A Proteomics-Based Analysis Reveals Predictive Biological Patterns in Fabry Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Blood Samples
2.2. Targeted Proteomic Analysis
2.3. Plasma lysoGb3 Analysis
2.4. Alpha-d-Galactopyranosidase Activity Analysis
2.5. Data Analysis
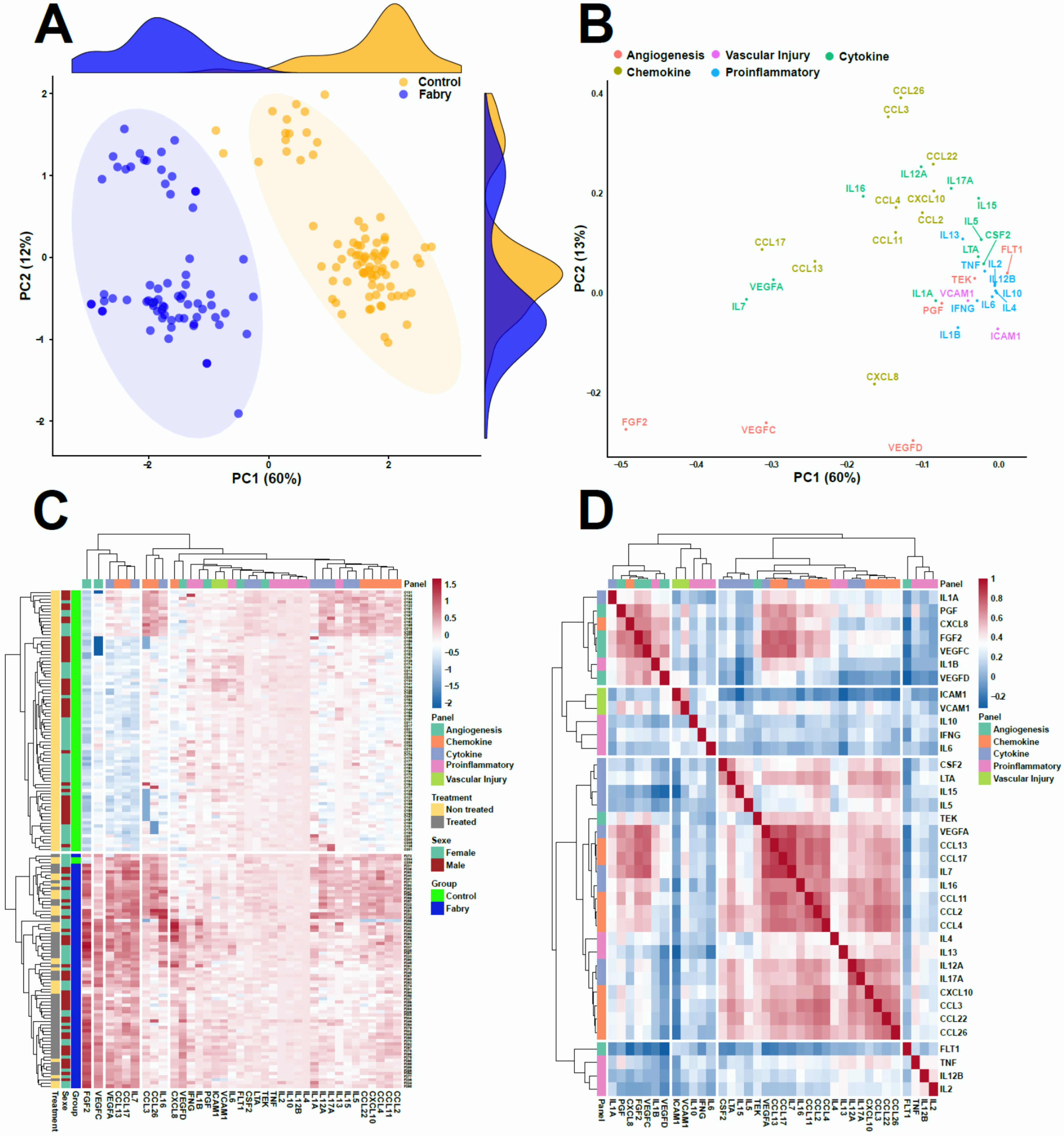
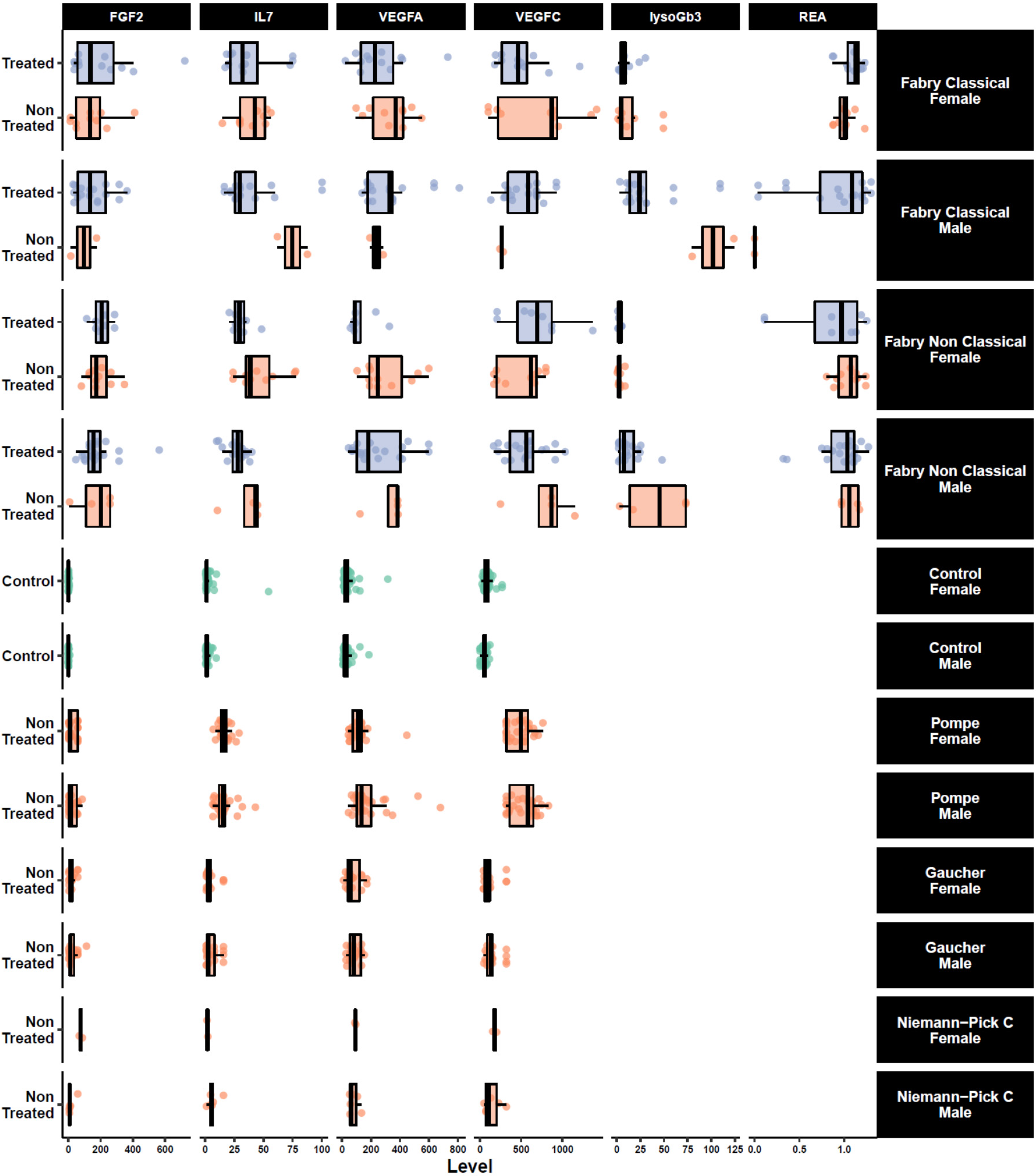
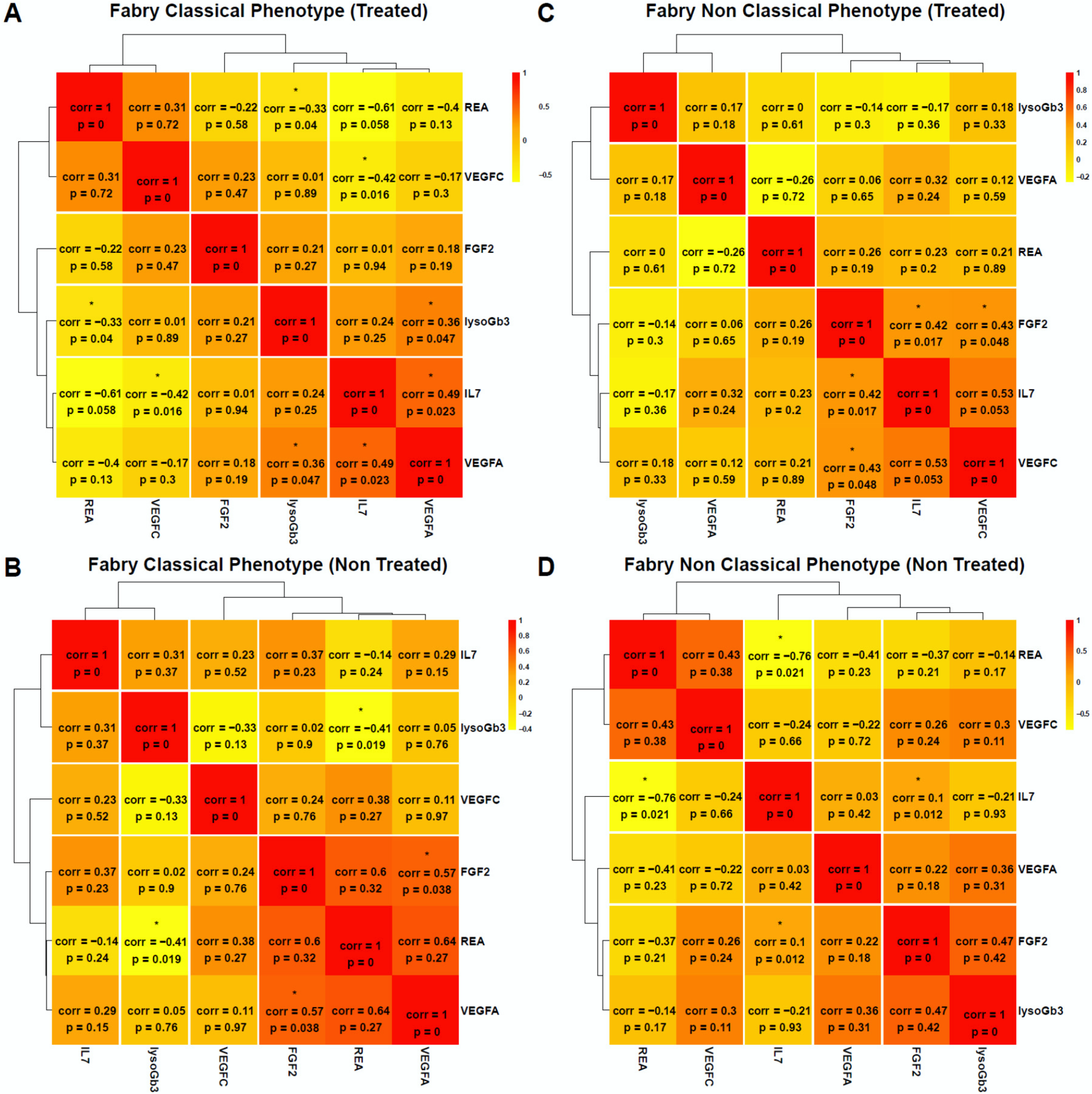
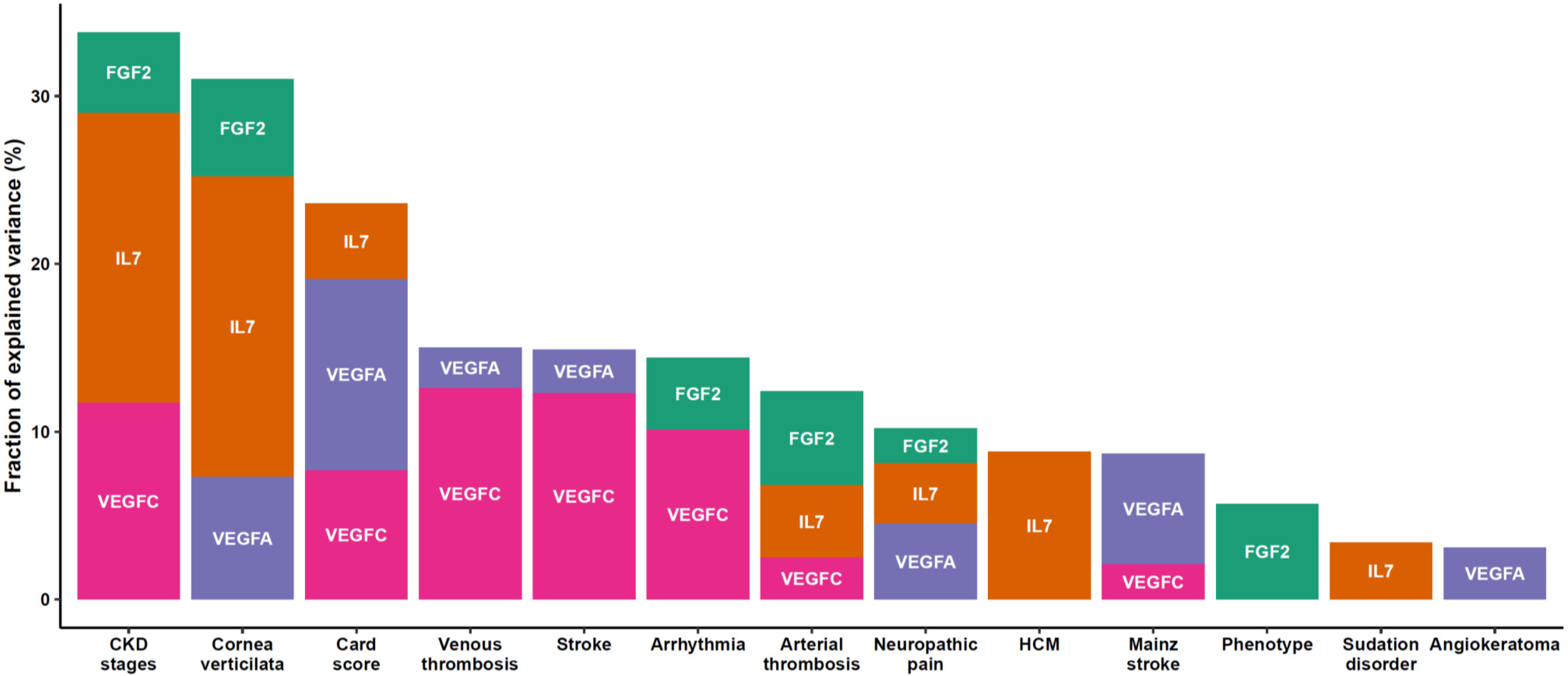
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Arends, M.; Wanner, C.; Hughes, D.; Mehta, A.; Oder, D.; Watkinson, O.T.; Elliott, P.M.; Linthorst, G.E.; Wijburg, F.A.; Biegstraaten, M.; et al. Characterization of classical and nonclassical fabry disease: A multicenter study. J. Am. Soc. Nephrol. 2017, 28, 1631–1641. [Google Scholar] [CrossRef] [PubMed]
- Meikle, P.J.; Hopwood, J.J.; Clague, A.E.; Carey, W.F. Prevalence of lysosomal storage disorders. Jama 1999, 281, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Inoue, T.; Hattori, K.; Ihara, K.; Ishii, A.; Nakamura, K.; Hirose, S. Newborn screening for Fabry disease in Japan: Prevalence and genotypes of Fabry disease in a pilot study. J. Hum. Genet. 2013, 58, 548. [Google Scholar] [CrossRef] [PubMed]
- Mechtler, T.P.; Stary, S.; Metz, T.F.; De Jesús, V.R.; Greber-Platzer, S.; Pollak, A.; Herkner, K.R.; Streubel, B.; Kasper, D.C. Neonatal screening for lysosomal storage disorders: Feasibility and incidence from a nationwide study in Austria. Lancet 2012, 379, 335–341. [Google Scholar] [CrossRef]
- Spada, M.; Pagliardini, S.; Yasuda, M.; Tukel, T.; Thiagarajan, G.; Sakuraba, H.; Ponzone, A.; Desnick, R.J. High incidence of later-onset Fabry disease revealed by newborn screening. Am. J. Hum. Genet. 2006, 79, 31–40. [Google Scholar] [CrossRef]
- Hopkins, P.V.; Campbell, C.; Klug, T.; Rogers, S.; Raburn-Miller, J.; Kiesling, J. Lysosomal storage disorder screening implementation: Findings from the first six months of full population pilot testing in Missouri. J. Pediatrics 2015, 166, 172–177. [Google Scholar] [CrossRef]
- Lin, H.Y.; Chong, K.W.; Hsu, J.H.; Yu, H.C.; Shih, C.C.; Huang, C.H.; Lin, S.J.; Chen, C.H.; Chiang, C.C.; Ho, H.J.; et al. High incidence of the cardiac variant of Fabry disease revealed by newborn screening in the Taiwan Chinese population. Circ. Cardiovasc. Genet. 2009, 2, 450–456. [Google Scholar] [CrossRef]
- Kramer, J.; Weidemann, F. Biomarkers for diagnosing and staging of Fabry disease. Curr. Med. Chem. 2018, 25, 1530–1537. [Google Scholar] [CrossRef]
- Waldek, S.; Patel, M.R.; Banikazemi, M.; Lemay, R.; Lee, P. Life expectancy and cause of death in males and females with Fabry disease: Findings from the Fabry Registry. Genet. Med. Off. J. Am. Coll. Med Genet. 2009, 11, 790–796. [Google Scholar] [CrossRef]
- Thomas, A.S.; Mehta, A.B. Difficulties and barriers in diagnosing Fabry disease: What can be learnt from the literature? Expert Opin. Med. Diagn. 2013, 7, 589–599. [Google Scholar] [CrossRef]
- El-Abassi, R.; Singhal, D.; England, J.D. Fabry’s disease. J. Neurol. Sci. 2014, 344, 5–19. [Google Scholar] [CrossRef]
- Duro, G.; Zizzo, C.; Cammarata, G.; Burlina, A.; Burlina, A.; Polo, G.; Scalia, S.; Oliveri, R.; Sciarrino, S.; Francofonte, D.; et al. Mutations in the GLA Gene and LysoGb3: Is it really Anderson-Fabry disease? Int. J. Mol. Sci. 2018, 19, 3726. [Google Scholar] [CrossRef]
- Winchester, B.; Young, E. Biochemical and genetic diagnosis of Fabry disease. In Fabry Disease: Perspectives from 5 Years of FOS; Oxford PharmaGenesis: Oxford, UK, 2006. [Google Scholar]
- Desnick, R.J.; Allen, K.Y.; Desnick, S.J.; Raman, M.K.; Bernlohr, R.W.; Krivit, W. Fabry’s disease: Enzymatic diagnosis of hemizygotes and heterozygotes: α-galactosidase activities in plasma, serum, urine, and leukocytes. J. Lab. Clin. Med. 1973, 81, 157–171. [Google Scholar] [PubMed]
- Chamoles, N.; Blanco, M.; Gaggioli, D. Fabry disease: Enzymatic diagnosis in dried blood spots on filter paper. Clin. Chim. Acta 2001, 1, 195–196. [Google Scholar] [CrossRef]
- Desnick, R.J.; Ioannou, Y.A.; Eng, C.M. α-Galactosidase A deficiency: Fabry disease. In The Metabolic and Molecular Bases of Inherited Disease; McGraw-Hill: New York, NY, USA, 2001; Volume 3, pp. 2741–2784. [Google Scholar]
- Mills, K.; Johnson, A.; Winchester, B. Synthesis of novel internal standards for the quantitative determination of plasma ceramide trihexoside in Fabry disease by tandem mass spectrometry. FEBS Lett. 2002, 515, 171–176. [Google Scholar] [CrossRef]
- Nowak, A.; Mechtler, T.; Kasper, D.C.; Desnick, R.J. Correlation of Lyso-Gb3 levels in dried blood spots and sera from patients with classic and Later-Onset Fabry disease. Mol. Genet. Metab. 2017, 121, 320–324. [Google Scholar] [CrossRef] [PubMed]
- Boscaro, F.; Pieraccini, G.; Marca, G.L.; Bartolucci, G.; Luceri, C.; Luceri, F.; Moneti, G. Rapid quantitation of globotriaosylceramide in human plasma and urine: A potential application for monitoring enzyme replacement therapy in Anderson-Fabry disease. Rapid Commun. Mass Spectrom. 2002, 16, 1507–1514. [Google Scholar] [CrossRef] [PubMed]
- Aerts, J.M.; Groener, J.E.; Kuiper, S.; Donker-Koopman, W.E.; Strijland, A.; Ottenhoff, R.; van Roomen, C.; Mirzaian, M.; Wijburg, F.A.; Linthorst, G.E. Elevated globotriaosylsphingosine is a hallmark of Fabry disease. Proc. Natl. Acad. Sci. USA 2008, 105, 2812–2817. [Google Scholar] [CrossRef]
- Talbot, A.; Nicholls, K.; Fletcher, J.M.; Fuller, M. A simple method for quantification of plasma globotriaosylsphingosine: Utility for Fabry disease. Mol. Genet. Metab. 2017, 122, 121–125. [Google Scholar] [CrossRef]
- Zarate, Y.A.; Hopkin, R.J. Fabry’s disease. Lancet 2008, 372, 1427–1435. [Google Scholar] [CrossRef]
- Auray-Blais, C.; Cyr, D.; Mills, K.; Giguere, R.; Drouin, R. Development of a filter paper method potentially applicable to mass and high-risk urinary screenings for Fabry disease. J. Inherit. Metab. Dis. 2007, 30, 106. [Google Scholar] [CrossRef] [PubMed]
- Mehta, A. Fabry disease: A review of current enzyme replacement strategies. Expert Opin. Orphan Drugs 2015, 3, 1319–1330. [Google Scholar] [CrossRef]
- Schiffmann, R.; Kopp, J.B.; Austin, H.A., 3rd; Sabnis, S.; Moore, D.F.; Weibel, T.; Balow, J.E.; Brady, R.O. Enzyme replacement therapy in Fabry disease: A randomized controlled trial. Jama 2001, 285, 2743–2749. [Google Scholar] [CrossRef] [PubMed]
- McCafferty, E.H.; Scott, L.J. Migalastat: A review in Fabry disease. Drugs 2019, 79, 543–554. [Google Scholar] [CrossRef] [PubMed]
- Arends, M.; Biegstraaten, M.; Hughes, D.A.; Mehta, A.; Elliott, P.M.; Oder, D.; Watkinson, O.T.; Vaz, F.M.; van Kuilenburg, A.B.P.; Wanner, C.; et al. Retrospective study of long-term outcomes of enzyme replacement therapy in Fabry disease: Analysis of prognostic factors. PLoS ONE 2017, 12, e0182379. [Google Scholar] [CrossRef]
- Citro, V.; Cammisa, M.; Liguori, L.; Cimmaruta, C.; Lukas, J.; Cubellis, M.V.; Andreotti, G. The large phenotypic spectrum of Fabry disease requires graduated diagnosis and personalized therapy: A meta-analysis can help to differentiate missense mutations. Int. J. Mol. Sci. 2016, 17, 2010. [Google Scholar] [CrossRef]
- Reisin, R.; Perrin, A.; Garcia-Pavia, P. Time delays in the diagnosis and treatment of Fabry disease. Int. J. Clin. Pract. 2017, 71, e12914. [Google Scholar] [CrossRef]
- Sudrie-Arnaud, B.; Marguet, F.; Patrier, S.; Martinovic, J.; Louillet, F.; Broux, F.; Charbonnier, F.; Dranguet, H.; Coutant, S.; Vezain, M.; et al. Metabolic causes of nonimmune hydrops fetalis: A next-generation sequencing panel as a first-line investigation. Clin. Chim. Acta Int. J. Clin. Chem. 2018, 481, 1–8. [Google Scholar] [CrossRef]
- Tebani, A.; Schmitz-Afonso, I.; Abily-Donval, L.; Heron, B.; Piraud, M.; Ausseil, J.; Brassier, A.; De Lonlay, P.; Zerimech, F.; Vaz, F.M.; et al. Urinary metabolic phenotyping of mucopolysaccharidosis type I combining untargeted and targeted strategies with data modeling. Clin. Chim. Acta Int. J. Clin. Chem. 2017, 475, 7–14. [Google Scholar] [CrossRef]
- Tebani, A.; Afonso, C.; Marret, S.; Bekri, S. Omics-based strategies in precision medicine: Toward a paradigm shift in inborn errors of metabolism investigations. Int. J. Mol. Sci. 2016, 17, 1555. [Google Scholar] [CrossRef]
- Tebani, A.; Abily-Donval, L.; Afonso, C.; Marret, S.; Bekri, S. Clinical metabolomics: The new metabolic window for inborn errors of metabolism investigations in the post-genomic era. Int. J. Mol. Sci. 2016, 17, 1167. [Google Scholar] [CrossRef]
- Cigna, D.; D’Anna, C.; Zizzo, C.; Francofonte, D.; Sorrentino, I.; Colomba, P.; Albeggiani, G.; Armini, A.; Bianchi, L.; Bini, L.; et al. Alteration of proteomic profiles in PBMC isolated from patients with Fabry disease: Preliminary findings. Mol. Biosyst. 2013, 9, 1162–1168. [Google Scholar] [CrossRef] [PubMed]
- Yogasundaram, H.; Nikhanj, A.; Putko, B.N.; Boutin, M.; Jain-Ghai, S.; Khan, A.; Auray-Blais, C.; West, M.L.; Oudit, G.Y. Elevated inflammatory plasma biomarkers in patients with Fabry disease: A critical link to heart failure with preserved ejection fraction. J. Am. Heart Assoc. 2018, 7, e009098. [Google Scholar] [CrossRef] [PubMed]
- Matafora, V.; Cuccurullo, M.; Beneduci, A.; Petrazzuolo, O.; Simeone, A.; Anastasio, P.; Mignani, R.; Feriozzi, S.; Pisani, A.; Comotti, C.; et al. Early markers of Fabry disease revealed by proteomics. Mol. Biosyst. 2015, 11, 1543–1551. [Google Scholar] [CrossRef] [PubMed]
- Manwaring, V.; Heywood, W.E.; Clayton, R.; Lachmann, R.H.; Keutzer, J.; Hindmarsh, P.; Winchester, B.; Heales, S.; Mills, K. The identification of new biomarkers for identifying and monitoring kidney disease and their translation into a rapid mass spectrometry-based test: Evidence of presymptomatic kidney disease in pediatric Fabry and Type-I diabetic patients. J. Proteome Res. 2013, 12, 2013–2021. [Google Scholar] [CrossRef]
- Heo, S.H.; Kang, E.; Kim, Y.M.; Go, H.; Kim, K.Y.; Jung, J.Y.; Kang, M.; Kim, G.H.; Kim, J.M.; Choi, I.H.; et al. Fabry disease: Characterisation of the plasma proteome pre- and post-enzyme replacement therapy. J. Med Genet. 2017, 54, 771–780. [Google Scholar] [CrossRef]
- Moore, D.F.; Krokhin, O.V.; Beavis, R.C.; Ries, M.; Robinson, C.; Goldin, E.; Brady, R.O.; Wilkins, J.A.; Schiffmann, R. Proteomics of specific treatment-related alterations in Fabry disease: A strategy to identify biological abnormalities. Proc. Natl. Acad. Sci. USA 2007, 104, 2873–2878. [Google Scholar] [CrossRef]
- Birket, M.J.; Raibaud, S.; Lettieri, M.; Adamson, A.D.; Letang, V.; Cervello, P.; Redon, N.; Ret, G.; Viale, S.; Wang, B.; et al. A human stem cell model of Fabry disease implicates LIMP-2 accumulation in cardiomyocyte pathology. Stem Cell Rep. 2019, 13, 380–393. [Google Scholar] [CrossRef]
- Song, H.-Y.; Chien, C.-S.; Yarmishyn, A.A.; Chou, S.-J.; Yang, Y.-P.; Wang, M.-L.; Wang, C.-Y.; Leu, H.-B.; Yu, W.-C.; Chang, Y.-L.; et al. Generation of GLA-knockout human embryonic stem cell lines to model autophagic dysfunction and exosome secretion in Fabry disease-associated hypertrophic cardiomyopathy. Cells 2019, 8, 327. [Google Scholar] [CrossRef]
- Weidemann, F.; Beer, M.; Kralewski, M.; Siwy, J.; Kampmann, C. Early detection of organ involvement in Fabry disease by biomarker assessment in conjunction with LGE cardiac MRI: Results from the SOPHIA study. Mol. Genet. Metab. 2019, 126, 169–182. [Google Scholar] [CrossRef]
- Doykov, I.D.; Heywood, W.E.; Nikolaenko, V.; Śpiewak, J.; Hällqvist, J.; Clayton, P.T.; Mills, P.; Warnock, D.G.; Nowak, A.; Mills, K. Rapid, proteomic urine assay for monitoring progressive organ disease in Fabry disease. J. Med. Genet. 2020, 57, 38–47. [Google Scholar] [CrossRef]
- Mauhin, W.; Lidove, O.; Amelin, D.; Lamari, F.; Caillaud, C.; Mingozzi, F.; Dzangue-Tchoupou, G.; Arouche-Delaperche, L.; Douillard, C.; Dussol, B.; et al. Deep characterization of the anti-drug antibodies developed in Fabry disease patients, a prospective analysis from the French multicenter cohort FFABRY. Orphanet J. Rare Dis. 2018, 13, 127. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, L.; Trygg, J.; Wold, S. A chemometrics toolbox based on projections and latent variables. J. Chemom. 2014, 28, 332–346. [Google Scholar] [CrossRef]
- Alharbi, F.J.; Baig, S.; Auray-Blais, C.; Boutin, M.; Ward, D.G.; Wheeldon, N.; Steed, R.; Dawson, C.; Hughes, D.; Geberhiwot, T. Globotriaosylsphingosine (Lyso-Gb3) as a biomarker for cardiac variant (N215S) Fabry disease. J. Inherit. Metab. Dis. 2018, 41, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Murakami, M.; Simons, M. Fibroblast growth factor regulation of neovascularization. Curr. Opin. Hematol. 2008, 15, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.H.; Choi, E.N.; Jeon, Y.J.; Jung, S.C. Possible role of transforming growth factor-beta1 and vascular endothelial growth factor in Fabry disease nephropathy. Int. J. Mol. Med. 2012, 30, 1275–1280. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Zhang, J.; Lao, X.; Jiang, H.; Yu, Y.; Deng, Y.; Zhong, J.; Liang, Y.; Xiong, L.; Deng, N. Construction of a disulfide-stabilized diabody against fibroblast growth factor-2 and the inhibition activity in targeting breast cancer. Cancer Sci. 2016, 107, 1141–1150. [Google Scholar] [CrossRef]
- Hamamoto, J.; Yasuda, H.; Nonaka, Y.; Fujiwara, M.; Nakamura, Y.; Soejima, K.; Betsuyaku, T. The FGF2 aptamer inhibits the growth of FGF2-FGFR pathway driven lung cancer cells. Biochem. Biophys. Res. Commun. 2018, 503, 1330–1334. [Google Scholar] [CrossRef]
- Satoh, K. Globotriaosylceramide induces endothelial dysfunction in fabry disease. Arterioscler. Thromb. Vasc. Biol. 2014, 34, 2–4. [Google Scholar] [CrossRef]
- Bird, S.; Hadjimichael, E.; Mehta, A.; Ramaswami, U.; Hughes, D. Fabry disease and incidence of cancer. Orphanet J. Rare Dis. 2017, 12, 150. [Google Scholar] [CrossRef]
- Thurberg, B.L.; Germain, D.P.; Perretta, F.; Jurca-Simina, I.E.; Politei, J.M. Fabry disease: Four case reports of meningioma and a review of the literature on other malignancies. Mol. Genet. Metab. Rep. 2017, 11, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Rozenfeld, P.; Feriozzi, S. Contribution of inflammatory pathways to Fabry disease pathogenesis. Mol. Genet. Metab. 2017, 122, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Liebau, M.C.; Braun, F.; Höpker, K.; Weitbrecht, C.; Bartels, V.; Müller, R.-U.; Brodesser, S.; Saleem, M.A.; Benzing, T.; Schermer, B. Dysregulated autophagy contributes to podocyte damage in Fabry’s disease. PLoS ONE 2013, 8, e63506. [Google Scholar] [CrossRef] [PubMed]
- Chévrier, M.; Brakch, N.; Céline, L.; Genty, D.; Ramdani, Y.; Moll, S.; Djavaheri-Mergny, M.; Brasse-Lagnel, C.; Annie Laquerrière, A.L.; Barbey, F. Autophagosome maturation is impaired in Fabry disease. Autophagy 2010, 6, 589–599. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.H.; Chien, Y.; Wang, K.L.; Leu, H.B.; Hsiao, C.Y.; Lai, Y.H.; Wang, C.Y.; Chang, Y.L.; Lin, S.J.; Niu, D.M.; et al. Evaluation of proinflammatory prognostic biomarkers for fabry cardiomyopathy with enzyme replacement therapy. Can. J. Cardiol. 2016, 32, 1221.e1–1221.e9. [Google Scholar] [CrossRef]
- Coats, C.J.; Parisi, V.; Ramos, M.; Janagarajan, K.; O’Mahony, C.; Dawnay, A.; Lachmann, R.H.; Murphy, E.; Mehta, A.; Hughes, D.; et al. Role of serum N-terminal pro-brain natriuretic peptide measurement in diagnosis of cardiac involvement in patients with anderson-fabry disease. Am. J. Cardiol. 2013, 111, 111–117. [Google Scholar] [CrossRef]
- Seydelmann, N.; Liu, D.; Krämer, J.; Drechsler, C.; Hu, K.; Nordbeck, P.; Schneider, A.; Störk, S.; Bijnens, B.; Ertl, G.; et al. High-sensitivity troponin: A clinical blood biomarker for staging cardiomyopathy in Fabry disease. J. Am. Heart Assoc. 2016, 5, e002839. [Google Scholar] [CrossRef]
- Hu, K.; Liu, D.; Salinger, T.; Oder, D.; Knop, S.; Ertl, G.; Weidemann, F.; Frantz, S.; Störk, S.; Nordbeck, P. Value of cardiac biomarker measurement in the differential diagnosis of infiltrative cardiomyopathy patients with preserved left ventricular systolic function. J. Thorac. Dis. 2018, 10, 4966–4975. [Google Scholar] [CrossRef]
- McLeod, I.X.; Zhou, X.; Li, Q.J.; Wang, F.; He, Y.W. The class III kinase Vps34 promotes T lymphocyte survival through regulating IL-7Ralpha surface expression. J. Immunol. 2011, 187, 5051–5061. [Google Scholar] [CrossRef]
- Lundstrom, W.; Fewkes, N.M.; Mackall, C.L. IL-7 in human health and disease. Semin. Immunol. 2012, 24, 218–224. [Google Scholar] [CrossRef]
- Stappers, F.; Scharnetzki, D.; Schmitz, B.; Manikowski, D.; Brand, S.M.; Grobe, K.; Lenders, M.; Brand, E. Neutralising anti-drug antibodies in Fabry disease can inhibit endothelial enzyme uptake and activity. J. Inherit. Metab. Dis. 2020, 43, 334–347. [Google Scholar] [CrossRef] [PubMed]

| Summary | n | Fabry Classical n = 34 (14%) 1 | Fabry Non-Classical n = 35 (14%) 1 | Control n = 83 (33%) 1 | Pompe n = 59 (24%) 1 | Gaucher n = 30 (12%) 1 | Niemann–Pick C n = 8 (3.2%) 1 | p-Value 2 | |
|---|---|---|---|---|---|---|---|---|---|
| Age (Years) | mean (SD) | 248 | 44 (15) | 48 (15) | 42 (13) | 54 (13) | 41 (18) | 45 (13) | 0.3 |
| Sex | 249 | 0.5 | |||||||
| Female | n/N (%) | 20/34 (59%) | 15/35 (43%) | 44/83 (53%) | 30/59 (51%) | 14/30 (47%) | 2/8 (25%) | ||
| Male | n/N (%) | 14/34 (41%) | 20/35 (57%) | 39/83 (47%) | 29/59 (49%) | 16/30 (53%) | 6/8 (75%) | ||
| BMI (kg/m2) | mean (SD) | 56 | 25 (7) | 29 (16) | 0.2 | ||||
| Cornea verticilata | n/N (%) | 57 | 26/26 (100%) | 3/31 (9.7%) | <0.001 | ||||
| Hypertrophic cardiomyopathy | n/N (%) | 67 | 14/33 (42%) | 19/34 (56%) | 0.3 | ||||
| Angiokeratoma | n/N (%) | 66 | 20/32 (62%) | 12/34 (35%) | 0.048 | ||||
| Arterial thrombosis | n/N (%) | 65 | 1/31 (3.2%) | 2/34 (5.9%) | >0.9 | ||||
| Venous thrombosis | n/N (%) | 66 | 6/32 (19%) | 1/34 (2.9%) | 0.051 | ||||
| Arrhythmia | n/N (%) | 69 | 19/34 (56%) | 14/35 (40%) | 0.2 | ||||
| Stroke | n/N (%) | 67 | 5/32 (16%) | 5/35 (14%) | >0.9 | ||||
| Neuropathic pain | n/N (%) | 66 | 31/31 (100%) | 15/35 (43%) | <0.001 | ||||
| Sudation disorder | n/N (%) | 69 | 24/34 (71%) | 13/35 (37%) | 0.008 | ||||
| Kidney Transplant | n/N (%) | 69 | 3/34 (8.8%) | 0/35 (0%) | 0.11 | ||||
| Dialysis | n/N (%) | 68 | 30/34 (88%) | 34/34 (100%) | 0.11 | ||||
| CKD stages | 66 | 0.2 | |||||||
| 0 | n/N (%) | 18/33 (55%) | 18/33 (55%) | ||||||
| 1 | n/N (%) | 2/33 (6.1%) | 3/33 (9.1%) | ||||||
| 2 | n/N (%) | 6/33 (18%) | 9/33 (27%) | ||||||
| 3 | n/N (%) | 1/33 (3.0%) | 3/33 (9.1%) | ||||||
| 4 | n/N (%) | 2/33 (6.1%) | 0/33 (0%) | ||||||
| 5 | n/N (%) | 4/33 (12%) | 0/33 (0%) | ||||||
| Treatment | 69 | >0.9 | |||||||
| Non-Treated | n/N (%) | 11/34 (32%) | 12/35 (34%) | ||||||
| Treated | n/N (%) | 23/34 (68%) | 23/35 (66%) | ||||||
| Therapy | 69 | 0.3 | |||||||
| Agalsidase α | n/N (%) | 9/34 (26%) | 3/35 (8.6%) | ||||||
| Agalsidase α/Agalsidase β | n/N (%) | 4/34 (12%) | 6/35 (17%) | ||||||
| Agalsidase α/Agalsidase β/Miglastat | n/N (%) | 0/34 (0%) | 1/35 (2.9%) | ||||||
| Agalsidase α/Miglastat | n/N (%) | 0/34 (0%) | 1/35 (2.9%) | ||||||
| Agalsidase β | n/N (%) | 10/34 (29%) | 11/35 (31%) | ||||||
| Miglastat | n/N (%) | 0/34 (0%) | 1/35 (2.9%) | ||||||
| Non-Treated | n/N (%) | 11/34 (32%) | 12/35 (34%) | ||||||
| Treatment Duration (Years) | mean (SD) | 45 | 6.8 (4.8) | 6.0 (5.2) | 0.6 | ||||
| Variant | 63 | 0.011 | |||||||
| Missense | n/N (%) | 12/29 (41%) | 25/34 (74%) | ||||||
| MTP | n/N (%) | 17/29 (59%) | 9/34 (26%) | ||||||
| Neutralizing Antibody (Positive) | n/N (%) | 69 | 30/34 (88%) | 33/35 (94%) | 0.4 | ||||
| lysoGb3 (ng/mL) | mean (SD) | 63 | 22 (31) | 10 (15) | 0.011 | ||||
| Residual Enzymatic Activity (%) | mean (SD) | 57 | 0.94 (0.37) | 0.98 (0.29) | >0.9 | ||||
| MDRD (mL/min) | mean (SD) | 63 | 85 (41) | 102 (30) | 0.14 |
| Protein | Comparison | Log Fold Change | p-Value |
|---|---|---|---|
| FGF2 | Fabry Non-Treated vs. Control | 2.30 | 7.49 × 10−26 |
| FGF2 | Fabry Treated vs. Control | 2.22 | 1.72 × 10−23 |
| FGF2 | Fabry Non-Treated vs. Gaucher | −1.33 | 1.87 × 10−6 |
| FGF2 | Fabry Treated vs. Gaucher | −1.24 | 7.46 × 10−6 |
| FGF2 | Fabry Treated Classic Female vs. Control Female | 2.21 | 2.81 × 10−17 |
| FGF2 | Fabry Non-Treated Non-Classic Female vs. Control Female | 2.26 | 1.45 × 10−15 |
| FGF2 | Fabry Treated Non-Classic Female vs. Control Female | 2.24 | 3.20 × 10−11 |
| FGF2 | Fabry Non-Treated Classic Male vs. Control Female | 2.00 | 1.45 × 10−3 |
| FGF2 | Fabry Non-Treated vs. Fabry Treated | −0.09 | 8.63 × 10−1 |
| IL-7 | Fabry Treated vs. Pompe | −1.48 | 4.47 × 10−11 |
| IL-7 | Fabry Non-Treated vs. Pompe | −1.29 | 5.55 × 10−9 |
| IL-7 | Fabry Treated vs. Niemann Pick C | −1.80 | 8.17 × 10−3 |
| IL-7 | Fabry Non-Treated vs. Niemann Pick C | −1.60 | 2.12 × 10−2 |
| IL-7 | Fabry Treated Classic Female vs. Control Female | 2.10 | 1.73 × 10−16 |
| IL-7 | Fabry Non-Treated Classic Female vs. Control Female | 1.94 | 2.21 × 10−12 |
| IL-7 | Fabry Non-Treated Non-Classic Female vs. Control Female | 1.89 | 7.67 × 10−12 |
| IL-7 | Fabry Treated Non-Classic Female vs. Control Female | 2.13 | 9.92 × 10−11 |
| IL-7 | Fabry Non-Treated vs. Fabry Treated | 0.19 | 8.63 × 10−1 |
| VEGFA | Fabry Non-Treated vs. Control | 1.82 | 6.38 × 10−15 |
| VEGFA | Fabry Treated vs. Control | 1.85 | 9.63 × 10−15 |
| VEGFA | Fabry Treated vs. Pompe | −1.49 | 4.19 × 10−9 |
| VEGFA | Fabry Non-Treated vs. Pompe | −1.47 | 5.31 × 10−9 |
| VEGFA | Fabry Treated vs. Niemann Pick C | −1.01 | 3.20 × 10−1 |
| VEGFA | Fabry Treated Classic Female vs. Control Female | 1.75 | 6.00 × 10−10 |
| VEGFA | Fabry Non-Treated Non-Classic Female vs. Control Female | 1.85 | 1.66 × 10−9 |
| VEGFA | Fabry Non-Treated Classic Female vs. Control Female | 1.79 | 6.00 × 10−9 |
| VEGFA | Fabry Treated Non-Classic Female vs. Control Female | 2.02 | 5.14 × 10−8 |
| VEGFA | Fabry Non-Treated Classic Male vs. Control Male | 1.62 | 4.92 × 10−2 |
| VEGFA | Fabry Non-Treated vs. Fabry Treated | 0.02 | 9.71 × 10−1 |
| VEGFC | Fabry Treated vs. Control | 1.82 | 1.69 × 10−15 |
| VEGFC | Fabry Treated vs. Pompe | −1.71 | 5.96 × 10−12 |
| VEGFC | Fabry Non-Treated vs. Pompe | −1.45 | 2.25 × 10−9 |
| VEGFC | Fabry Treated Classic Female vs. Control Female | 2.02 | 8.04 × 10−14 |
| VEGFC | Fabry Non-Treated Non-Classic Female vs. Control Female | 1.71 | 3.71 × 10−9 |
| VEGFC | Fabry Non-Treated vs. Fabry Treated | 0.26 | 8.63 × 10−1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tebani, A.; Mauhin, W.; Abily-Donval, L.; Lesueur, C.; Berger, M.G.; Nadjar, Y.; Berger, J.; Benveniste, O.; Lamari, F.; Laforêt, P.; et al. A Proteomics-Based Analysis Reveals Predictive Biological Patterns in Fabry Disease. J. Clin. Med. 2020, 9, 1325. https://doi.org/10.3390/jcm9051325
Tebani A, Mauhin W, Abily-Donval L, Lesueur C, Berger MG, Nadjar Y, Berger J, Benveniste O, Lamari F, Laforêt P, et al. A Proteomics-Based Analysis Reveals Predictive Biological Patterns in Fabry Disease. Journal of Clinical Medicine. 2020; 9(5):1325. https://doi.org/10.3390/jcm9051325
Chicago/Turabian StyleTebani, Abdellah, Wladimir Mauhin, Lenaig Abily-Donval, Céline Lesueur, Marc G. Berger, Yann Nadjar, Juliette Berger, Oliver Benveniste, Foudil Lamari, Pascal Laforêt, and et al. 2020. "A Proteomics-Based Analysis Reveals Predictive Biological Patterns in Fabry Disease" Journal of Clinical Medicine 9, no. 5: 1325. https://doi.org/10.3390/jcm9051325
APA StyleTebani, A., Mauhin, W., Abily-Donval, L., Lesueur, C., Berger, M. G., Nadjar, Y., Berger, J., Benveniste, O., Lamari, F., Laforêt, P., Noel, E., Marret, S., Lidove, O., & Bekri, S. (2020). A Proteomics-Based Analysis Reveals Predictive Biological Patterns in Fabry Disease. Journal of Clinical Medicine, 9(5), 1325. https://doi.org/10.3390/jcm9051325





