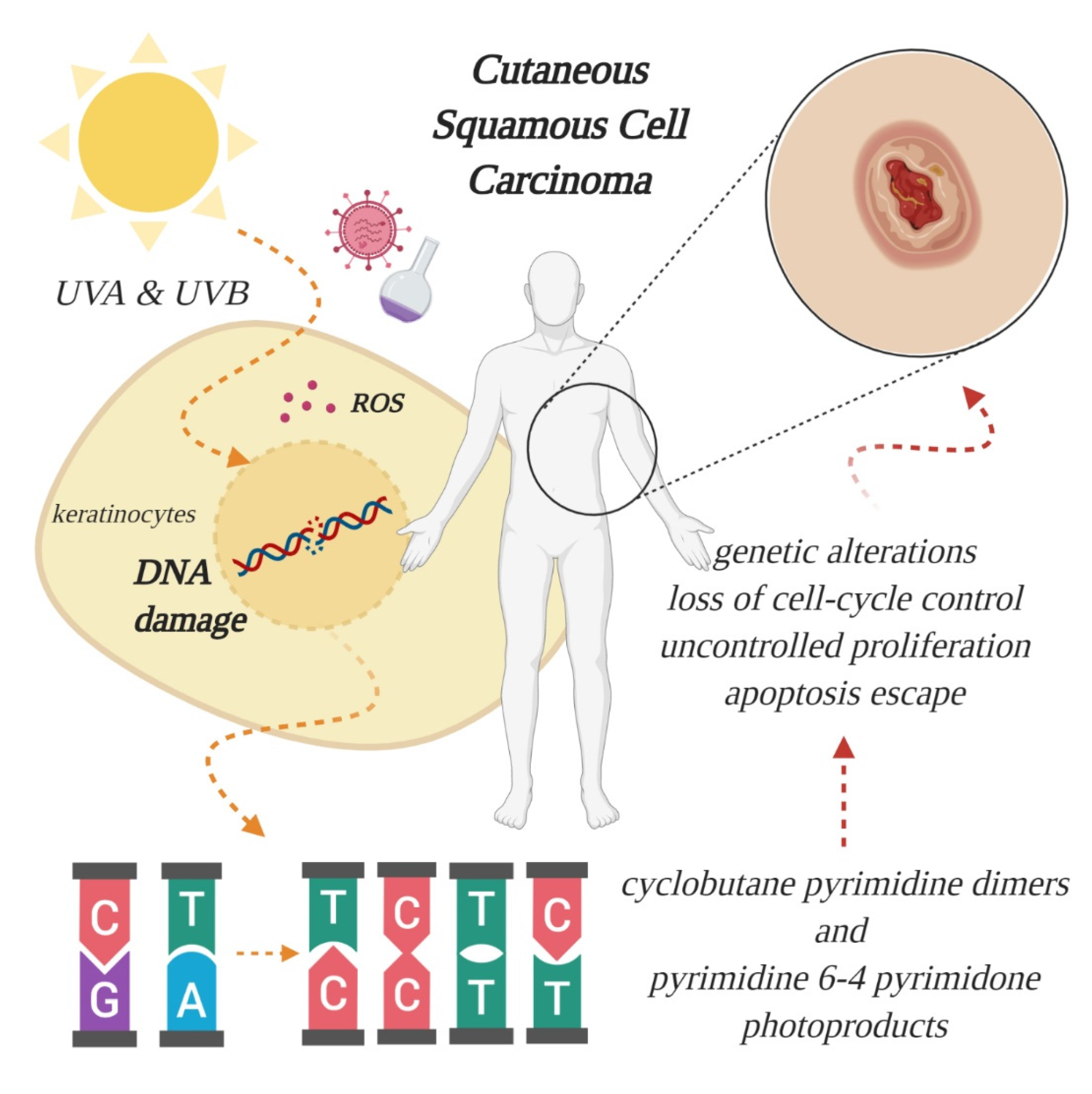
1. Introduction
2. Etiology, Prevention and Early Diagnosis of Cutaneous SCC
3. Established SCC-Associated Markers
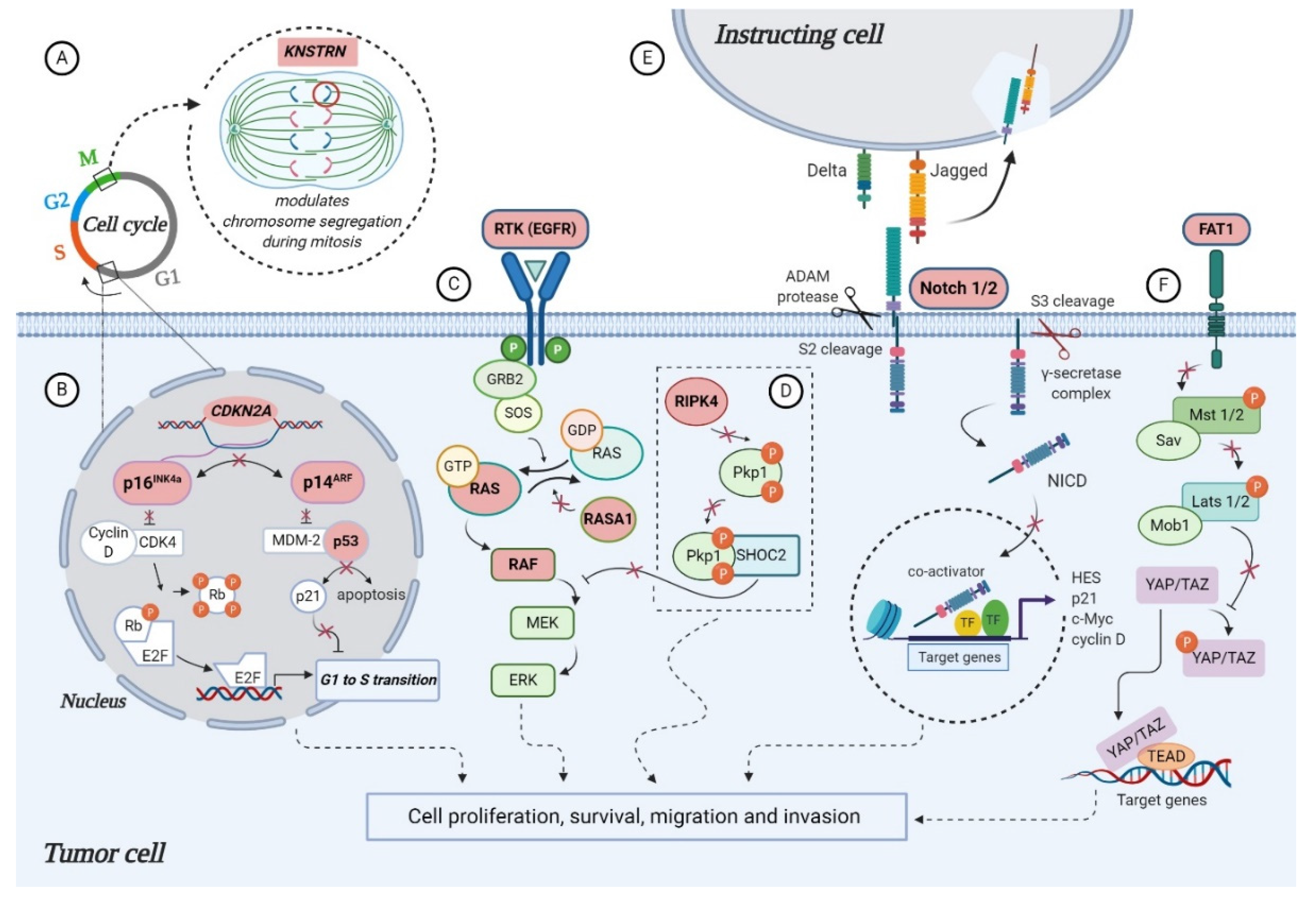
3.1. TP53
3.2. CDKN2A
3.3. RAS Signaling Genes
3.4. NOTCH Signaling Genes
3.5. FAT1
4. Novel SCC-Associated Markers
4.1. KNSTRN
4.2. RASA1
4.3. RIPK4
4.4. Chromatin Remodeling Genes
4.5. Other Potential Cutaneous SCC-Associated Genes
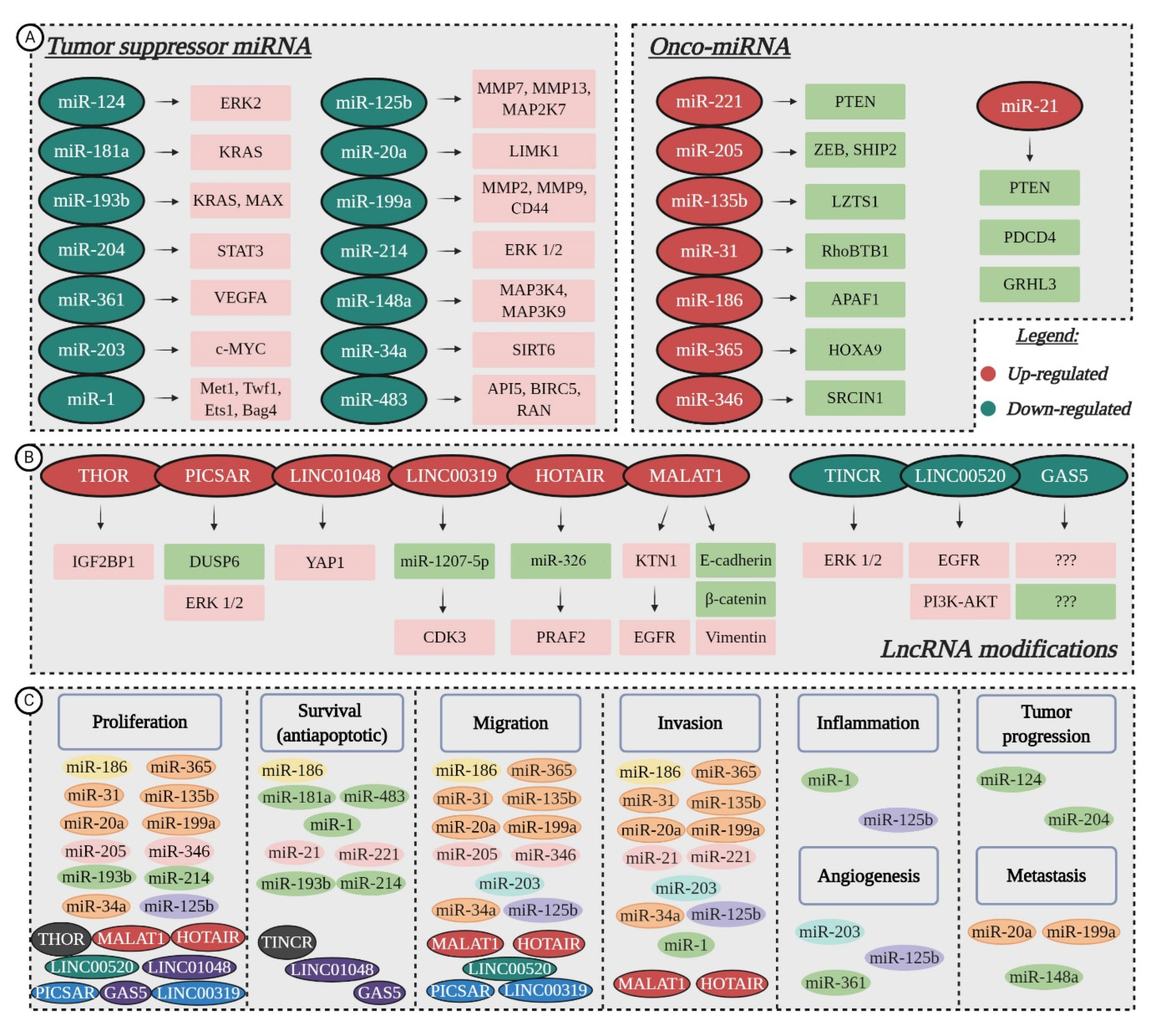
5. Non-Coding RNA Modifications in Cutaneous SCC
5.1. Tumor Suppressor miRNAs Downregulated in cSCC
5.2. Oncogenic miRNAs Upregulated in cSCC
5.3. Aberrant Activity of lncRNAs
6. Novel Therapeutic Approaches for Cutaneous SCC
6.1. Targeted Therapy
6.2. Immunotherapy
6.3. Topical Treatment
7. Discussion
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Apalla, Z.; Nashan, D.; Weller, R.B.; Castellsagué, X. Skin cancer: Epidemiology, disease burden, pathophysiology, diagnosis, and therapeutic approaches. Dermatol. Ther. 2017, 7, 5–19. [Google Scholar] [CrossRef]
- Greinert, R. Skin cancer: New markers for better prevention. Pathobiology 2009, 76, 64–81. [Google Scholar] [CrossRef] [PubMed]
- Global Burden of Disease Cancer Collaboration. Global, regional and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2017. JAMA Oncol. 2019, 5, 1749–1768. [Google Scholar] [CrossRef] [PubMed]
- Gloster, H.M.; Neal, K. Skin cancer in skin of color. J. Am. Acad. Dermatol. 2006, 55, 741–760. [Google Scholar] [CrossRef] [PubMed]
- Lopez, A.T.; Liu, L.; Geskin, L. Molecular Mechanisms and Biomarkers of Skin Photocarcinogenesis. In Human Skin Cancers: Pathways, Mechanisms, Targets and Treatments; Blumenberg, M., Ed.; IntechOpen: London, UK, 2017; pp. 175–200. [Google Scholar]
- Padilla, R.S.; Sebastian, S.; Jiang, Z.; Nindl, I.; Larson, R. Gene expression patterns of normal human skin, actinic keratosis, and squamous cell carcinoma: A spectrum of disease progression. Arch. Dermatol. 2010, 146, 288–293. [Google Scholar] [CrossRef]
- Brougham, N.D.; Dennett, E.R.; Cameron, R.; Tan, S.T. The incidence of metastasis from cutaneous squamous cell carcinoma and the impact of its risk factors. J. Surg. Oncol. 2012, 106, 811–815. [Google Scholar] [CrossRef]
- Palyca, P.; Koshenkov, V.P.; Mehnert, J.M. Developments in the treatment of locally advanced and metastatic squamous cell carcinoma of the skin: A rising unmet need. Am. Soc. Clin. Oncol. Educ. Book 2014, 2014, e397–e404. [Google Scholar] [CrossRef]
- Durinck, S.; Ho, C.; Wang, N.J.; Liao, W.; Jakkula, L.R.; Collisson, E.A.; Pons, J.; Chan, S.W.; Lam, E.T.; Chu, C.; et al. Temporal dissection of tumorigenesis in primary cancers. Cancer Discov. 2011, 1, 137–143. [Google Scholar] [CrossRef]
- Ashton, K.J.; Weinstein, S.R.; Maguire, D.J.; Griffiths, L.R. Chromosomal aberrations in squamous cell carcinoma and solar keratoses revealed by comparative genomic hybridization. Arch. Dermatol. 2003, 139, 876–882. [Google Scholar] [CrossRef]
- Ratushny, V.; Gober, M.D.; Hick, R.; Ridky, T.W.; Seykora, J.T. From keratinocyte to cancer: The pathogenesis and modeling of cutaneous squamous cell carcinoma. J. Clin. Investig. 2012, 122, 464–472. [Google Scholar] [CrossRef]
- Parekh, V.; Seykora, J.T. Cutaneous Squamous Cell Carcinoma. Clin. Lab. Med. 2017, 37, 503–525. [Google Scholar] [CrossRef] [PubMed]
- García-Sancha, N.; Corchado-Cobos, R.; Pérez-Losada, J.; Cañueto, J. MicroRNA Dysregulation in Cutaneous Squamous Cell Carcinoma. Int. J. Mol. Sci. 2019, 20, 2181. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, B.; Wen, X.; Hao, D.; Du, D.; He, G.; Jiang, X. The Roles of lncRNA in Cutaneous Squamous Cell Carcinoma. Front. Oncol. 2020, 10, 158. [Google Scholar] [CrossRef] [PubMed]
- Gordon, R. Skin cancer: An overview of epidemiology and risk factors. Semin. Oncol. Nurs. 2013, 29, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Pesch, B.; Ranft, U.; Jakubis, P.; Nieuwenhuijsen, M.J.; Hergemoller, A.; Unfried, K.; Jakubis, M.; Miskovic, P.; Keegan, T. Environmental arsenic exposure from a coal-burning power plant as a potential risk factor for nonmelanoma skin carcinoma: Results from a case-control study in the district of Prievidza, Slovakia. Am. J. Epidemiol. 2002, 155, 798–809. [Google Scholar] [CrossRef]
- Nindl, I.; Gottschling, M.; Stockfleth, E. Human papillomaviruses and non-melanoma skin cancer: Basic virology and clinical manifestations. Dis. Markers 2007, 23, 247–259. [Google Scholar] [CrossRef]
- Brenner, M.; Hearing, V.J. The protective role of melanin against UV damage in human skin. Photochem. Photobiol. 2008, 84, 539–549. [Google Scholar] [CrossRef]
- Hussein, M.R. Ultraviolet radiation and skin cancer: Molecular mechanisms. J. Cutan. Pathol. 2005, 32, 191–205. [Google Scholar] [CrossRef]
- Beani, J.C. Ultraviolet A-induced DNA damage: Role in skin cancer. Bull. Acad. Natl. Med. 2014, 198, 273–295. [Google Scholar]
- Nilsen, L.T.; Hannevik, M.; Veierod, M.B. Ultraviolet exposure from indoor tanning devices: A systematic review. BJD 2016, 174, 730–740. [Google Scholar] [CrossRef]
- Wehner, M.R.; Shive, M.L.; Chren, M.M.; Han, J.; Qureshi, A.A.; Linos, E. Indoor tanning and non-melanoma skin cancer: Systematic review and meta-analysis. BMJ 2012, 345, e5909. [Google Scholar] [CrossRef] [PubMed]
- Breitbart, E.W.; Greinert, R.; Volkmer, B. Effectiveness of information campaigns. Prog. Biophys. Mol. Biol. 2006, 92, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Green, A.; Williams, G.; Neale, R.; Hart, V.; Leslie, D.; Parsons, P.; Marks, G.C.; Gaffney, P.; Battistutta, D.; Frost, C.; et al. Daily sunscreen application and betacarotene supplementation in prevention of basal-cell and squamous-cell carcinomas of the skin: A randomized controlled trial. Lancet 1999, 354, 723–729. [Google Scholar] [CrossRef]
- Green, A.C.; Williams, G.M.; Logan, V.; Strutton, G.M. Reduced melanoma after regular sunscreen use: Randomized trial follow-up. J. Clin. Oncol. 2011, 29, 257–263. [Google Scholar] [CrossRef]
- Gandini, S.; Sera, F.; Cattaruzza, M.S.; Pasquini, P.; Zanetti, R.; Masini, C.; Boyle, P.; Melchi, C.F. Meta-analysis of risk factors for cutaneous melanoma: III. Family history, actinic damage and phenotypic factors. Eur. J. Cancer 2005, 41, 2040–2059. [Google Scholar] [CrossRef]
- South, A.P.; Purdie, K.J.; Watt, S.A.; Haldenby, S.; den Breems, N.; Dimon, M.; Arron, S.T.; Kluk, M.J.; Aster, J.C.; McHugh, A.; et al. NOTCH1 mutations occur early during cutaneous squamous cell carcinogenesis. J. Investig. Dermatol. 2014, 134, 2630–2638. [Google Scholar] [CrossRef]
- Pickering, C.R.; Zhou, J.H.; Lee, J.J.; Drummond, J.A.; Peng, S.A.; Saade, R.E.; Tsai, K.Y.; Curry, J.L.; Tetzlaff, M.T.; Lai, S.Y.; et al. Mutational landscape of aggressive cutaneous squamous cell carcinoma. Clin. Cancer Res. J. Am. Assoc. Cancer Res. 2014, 20, 6582–6592. [Google Scholar] [CrossRef]
- Li, Y.Y.; Hanna, G.J.; Laga, A.C.; Haddad, R.I.; Lorch, J.H.; Hammerman, P.S. Genomic analysis of metastatic cutaneous squamous cell carcinoma. Clin. Cancer Res. J. Am. Assoc. Cancer Res. 2015, 21, 1447–1456. [Google Scholar] [CrossRef]
- Yilmaz, A.S.; Ozer, H.G.; Gillespie, J.L.; Allain, D.C.; Bernhardt, M.N.; Furlan, K.C.; Castro, L.T.F.; Peteres, S.B.; Nagarajan, P.; Kang, S.Y.; et al. Differential mutation frequencies in metastatic cutaneous squamous cell carcinomas versus primary tumors. Cancer 2017, 123, 1184–1193. [Google Scholar] [CrossRef]
- Inman, G.J.; Wang, J.; Nagano, A.; Alexandrov, L.; Purdie, K.J.; Taylor, R.G.; Sherwood, V.; Thomson, J.; Hogan, S.; Spender, L.C.; et al. The genomic landscape of cutaneous SCC reveals drivers and a novel azathioprine associated mutational signature. Nat. Commun. 2018, 9, 3667. [Google Scholar] [CrossRef]
- Kim, E.J.; Park, J.S.; Um, S.J. Identification and characterization of HIPK2 interacting with p73 and modulating functions of the p53 family in vivo. J. Biol. Chem. 2002, 277, 32020–32028. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.S.; Bhaduri, A.; Mah, A.; Johnson, W.L.; Ungewickell, A.; Aros, C.J.; Nguyen, C.B.; Rios, E.J.; Siprashvili, Z.; Straight, A.; et al. Recurrent point mutations in the kinetochore gene KNSTRN in cutaneous squamous cell carcinoma. Nat. Genet. 2014, 46, 1060–1062. [Google Scholar] [CrossRef] [PubMed]
- Brown, V.L.; Harwood, C.A.; Crook, T.; Cronin, J.G.; Kelsell, D.P.; Proby, C.M. p16INK4a and p14ARF tumor suppressor genes are commonly inactivated in cutaneous squamous cell carcinoma. J. Investig. Dermatol. 2004, 122, 1284–1292. [Google Scholar] [CrossRef] [PubMed]
- Nagarajan, P.; Ivan, D. Cutaneous squamous cell carcinomas: Focus on high-risk features and molecular alterations. Glob Derm. 2016, 3, 359–365. [Google Scholar]
- Kern, F.; Niault, T.; Baccarini, M. Ras and Raf pathways in epidermis development and carcinogenesis. Br. J. Cancer 2011, 104, 229–234. [Google Scholar] [CrossRef]
- Toll, A.; Salgado, R.; Yébenes, M.; Martín-Ezquerra, G.; Gilaberte, M.; Baró, T.; Solé, F.; Alameda, F.; Espinet, B.; Pujol, R.M. Epidermal growth factor receptor gene numerical aberrations are frequent events in actinic keratoses and invasive cutaneous squamous cell carcinomas. Exp. Dermatol. 2010, 19, 151–153. [Google Scholar] [CrossRef]
- Su, F.; Viros, A.; Milagre, C.; Trunzer, K.; Bollag, G.; Spleiss, O.; Reis-Filho, J.S.; Kong, X.; Koya, R.C.; Flaherty, K.T.; et al. RAS mutations in cutaneous squamous-cell carcinomas in patients treated with BRAF inhibitors. N. Engl. J. Med. 2012, 366, 207–215. [Google Scholar] [CrossRef]
- Ashford, B.G.; Clark, J.; Gupta, R.; Iyer, N.G.; Yu, B.; Ranson, M. Reviewing the genetic alterations in high-risk cutaneous squamous cell carcinoma: A search for prognostic markers and therapeutic targets. Head Neck 2017, 39, 1462–1469. [Google Scholar] [CrossRef]
- Moriyama, M.; Durham, A.D.; Moriyama, H.; Hasegawa, K.; Nishikawa, S.I.; Radtke, F.; Osawa, M. Multiple roles of Notch signaling in the regulation of epidermal development. Dev. Cell 2008, 14, 594–604. [Google Scholar] [CrossRef]
- Okuyama, R.; Tagami, H.; Aiba, S. Notch signaling: Its role in epidermal homeostasis and in the pathogenesis of skin diseases. J. Dermatol. Sci. 2008, 49, 187–194. [Google Scholar] [CrossRef]
- Santos-de-Frutos, K.; Segrelles, C.; Lorz, C. Hippo Pathway and YAP Signaling Alterations in Squamous Cancer of the Head and Neck. J. Clin. Med. 2019, 8, 2131. [Google Scholar] [CrossRef] [PubMed]
- Harwood, C.A.; Proby, C.M.; Inman, G.J.; Leigh, I.M. The promise of genomics and the development of targeted therapies for cutaneous squamous cell carcinoma. Acta Derm. Venereol. 2016, 96, 3–16. [Google Scholar] [CrossRef]
- Lawrence, M.S.; Stojanov, P.; Mermel, C.H.; Robinson, J.T.; Garraway, L.A.; Golub, T.R.; Meyerson, M.; Gabriel, S.B.; Lander, E.S.; Getz, G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 2014, 505, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Maertens, O.; Cichowski, K. An expanding role for RAS GTPase activating proteins (RAS GAPs) in cancer. Adv. Biol. Regul. 2014, 55, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Davoli, T.; Xu, A.W.; Mengwasser, K.E.; Sack, L.M.; Yoon, J.C.; Park, P.J.; Elledge, S.J. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 2013, 155, 948–962. [Google Scholar] [CrossRef] [PubMed]
- Holland, P.; Willis, C.; Kanaly, S.; Glaccum, M.; Warren, A.; Charrier, K.; Murison, J.; Derry, J.; Virca, G.; Bird, T.; et al. RIP4 is an ankyrin repeat-containing kinase essential for keratinocyte differentiation. Curr. Biol. 2002, 12, 1424–1428. [Google Scholar] [CrossRef]
- Stransky, N.; Egloff, A.M.; Tward, A.D.; Kostic, A.D.; Cibulskis, K.; Sivachenko, A.; Kryukov, G.V.; Lawrence, M.S.; Sougnez, C.; McKenna, A.; et al. The mutational landscape of head and neck squamous cell carcinoma. Science 2011, 333, 1157–1160. [Google Scholar] [CrossRef]
- Kalay, E.; Sezgin, O.; Chellappa, V.; Mutlu, M.; Morsy, H.; Kayserili, H.; Kreiger, E.; Cansu, A.; Toraman, B.; Abdalla, E.M.; et al. Mutations in RIPK4 cause the autosomal-recessive form of popliteal pterygium syndrome. Am. J. Hum. Genet. 2012, 90, 76–85. [Google Scholar] [CrossRef]
- Lee, P.; Jiang, S.; Li, Y.; Yue, J.; Gou, X.; Chen, S.Y.; Zhao, Y.; Schober, M.; Tan, M.; Wu, X. Phosphorylation of Pkp1 by RIPK 4 regulates epidermal differentiation and skin tumorigenesis. EMBO J. 2017, 36, 1963–1980. [Google Scholar] [CrossRef]
- Gui, Y.; Guo, G.; Huang, Y.; Hu, X.; Tang, A.; Gao, S.; Wu, R.; Chen, C.; Li, X.; Zhou, L.; et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat. Genet. 2011, 43, 875–878. [Google Scholar] [CrossRef]
- Ellis, M.J.; Ding, L.; Shen, D.; Luo, J.; Suman, V.J.; Wallis, J.W.; Van Tine, B.A.; Hoog, J.; Goiffon, R.J.; Goldstein, T.C.; et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature 2012, 486, 353–360. [Google Scholar] [CrossRef]
- Je, E.M.; Lee, S.H.; Yoo, N.J.; Lee, S.H. Mutational and expressional analysis of MLL genes in gastric and colorectal cancers with microsatellite instability. Neoplasma 2013, 60, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.J.; Sholl, L.M.; Lindeman, N.I.; Lee, J.J.; Sholl, L.M.; Lindeman, N.I.; Granter, S.R.; Laga, A.C.; Shivdasani, P.; Chin, G.; et al. Targeted next-generation sequencing reveals high frequency of mutations in epigenetic regulators across treatment-naïve patient melanomas. Clin. Epigenet. 2015, 7, 59. [Google Scholar] [CrossRef]
- Gao, Y.B.; Chen, Z.L.; Li, J.G.; Hu, X.D.; Shi, X.J.; Sun, Z.M.; Zhang, F.; Zhao, Z.R.; Li, Z.T.; Liu, Z.Y.; et al. Genetic landscape of esophageal squamous cell carcinoma. Nat. Genet. 2014, 46, 1097–1102. [Google Scholar] [CrossRef]
- Chung, C.H.; Guthrie, V.B.; Masica, D.L.; Tokheim, C.; Kang, H.; Richmon, J.; Agrawal, N.; Fakhry, C.; Quon, H.; Subramaniam, R.M.; et al. Genomic alterations in head and neck squamous cell carcinoma determined by cancer gene-targeted sequencing. Ann. Oncol. 2015, 26, 1216–1223. [Google Scholar] [CrossRef] [PubMed]
- Peacock, J.W.; Takeuchi, A.; Hayashi, N.; Liu, L.; Tam, K.J.; Nakouzi, N.A.; Khazamipour, N.; Tombe, T.; Dejima, T.; Lee,, K.C.K.; et al. SEMA3C drives cancer growth by transactivating multiple receptor tyrosine kinases via Plexin B1. EMBO Mol. Med. 2018, 10, 219–238. [Google Scholar] [CrossRef]
- Hui, D.H.F.; Tam, K.J.; Jiao, I.Z.F.; Ong, C.J. Semaphorin 3C as a Therapeutic Target in Prostate and Other Cancers. Int. J. Mol. Sci. 2019, 20, 774. [Google Scholar] [CrossRef] [PubMed]
- Gomes, I.M.; Maia, C.J.; Santos, C.R. STEAP proteins: From structure to applications in cancer therapy. Mol. Cancer Res. 2012, 10, 573–587. [Google Scholar] [CrossRef]
- Xue, X.; Bredell, B.X.; Anderson, E.R.; Martin, A.; Mays, C.; Nagao-Kitamoto, H.; Huang, S.; Győrffy, B.; Greenson, J.K.; Hardiman, K.; et al. Quantitative proteomics identifies STEAP4 as a critical regulator of mitochondrial dysfunction linking inflammation and colon cancer. Proc. Natl. Acad. Sci. USA 2017, 114, E9608–E9617. [Google Scholar] [CrossRef] [PubMed]
- Briso, E.M.; Guinea-Viniegra, J.; Bakiri, L.; Rogon, Z.; Petzelbauer, P.; Eils, R.; Wolf, R.; Rincón, M.; Angel, P.; Wagner, E.F. Inflammation-mediated skin tumorigenesis induced by epidermal c-Fos. Genes Dev. 2013, 27, 1959–1973. [Google Scholar] [CrossRef] [PubMed]
- Justilien, V.; Regala, R.P.; Tseng, I.C.; Walsh, M.P.; Batra, J.; Radisky, E.S.; Murray, N.R.; Fields, A.P. Matrix metalloproteinase-10 is required for lung cancer stem cell maintenance, tumor initiation and metastatic potential. PLoS ONE 2012, 7, e35040. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Di, J.; Lu, Z.; Gao, K.; Zheng, J. Rap2BGTPase: Structure, functions, and regulation. Tumor Biol. J. Int. Soc. Oncodev. Biol. Med. 2016, 37, 7085–7093. [Google Scholar] [CrossRef] [PubMed]
- Le Duff, M.; Gouju, J.; Jonchere, B.; Guillon, J.; Toutain, B.; Boissard, A.; Henry, C.; Guette, C.; Lelievre, E.; Coqueret, O. Regulation of senescence escape by the cdk4-EZH2-AP2M1 pathway in response to chemotherapy. Cell Death Dis. 2018, 9, 1–15. [Google Scholar] [CrossRef]
- Croce, C.M.; Calin, G.A. miRNAs, cancer, and stem cell division. Cell 2005, 122, 6–7. [Google Scholar] [CrossRef] [PubMed]
- Gregory, R.I.; Shiekhattar, R. MicroRNA biogenesis and cancer. Cancer Res. 2005, 65, 3509–3512. [Google Scholar] [CrossRef]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and functions of long noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Fullwood, M.J. Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer. Genom. Proteom. Bioinf. 2016, 14, 42–54. [Google Scholar] [CrossRef] [PubMed]
- Yamane, K.; Jinnin, M.; Etoh, T.; Kobayashi, Y.; Shimozono, N.; Fukushima, S.; Masuguchi, S.; Maruo, K.; Inoue, Y.; Ishihara, T.; et al. Down-regulation of miR-124/-214 in cutaneous squamous cell carcinoma mediates abnormal cell proliferation via the induction of ERK. J. Mol. Med. 2013, 91, 69–81. [Google Scholar] [CrossRef]
- Roskoski, R., Jr. Targeting ERK1/2 protein-serine/threonine kinases in human cancers. Pharmacol. Res. 2019, 142, 151–168. [Google Scholar] [CrossRef]
- Suiqing, C.; Min, Z.; Lirong, C. Overexpression of phosphorylated-STAT3 correlated with the invasion and metastasis of cutaneous squamous cell carcinoma. J. Dermatol. 2005, 32, 354–360. [Google Scholar] [CrossRef]
- Toll, A.; Salgado, R.; Espinet, B.; Diaz-Lagares, A.; Hernandez-Ruiz, E.; Andrades, E.; Sandoval, J.; Esteller, M.; Pujol, R.M.; Hernandez-Munoz, I. MiR-204 silencing in intraepithelial to invasive cutaneous squamous cell carcinoma progression. Mol. Cancer 2016, 15, 53. [Google Scholar] [CrossRef] [PubMed]
- Gastaldi, C.; Bertero, T.; Xu, N.; Bourget-Ponzio, I.; Lebrigand, K.; Fourre, S.; Popa, A.; Cardot-Leccia, N.; Meneguzzi, G.; Sonkoly, E.; et al. miR-193b/365a cluster controls progression of epidermal squamous cell carcinoma. Carcinogenesis 2014, 35, 1110–1120. [Google Scholar] [CrossRef] [PubMed]
- Neu, J.; Dziunycz, P.J.; Dzung, A.; Lefort, K.; Falke, M.; Denzler, R.; Freiberger, S.N.; Iotzova-Weiss, G.; Kuzmanov, A.; Levesque, M.P.; et al. miR-181a decelerates proliferation in cutaneous squamous cell carcinoma by targeting the proto-oncogene KRAS. PLoS ONE 2017, 12, e0185028. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Li, W.; Zhao, T.; Tian, X.; Liu, Y.; Zhang, X. Role of miR-148a in cutaneous squamous cell carcinoma by repression of MAPK pathway. Arch. Biochem. Biophys. 2015, 583, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Bertero, T.; Bourget-Ponzio, I.; Puissant, A.; Loubat, A.; Mari, B.; Meneguzzi, G.; Auberger, P.; Barbry, P.; Ponzio, G.; Rezzonico, R. Tumor suppressor function of miR-483-3p on squamous cell carcinomas due to its pro-apoptotic properties. Cell Cycle 2013, 12, 2183–2193. [Google Scholar] [CrossRef]
- Fleming, J.L.; Gable, D.L.; Samadzadeh-Tarighat, S.; Cheng, L.; Yu, L.; Gillespie, J.L.; Toland, A.E. Differential expression of miR-1, a putative tumor suppressing microRNA, in cancer resistant and cancer susceptible mice. PeerJ 2013, 1, e68. [Google Scholar] [CrossRef][Green Version]
- Yu, X.; Li, Z. The role of miRNAs in cutaneous squamous cell carcinoma. J. Cell. Mol. Med. 2016, 20, 3–9. [Google Scholar] [CrossRef]
- Lefort, K.; Brooks, Y.; Ostano, P.; Cario-Andre, M.; Calpini, V.; Guinea-Viniegra, J.; Albinger-Hegyi, A.; Hoetzenecker, W.; Kolfschoten, I.; Wagner, E.F.; et al. A miR-34a-SIRT6 axis in the squamous cell differentiation network. EMBO J. 2013, 32, 2248–2263. [Google Scholar] [CrossRef]
- Li, S.; Luo, C.; Zhou, J.; Zhang, Y. MicroRNA-34a directly targets high-mobility group box 1 and inhibits the cancer cell proliferation, migration and invasion in cutaneous squamous cell carcinoma. Exp. Ther. Med. 2017, 14, 5611–5618. [Google Scholar] [CrossRef]
- Jia, G.; Su, L.; Singhal, S.; Liu, X. Emerging roles of SIRT6 on telomere maintenance, DNA repair, metabolism and mammalian aging. Mol. Cell. Biochem. 2012, 364, 345–350. [Google Scholar] [CrossRef]
- Bouchie, A. First microRNA mimic enters clinic. Nat. Biotechnol. 2013, 31, 577. [Google Scholar] [CrossRef] [PubMed]
- Beg, M.S.; Brenner, A.J.; Sachdev, J.; Borad, M.; Kang, Y.K.; Stoudemire, J.; Smith, S.; Bader, A.G.; Kim, S.; Hong, D.S. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Investig. New Drugs 2017, 35, 180–188. [Google Scholar] [CrossRef]
- Folini, M.; Gandellini, P.; Longoni, N.; Profumo, V.; Callari, M.; Pennati, M.; Colecchia, M.; Supino, R.; Veneroni, S.; Salvioni, R.; et al. miR-21: An oncomir on strike in prostate cancer. Mol. Cancer 2010, 9, 12. [Google Scholar] [CrossRef] [PubMed]
- Lohcharoenkal, W.; Harada, M.; Loven, J.; Meisgen, F.; Landen, N.X.; Zhang, L.; Lapins, J.; Mahapatra, K.D.; Shi, H.; Nissinen, L.; et al. MicroRNA-203 Inversely Correlates with Differentiation Grade, Targets c-MYC, and Functions as a Tumor Suppressor in cSCC. J. Investig. Dermatol. 2016, 136, 2485–2494. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Liu, R.; Luo, C.; Zhou, X.; Xia, K.; Chen, X.; Zhou, M.; Zou, Q.; Cao, P.; Cao, K. MiR-20a inhibits cutaneous squamous cell carcinoma metastasis and proliferation by directly targeting LIMK1. Cancer Biol. Ther. 2014, 15, 1340–1349. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.H.; Zhou, J.D.; He, Q.Y.; Yin, Z.Q.; Cao, K.; Luo, C.Q. MiR-199a inhibits the ability of proliferation and migration by regulating CD44-Ezrin signaling in cutaneous squamous cell carcinoma cells. Int. J. Clin. Exp. Pathol. 2014, 7, 7131–7141. [Google Scholar] [PubMed]
- Xu, N.; Zhang, L.; Meisgen, F.; Harada, M.; Heilborn, J.; Homey, B.; Grander, D.; Stahle, M.; Sonkoly, E.; Pivarcsi, A. MicroRNA-125b down-regulates matrix metallopeptidase 13 and inhibits cutaneous squamous cell carcinoma cell proliferation, migration, and invasion. J. Biol. Chem. 2012, 287, 29899–29908. [Google Scholar] [CrossRef]
- Martin, T.A.; Harrison, G.; Mansel, R.E.; Jiang, W.G. The role of the CD44/ezrin complex in cancer metastasis. Crit. Rev. Oncol. Hematol. 2003, 46, 165–186. [Google Scholar] [CrossRef]
- Chen, C.; Zhao, S.; Karnad, A.; Freeman, J.W. The biology and role of CD44 in cancer progression: Therapeutic implications. J. Hematol. Oncol. 2018, 11, 64. [Google Scholar] [CrossRef]
- Konicke, K.; Lopez-Luna, A.; Munoz-Carrillo, J.L.; Servin-Gonzalez, L.S.; Flores-de la Torre, A.; Olasz, E.; Lazarova, Z. The microRNA landscape of cutaneous squamous cell carcinoma. Drug Discov. Today 2018, 23, 864–870. [Google Scholar] [CrossRef]
- Gong, Z.; Zhou, F.; Shi, C.; Xiang, T.; Zhou, C.K.; Wang, Q.Q.; Jiang, Y.S.; Gao, S.F. miRNA-221 promotes cutaneous squamous cell carcinoma progression by targeting PTEN. Cell. Mol. Biol. Lett. 2019, 24, 9. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Huang, K.; Yu, J. Inhibition of microRNA-21 upregulates the expression of programmed cell death 4 and phosphatase tensin homologue in the A431 squamous cell carcinoma cell line. Oncol. Lett. 2014, 8, 203–207. [Google Scholar] [CrossRef]
- Darido, C.; Georgy, S.R.; Wilanowski, T.; Dworkin, S.; Auden, A.; Zhao, Q.; Rank, G.; Srivastava, S.; Finlay, M.J.; Papenfuss, A.T.; et al. Targeting of the tumor suppressor GRHL3 by a miR-21-dependent proto-oncogenic network results in PTEN loss and tumorigenesis. Cancer Cell 2011, 20, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Olasz, E.B.; Seline, L.N.; Schock, A.M.; Duncan, N.E.; Lopez, A.; Lazar, J.; Flister, M.J.; Lu, Y.; Liu, P.; Sokumbi, O.; et al. MicroRNA-135b Regulates Leucine Zipper Tumor Suppressor 1 in Cutaneous Squamous Cell Carcinoma. PLoS ONE 2015, 10, e0125412. [Google Scholar] [CrossRef] [PubMed]
- Vecchione, A.; Baldassarre, G.; Ishii, H.; Nicoloso, M.S.; Belletti, B.; Petrocca, F.; Zanesi, N.; Fong, L.Y.; Battista, S.; Guarnieri, D.; et al. Fez1/Lzts1 absence impairs Cdk1/Cdc25C interaction during mitosis and predisposes mice to cancer development. Cancer Cell 2007, 11, 275–289. [Google Scholar] [CrossRef][Green Version]
- Chen, B.; Pan, W.; Lin, X.; Hu, Z.; Jin, Y.; Chen, H.; Ma, G.; Qiu, Y.; Chang, L.; Hua, C.; et al. MicroRNA-346 functions as an oncogene in cutaneous squamous cell carcinoma. Tumor Biol. 2015, 37, 2765–2771. [Google Scholar] [CrossRef]
- Bruegger, C.; Kempf, W.; Spoerri, I.; Arnold, A.W.; Itin, P.H.; Burger, B. MicroRNA expression differs in cutaneous squamous cell carcinomas and healthy skin of immunocompetent individuals. Exp. Dermatol. 2013, 22, 426–428. [Google Scholar] [CrossRef]
- Yu, J.; Peng, H.; Ruan, Q.; Fatima, A.; Getsios, S.; Lavker, R.M. MicroRNA-205 promotes keratinocyte migration via the lipid phosphatase SHIP2. FASEB J. 2010, 24, 3950–3959. [Google Scholar] [CrossRef]
- Tian, J.; Shen, R.; Yan, Y.; Deng, L. miR-186 promotes tumor growth in cutaneous squamous cell carcinoma by inhibiting apoptotic protease activating factor-1. Exp. Ther. Med. 2018, 16, 4010–4018. [Google Scholar] [CrossRef]
- Shakeri, R.; Kheirollahi, A.; Davoodi, J. Apaf-1: Regulation and function in cell death. Biochimie 2017, 135, 111–125. [Google Scholar] [CrossRef]
- Wang, A.; Landen, N.X.; Meisgen, F.; Lohcharoenkal, W.; Stahle, M.; Sonkoly, E.; Pivarcsi, A. MicroRNA-31 is overexpressed in cutaneous squamous cell carcinoma and regulates cell motility and colony formation ability of tumor cells. PLoS ONE 2014, 9, e103206. [Google Scholar] [CrossRef] [PubMed]
- Lin, N.; Zhou, Y.; Lian, X.; Tu, Y. MicroRNA-31 functions as an oncogenic microRNA in cutaneous squamous cell carcinoma cells by targeting RhoTBT1. Oncol. Lett. 2017, 13, 1078–1082. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Wang, Y.; Zhou, M.; Zhang, Y.; Wang, P.; Li, X.; Yang, J.; Wang, H.; Ding, Z. HOXA9 inhibits HIF-1-mediated glycolysis through interacting with CRIP2 to repress cutaneous squamous cell carcinoma development. Nat. Commun. 2018, 9, 1480. [Google Scholar] [CrossRef] [PubMed]
- Dinescu, S.; Ignat, S.; Lazar, A.D.; Constantin, C.; Neagu, M.; Costache, M. Epitranscriptomic Signatures in lncRNAs and Their Possible Roles in Cancer. Genes 2019, 10, 52. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Zhu, Y.; Sun, D.; Zhang, Q. Emerging Roles of Long non-coding RNAs in The Tumor Microenvironment. Int. J. Biol. Sci. 2020, 16, 2094–2103. [Google Scholar] [CrossRef]
- Zhang, Y.; Gao, L.; Ma, S.; Ma, J.; Wang, Y.; Li, S.; Hu, X.; Han, S.; Zhou, M.; Zhou, L.; et al. MALAT1-KTN1-EGFR regulatory axis promotes the development of cutaneous squamous cell carcinoma. Cell Death Differ. 2019, 26, 2061–2073. [Google Scholar] [CrossRef] [PubMed]
- Li, S.S.; Zhou, L.; Gao, L.; Wang, Y.H.; Ding, Z.H. Role of long noncoding RNA MALAT1 promotes the occurrence and progression of cutaneous squamous cell carcinoma. J. South. Med. Univ. 2018, 38, 421–427. [Google Scholar]
- Yu, G.-J.; Sun, Y.; Zhang, D.-W.; Zhang, P. Long non-coding RNA HOTAIR functions as a competitive endogenous RNA to regulate PRAF2 expression by sponging miR-326 in cutaneous squamous cell carcinoma. Cancer Cell Int. 2019, 19, 270. [Google Scholar] [CrossRef]
- Sun, G.; Wang, Y.; Zhang, J.; Lin, N.; You, Y. MiR-15b/HOTAIR/p53 form a regulatory loop that affects the growth of glioma cells. J. Cell. Biochem. 2018, 119, 4540–4547. [Google Scholar] [CrossRef]
- Hosono, Y.; Niknafs, Y.S.; Prensner, J.R.; Iyer, M.K.; Dhanasekaran, S.M.; Mehra, R.; Pitchiaya, S.; Tien, J.; Escara-Wilke, J.; Poliakov, A.; et al. Oncogenic role of THOR, a conserved cancer/testis long non-coding RNA. Cell 2017, 171, 1559. [Google Scholar] [CrossRef]
- Liu, Z.; Wu, G.; Lin, C.; Guo, H.; Xu, J.; Zhao, T. IGF2BP1 over-expression in skin squamous cell carcinoma cells is essential for cell growth. Biochem. Biophys. Res. Commun. 2018, 501, 731–738. [Google Scholar] [CrossRef] [PubMed]
- Piipponen, M.; Heino, J.; Kahari, V.-M.; Nissinen, L. Long non-coding RNA PICSAR decreases adhesion and promotes migration of squamous carcinoma cells by downregulating alpha 2 beta 1 and alpha 5 beta 1 integrin expression. Biol. Open 2018, 7, bio037044. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chen, Q.; Kuang, S.; Zhao, C.; Yang, L.; Zhang, Y.; Zhu, H.; Yang, R. USF1-induced upregulation of LINC01048 promotes cell proliferation and apoptosis in cutaneous squamous cell carcinoma by binding to TAF15 to transcriptionally activate YAP1. Cell Death Dis. 2019, 10, 296. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Liao, J.; Duan, X.; He, Y.; Liao, Y. Upregulation of LINC00319 indicates a poor prognosis and promotes cell proliferation and invasion in cutaneous squamous cell carcinoma. J. Cell. Biochem. 2018, 119, 10393–10405. [Google Scholar] [CrossRef] [PubMed]
- Kretz, M.; Siprashvili, Z.; Chu, C.; Webster, D.E.; Zehnder, A.; Qu, K.; Lee, C.S.; Flockhart, R.J.; Groff, A.F.; Chow, J.; et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 2013, 493, 231–245. [Google Scholar] [CrossRef]
- Zhou, W.; Zhang, S.; Li, J.; Li, Z.; Wang, Y.; Li, X. lncRNA TINCR participates in ALA-PDT-induced apoptosis and autophagy in cutaneous squamous cell carcinoma. J. Cell. Biochem. 2019, 120, 13893–13902. [Google Scholar] [CrossRef]
- Mei, X.-L.; Zhong, S. Long noncoding RNA LINC00520 prevents the progression of cutaneous squamous cell carcinoma through the inactivation of the PI3K/Akt signaling pathway by downregulating EGFR. Chin. Med. J. 2019, 132, 454–465. [Google Scholar] [CrossRef]
- Qiao, H.-P.; Gao, W.-S.; Huo, J.-X.; Yang, Z.-S. Long non-coding RNA GAS5 functions as a tumor suppressor in renal cell carcinoma. Asian Pac. J. Cancer Prev. 2013, 14, 1077–1082. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhu, Z.; Watabe, K.; Zhang, X.; Bai, C.; Xu, M.; Wu, F.; Mo, Y.-Y. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ. 2013, 20, 1558–1568. [Google Scholar] [CrossRef]
- Wang, T.-H.; Chan, C.-W.; Fang, J.-Y.; Shih, Y.-M.; Liu, Y.-W.; Wang, T.-C.V.; Chen, C.-Y. 2-O-Methylmagnolol upregulates the long non-coding RNA, GAS5, and enhances apoptosis in skin cancer cells. Cell Death Dis. 2017, 8, e2638. [Google Scholar] [CrossRef]
- Rundhaug, J.E.; Fischer, S.M. Molecular mechanisms of mouse skin tumor promotion. Cancers 2010, 2, 436–482. [Google Scholar] [CrossRef] [PubMed]
- Lewis, C.M.; Glisson, B.S.; Feng, L.; Wan, F.; Tang, X.; Wistuba, I.I.; El-Naggar, A.K.; Rosenthal, D.I.; Chambers, M.S.; Lustig, R.A.; et al. A phase II study of gefitinib for aggressive cutaneous squamous cell carcinoma of the head and neck. Clin. Cancer Res. 2012, 18, 1435–1446. [Google Scholar] [CrossRef] [PubMed]
- Preneau, S.; Rio, E.; Brocard, A.; Peuvrel, L.; Nguyen, J.M.; Quéreux, G.; Dreno, B. Efficacy of cetuximab in the treatment of squamous cell carcinoma. J. Dermatol. Treat. 2014, 25, 424–427. [Google Scholar] [CrossRef] [PubMed]
- Reigneau, M.; Robert, C.; Routier, E.; Mamelle, G.; Moya-Plana, A.; Tomasic, G.; Mateus, C. Efficacy of neoadjuvant cetuximab alone or with platinum salt for the treatment of unresectable advanced nonmetastatic cutaneous squamous cell carcinomas. Br. J. Dermatol. 2015, 173, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, Y.; Nakamura, K.; Nishibu, A.; Yanagihori, H.; Kimura, H.; Yamamoto, T. Regression of cutaneous squamous cell carcinoma in a patient with chronic myeloid leukaemia on imatinib mesylate treatment. Acta Derm. Venereol. 2008, 88, 185–186. [Google Scholar] [CrossRef]
- William, W.N., Jr.; Feng, L.; Ferrarotto, R.; Ginsberg, L.; Kies, M.; Lippman, S.; Glisson, B.; Kim, E.S. Gefitinib for patients with incurable cutaneous squamous cell carcinoma: A single-arm phase II clinical trial. J. Am. Acad. Dermatol. 2017, 77, 1110–1113. [Google Scholar] [CrossRef]
- Yanagi, T.; Kitamura, S.; Hata, H. Novel Therapeutic Targets in Cutaneous Squamous Cell Carcinoma. Front. Oncol. 2018, 8, 79. [Google Scholar] [CrossRef]
- De Lima, P.O.; Joseph, S.; Panizza, B.; Simpson, F. Epidermal growth factor receptor’s function in cutaneous squamous cell carcinoma and its role as a therapeutic target in the age of immunotherapies. Curr. Treat. Options Oncol. 2020, 21, 9. [Google Scholar] [CrossRef]
- Patel, R.; Chang, A.L.S. Immune Checkpoint Inhibitors for Treating Advanced Cutaneous Squamous Cell Carcinoma. Am. J. Clin. Dermatol. 2019, 20, 477–482. [Google Scholar] [CrossRef]
- US Food and Drug Administration. FDA Approves Cemiplimab-Rwlc for Metastatic or Locally Advanced Cutaneous Squamous Cell Carcinoma. FDA Website. Available online: https://www.fda.gov/drugs/drug-approvals-and-databases (accessed on 22 June 2020).
- Love, W.E.; Bernhard, J.D.; Bordeaux, J.S. Topical imiquimod or fluorouracil therapy for basal and squamous cell carcinoma: A systematic review. Arch. Dermatol. 2009, 145, 1431–1438. [Google Scholar] [CrossRef]
- Fayne, R.; Nanda, S.; Nichols, A.; Shen, J. Combination Topical Chemotherapy for the Treatment of an Invasive Cutaneous Squamous Cell Carcinoma. J. Drugs Dermatol. 2020, 19, 202–204. [Google Scholar] [CrossRef] [PubMed]
| Gene | No. of Analyzed Samples | Mutations (%) | References |
|---|---|---|---|
| Cell-cycle control | |||
| TP53 | 100 | 42 | [33] |
| 91 | 64 | [27] | |
| 39 | 94.9 | [28] | |
| 29 | 79 | [29] | |
| 28 | 54–85 | [30] | |
| 40 | 70 | [31] | |
| CDKN2A | 100 | 28 | [33] |
| 91 | 23 | [27] | |
| 39 | 43.6 | [28] | |
| 29 | 45 | [29] | |
| 28 | 29–42 | [30] | |
| 40 | 45 | [31] | |
| Keratinocyte differentiation | |||
| NOTCH 1 | 100 | 54 | [33] |
| 91 | 75 | [27] | |
| 39 | 59 | [28] | |
| 29 | 48 | [29] | |
| 28 | 50–63 | [30] | |
| 40 | 75 | [31] | |
| NOTCH 2 | 100 | 34 | [33] |
| 91 | 63 | [27] | |
| 39 | 51.3 | [28] | |
| 29 | 31 | [29] | |
| 28 | 41–48 | [30] | |
| 40 | 50 | [31] | |
| FAT1 | 39 | 43.6 | [28] |
| 170 | 40 | [27] | |
| 28 | 22–37 | [30] | |
| 40 | 60 | [31] | |
| RIPK4 | 39 | 28 | [28] |
| 29 | 24 | [29] | |
| RAS signaling | |||
| HRAS | 100 | 6 | [33] |
| 91 | 16 | [27] | |
| 39 | 20.5 | [28] | |
| 29 | 13 | [29] | |
| 28 | 12–13 | [30] | |
| 40 | 22.5 | [31] | |
| KRAS | 91 | 13 | [27] |
| 29 | 10 | [29] | |
| BRAF | 39 | 17.9 | [28] |
| 29 | 13 | [29] | |
| 28 | 5–13 | [30] | |
| RASA1 | 39 | 13 | [28] |
| Chromatin segregation/remodeling | |||
| KNSTRN | 100 | 19 | [33] |
| KMT2C | 39 | 38.5 | [28] |
| 28 | 36–43 | [30] | |
| KMT2D | 39 | 69.2 | [28] |
| 28 | 31–62 | [30] | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lazar, A.D.; Dinescu, S.; Costache, M. Deciphering the Molecular Landscape of Cutaneous Squamous Cell Carcinoma for Better Diagnosis and Treatment. J. Clin. Med. 2020, 9, 2228. https://doi.org/10.3390/jcm9072228
Lazar AD, Dinescu S, Costache M. Deciphering the Molecular Landscape of Cutaneous Squamous Cell Carcinoma for Better Diagnosis and Treatment. Journal of Clinical Medicine. 2020; 9(7):2228. https://doi.org/10.3390/jcm9072228
Chicago/Turabian StyleLazar, Andreea D., Sorina Dinescu, and Marieta Costache. 2020. "Deciphering the Molecular Landscape of Cutaneous Squamous Cell Carcinoma for Better Diagnosis and Treatment" Journal of Clinical Medicine 9, no. 7: 2228. https://doi.org/10.3390/jcm9072228
APA StyleLazar, A. D., Dinescu, S., & Costache, M. (2020). Deciphering the Molecular Landscape of Cutaneous Squamous Cell Carcinoma for Better Diagnosis and Treatment. Journal of Clinical Medicine, 9(7), 2228. https://doi.org/10.3390/jcm9072228







