The Clinical Significance of PIWIL3 and PIWIL4 Expression in Pancreatic Cancer
Abstract
1. Introduction
2. Experimental Section
2.1. Cell Lines and Cell Culture
2.2. Patient Samples
3. Results
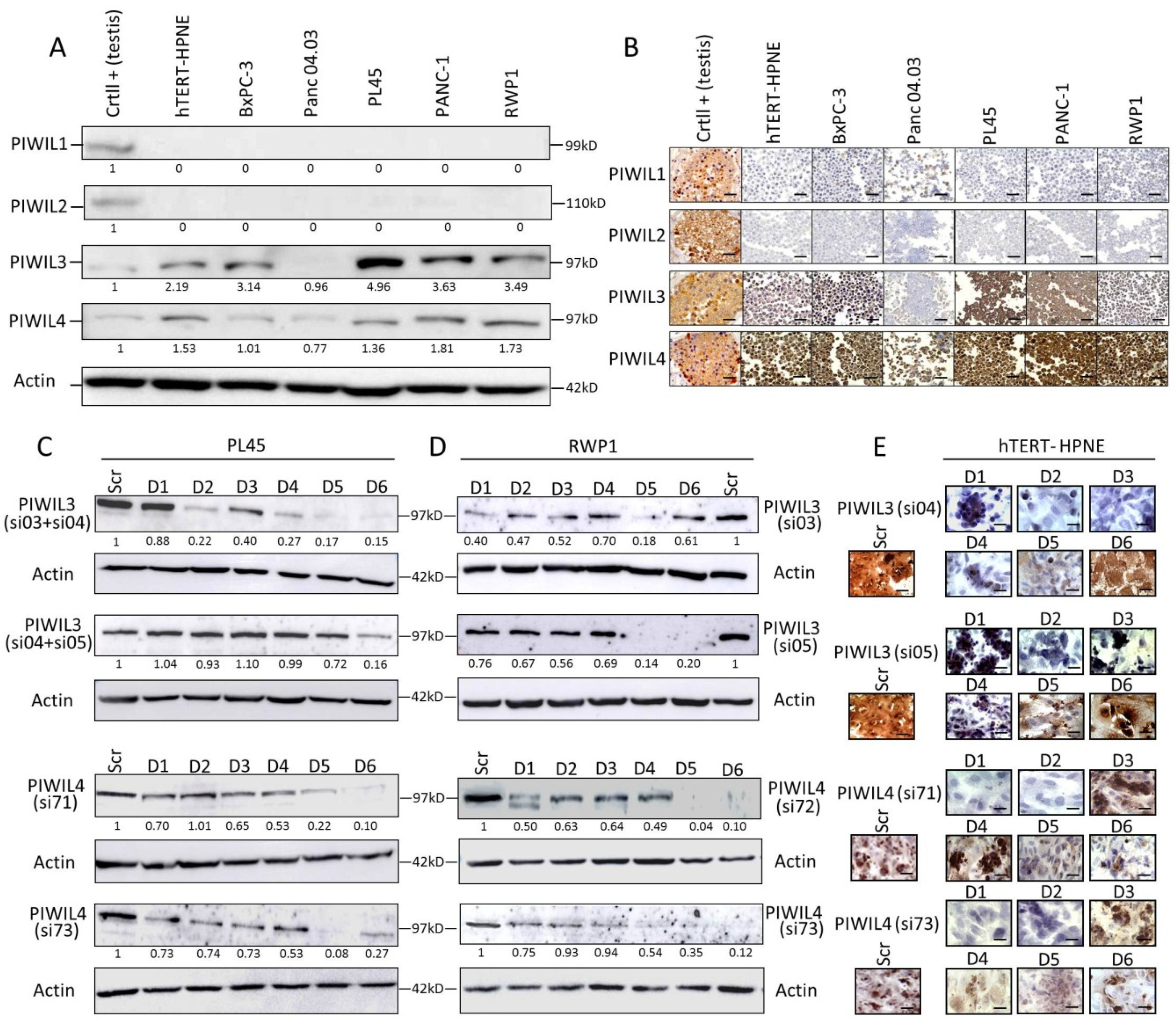
3.1. PIWIL3 and PIWIL4 Are Overexpressed in Non-Tumor and Tumor-Derived Cell Lines
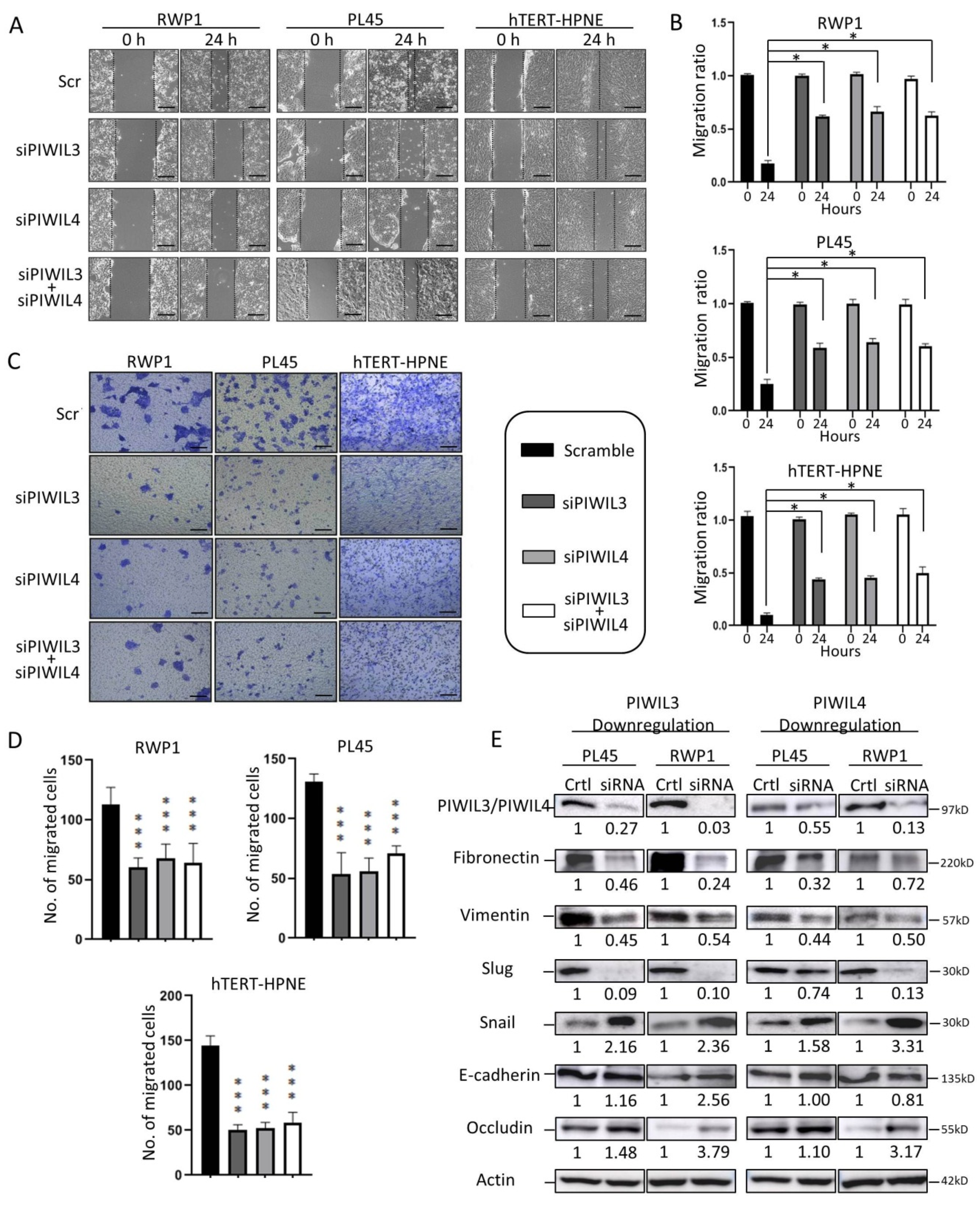
3.2. PIWIL3 and/or PIWIL4 Are Necessary for Cell Motility of Both Non-Tumor and Tumor-Derived Cell Lines
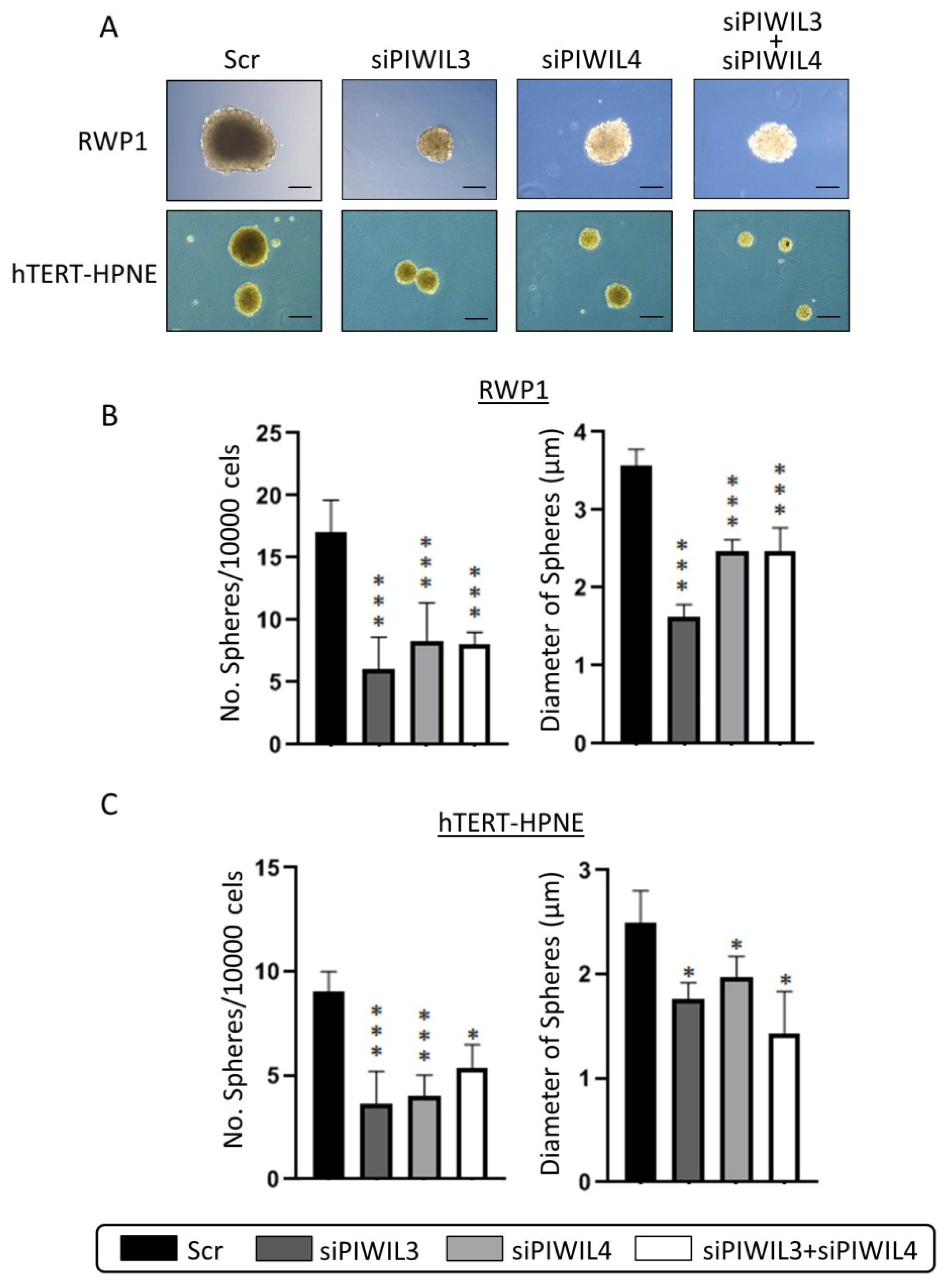
3.3. Downregulation of PIWIL3 and/or PIWIL4 Impairs Undifferentiated Phenotype
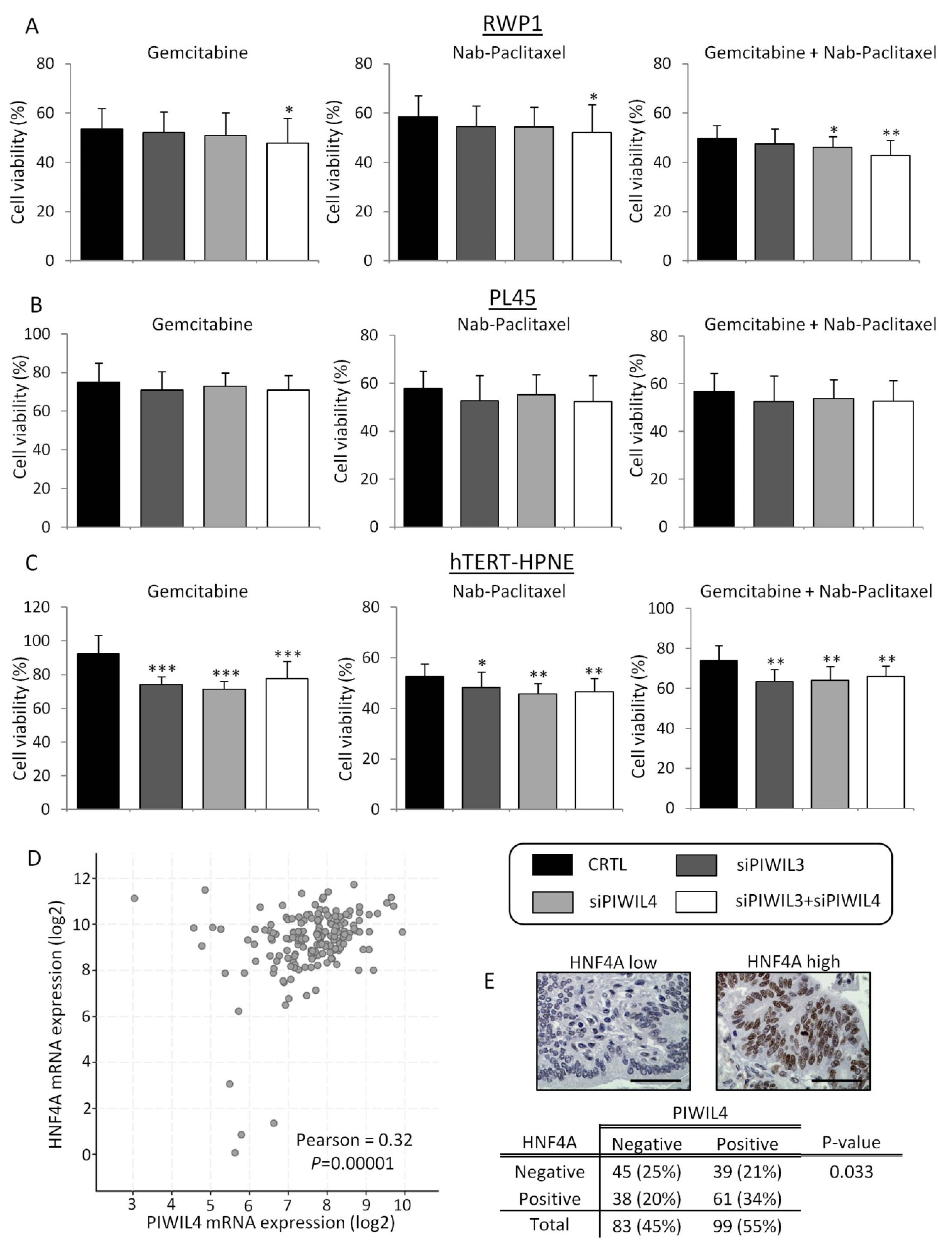
3.4. PIWIL3 and PIWIL4 Downregulation Potentiate the Cytotoxic Effect of Chemotherapies
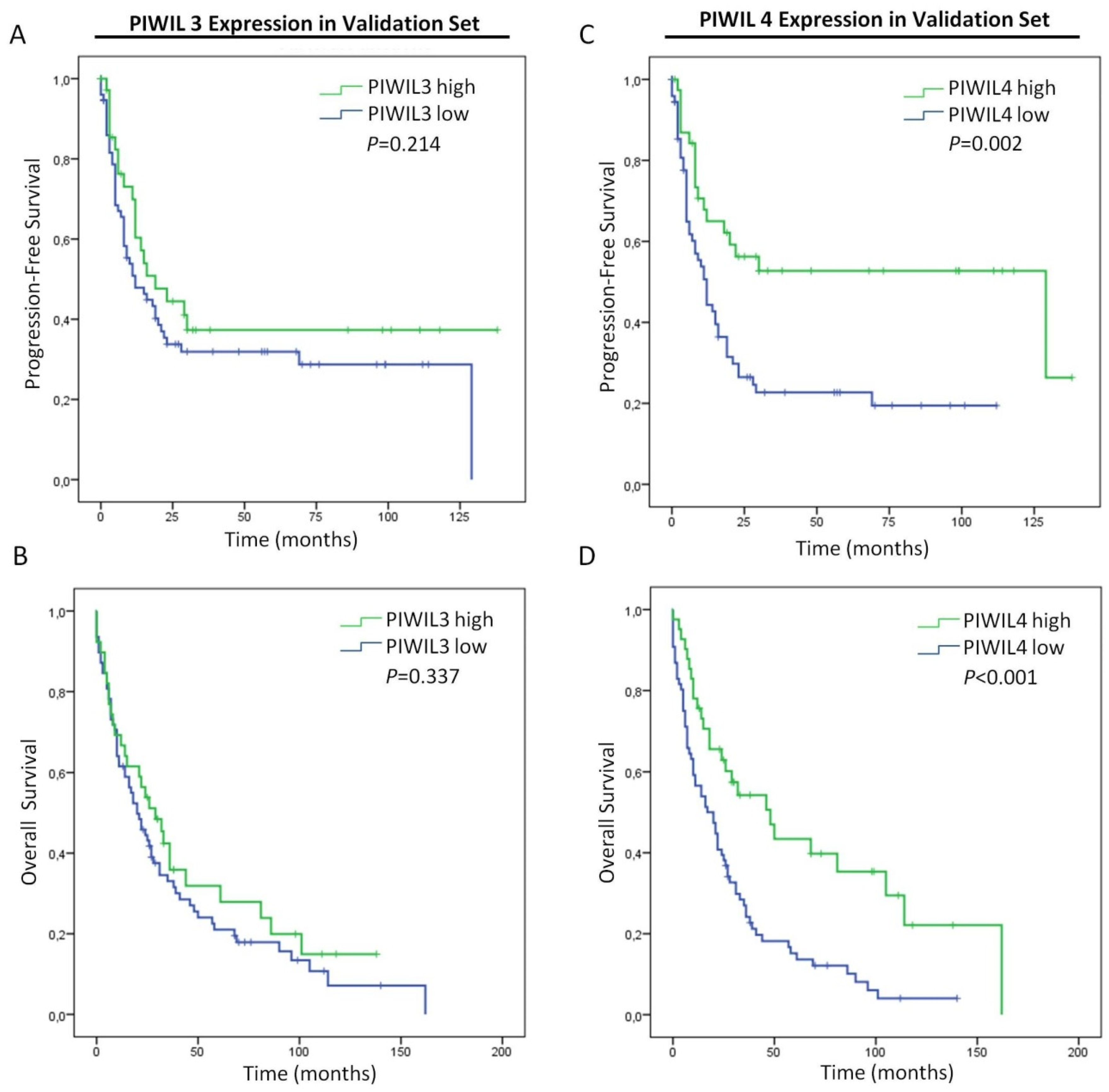
3.5. Low Expression of PIWIL4 Is a Poor Prognosis Factor of Pancreatic Cancer Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef] [PubMed]
- Tamm, E.P.; Bhosale, P.R.; Vikram, R.; de Almeida Marcal, L.P.; Balachandran, A. Imaging of pancreatic ductal adenocarcinoma: State of the art. World J. Radiol. 2013, 5, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Kelsen, D.P.; Portenoy, R.; Thaler, H.; Tao, Y.; Brennan, M. Pain as a predictor of outcome in patients with operable pancreatic carcinoma. Surgery 1997, 122, 53–59. [Google Scholar] [CrossRef]
- Oettle, H.; Post, S.; Neuhaus, P.; Gellert, K.; Langrehr, J.; Ridwelski, K.; Schramm, H.; Fahlke, J.; Zuelke, C.; Riess, H.; et al. Adjuvant chemotherapy with gemcitabine vs observation in patients undergoing curative-intent resection of pancreatic cancer: A randomized controlled trial. JAMA 2007, 297, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Neoptolemos, J.P.; Stocken, D.D.; Bassi, C.; Ghaneh, P.; Cunningham, D.; Goldstein, D.; Valle, J.W.; Palmer, D.H.; Mckay, C.J.; Doi, R.; et al. Adjuvant chemotherapy with fluorouracil plus folinic acid vs gemcitabine following pancreatic cancer resection: A randomized controlled trial. JAMA 2010, 304, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Vera, R.; Dotor, E.; Feliu, J.; González, E.; Laquente, B.; Macarulla, T.; Maurel, J. SEOM Clinical Guideline for the treatment of pancreatic cancer (2016). Clin. Trans. Oncol. 2016, 18, 1172–1178. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zeng, S.; Pöttler, M.; Lan, B.; Grützmann, R.; Pilarsky, C.; Yang, H. Chemoresistance in Pancreatic Cancer. Int. J. Mol. Sci. 2019, 20, 4504. [Google Scholar] [CrossRef]
- Vagin, V.V.; Sigova, A.; Li, C.; Seitz, H.; Gvozdev, V.; Zamore, P.D. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 2006, 313, 320–324. [Google Scholar] [CrossRef]
- Sasaki, T.; Shiohama, A.; Minoshima, S.; Shimizu, N. Identification of eight members of the Argonaute family in the human genome. Genomics 2003, 82, 323–330. [Google Scholar] [CrossRef]
- Farazi, T.A.; Juranek, S.A.; Tuschl, T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development 2008, 135, 1201–1214. [Google Scholar] [CrossRef]
- Gomes Fernandes, M.; He, N.; Wang, F.; Van Iperen, L.; Eguizabal, C.; Matorras, R.; Roelen, B.A.J. Human-specific subcellular compartmentalization of P-element induced wimpy testis-like (PIWIL) granules during germ cell development and spermatogenesis. Hum. Reprod. 2018, 33, 258–269. [Google Scholar] [CrossRef] [PubMed]
- Siomi, M.C.; Sato, K.; Pezic, D.; Aravin, A.A. PIWI-interacting small RNAs: The vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 2011, 12, 246–258. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.-N.; Li, Y.; Xia, S.-Q.; Zhang, Y.-Y.; Zheng, J.-H.; Li, W. PIWI Proteins and PIWI-Interacting RNA: Emerging Roles in Cancer. Cell Physiol. Biochem. 2017, 44, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Yang, Y.; Ji, D.; Zhang, D.; Yao, X.; Zhang, X. Hiwi downregulation, mediated by shRNA, reduces the proliferation and migration of human hepatocellular carcinoma cells. Mol. Med. Rep. 2015, 11, 1455–1461. [Google Scholar] [CrossRef] [PubMed]
- Siddiqi, S.; Terry, M.; Matushansky, I. Hiwi Mediated Tumorigenesis Is Associated with DNA Hypermethylation. PLoS ONE 2012, 7, e33711. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Sun, R.; Li, D.; Gong, F. PIWI-like protein 1 upregulation promotes gastric cancer invasion and metastasis. Onco Targets Ther. 2018, 11, 8783–8789. [Google Scholar] [CrossRef]
- Xie, K.; Zhang, K.; Kong, J.; Wang, C.; Gu, Y.; Liang, C.; Qin, N.; Liu, M.; Ma, H.; Dai, J.; et al. Cancer-testis gene PIWIL1 promotes cell proliferation, migration, and invasion in lung adenocarcinoma. Cancer Med. 2017, 7, 157–166. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, J.; Wu, G.; Yang, F. Manipulations in HIWI level exerts influence on the proliferation of human non-small cell lung cancer cells. Exp. Ther. Med. 2016, 11, 1971–1976. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, Y.; Ma, N.; Sang, B.; Hu, X.; Cong, X. Overexpression of Hiwi Inhibits the Growth and Migration of Chronic Myeloid Leukemia Cells. Cell Biochem. Biophys. 2015, 73, 117–124. [Google Scholar] [CrossRef]
- Li, W.; Martinez-Useros, J.; Garcia-Carbonero, N.; Fernandez-Aceñero, M.J.; Ortega-Medina, L.; Garcia-Botella, S. The Prognosis Value of PIWIL1 and PIWIL2 Expression in Pancreatic Cancer. J. Clin. Med. 2019, 8, 1275. [Google Scholar] [CrossRef]
- Chen, C.; Liu, J.; Xu, G. Overexpression of PIWI proteins in human stage III epithelial ovarian cancer with lymph node metastasis. Cancer Biomark. 2013, 13, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, P.; Ghosh, S.; Graham, K.; Mackey, J.R.; Kovalchuk, O.; Damaraju, S. Piwi-interacting RNAs and PIWI genes as novel prognostic markers for breast cancer. Oncotarget 2016, 7, 37944–37956. [Google Scholar] [CrossRef] [PubMed]
- Gambichler, T.; Kohsik, C.; Höh, A.-K.; Lang, K.; Käfferlein, H.U.; Brüning, T. Expression of PIWIL3 in primary and metastatic melanoma. J. Cancer Res. Clin. Oncol. 2017, 143, 433–437. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Wang, W.-J.; Li, Z.-W.; Wang, X.-Z. Downregulation of Piwil3 suppresses cell proliferation, migration and invasion in gastric cancer. Cancer Biomark. 2017, 20, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zheng, J.; Xue, Y.; Yu, H.; Gong, W.; Wang, P.; Li, Z. PIWIL3/OIP5-AS1/miR-367-3p/CEBPA feedback loop regulates the biological behavior of glioma cells. Theranostics 2018, 8, 1084–1105. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, K.; Kage, H.; Aki, N.; Sano, A.; Kitagawa, H.; Nagase, T. The induction of H3K9 methylation by PIWIL4 at the p16Ink4a locus. Biochem. Biophys. Res. Commun. 2007, 359, 497–502. [Google Scholar] [CrossRef]
- Coley, W.; Van Duyne, R.; Carpio, L.; Guendel, I.; Kehn-Hall, K.; Chevalier, S. Absence of DICER in monocytes and its regulation by HIV-1. J. Biol. Chem. 2010, 285, 31930–31943. [Google Scholar] [CrossRef]
- Henaoui, I.S.; Jacovetti, C.; Guerra Mollet, I.; Guay, C.; Sobel, J.; Eliasson, L. PIWI-interacting RNAs as novel regulators of pancreatic beta cell function. Diabetologia 2017, 60, 1977–1986. [Google Scholar] [CrossRef]
- Li, L.; Yu, C.; Gao, H.; Li, Y. Argonaute proteins: Potential biomarkers for human colon cancer. BMC Cancer 2010, 10, 38. [Google Scholar] [CrossRef]
- Su, C.; Ren, Z.-J.; Wang, F.; Liu, M.; Li, X.; Tang, H. PIWIL4 regulates cervical cancer cell line growth and is involved in down-regulating the expression of p14ARF and p53. FEBS Lett. 2012, 586, 1356–1362. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, Y.; Shen, X.; Zhang, X.; Chen, X.; Yang, C.; Chen, X.M. The PIWI protein acts as a predictive marker for human gastric cancer. Int. J. Clin. Exp. Pathol. 2012, 5, 315–325. [Google Scholar] [PubMed]
- Wang, Q.; Hao, J.; Pu, J.; Zhao, L.; Lü, Z.; Hu, J. Icariin induces apoptosis in mouse MLTC-10 Leydig tumor cells through activation of the mitochondrial pathway and down-regulation of the expression of piwil4. Int. J. Oncol. 2011, 39, 973–980. [Google Scholar] [PubMed]
- Heng, Z.S.L.; Lee, J.Y.; Subhramanyam, C.S.; Wang, C.; Thanga, L.Z.; Hu, Q. The role of 17β-estradiol-induced upregulation of Piwi-like 4 in modulating gene expression and motility in breast cancer cells. Oncol. Rep. 2018, 40, 2525–2535. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liu, N.; Shi, S.; Liu, S.; Lin, H. The Role of PIWIL4, an Argonaute Family Protein, in Breast Cancer. J. Biol. Chem. 2016, 291, 10646–10658. [Google Scholar] [CrossRef]
- Zeng, G.; Zhang, D.; Liu, X.; Kang, Q.; Fu, Y.; Tang, B.; Guo, W.; Zhang, Y.; Wei, G.; He, D.; et al. Co-expression of Piwil2/Piwil4 in nucleus indicates poor prognosis of hepatocellular carcinoma. Oncotarget 2017, 8, 4607–4617. [Google Scholar] [CrossRef][Green Version]
- Kitagawa, N.; Ojima, H.; Shirakihara, T.; Shimizu, H.; Kokubu, A.; Urushidate, T. Downregulation of the microRNA biogenesis components and its association with poor prognosis in hepatocellular carcinoma. Cancer Sci. 2013, 104, 543–551. [Google Scholar] [CrossRef]
- Greither, T.; Koser, F.; Kappler, M.; Bache, M.; Lautenschläger, C.; Göbel, S. Expression of human Piwi-like genes is associated with prognosis for soft tissue sarcoma patients. BMC Cancer 2012, 12, 272. [Google Scholar] [CrossRef]
- Navarro, A.; Tejero, R.; Viñolas, N.; Cordeiro, A.; Marrades, R.M.; Fuster, D.; Caritg, O.; Molins, L. The significance of PIWI family expression in human lung embryogenesis and non-small cell lung cancer. Oncotarget 2015, 6, 31544–31556. [Google Scholar] [CrossRef]
- Iliev, R.; Stanik, M.; Fedorko, M.; Poprach, A.; Vychytilova-Faltejskova, P.; Slaba, K. Decreased expression levels of PIWIL1, PIWIL2, and PIWIL4 are associated with worse survival in renal cell carcinoma patients. Onco Targets Ther. 2016, 9, 217–222. [Google Scholar]
- Ferreira, H.J.; Heyn, H.; Garcia del Muro, X.; Vidal, A.; Larriba, S.; Muñoz, C. Epigenetic loss of the PIWI/piRNA machinery in human testicular tumorigenesis. Epigenetics 2014, 9, 113–118. [Google Scholar] [CrossRef]
- Chen, H.-C. Boyden Chamber Assay. Methods Mol. Biol. 2005, 294, 15–22. [Google Scholar] [PubMed]
- Awasthi, N.; Zhang, C.; Schwarz, A.M.; Hinz, S.; Wang, C.; Williams, N.S.; Schwarz, M.N.; Schwarz, R.E. Comparative benefits of Nab-paclitaxel over gemcitabine or polysorbate-based docetaxel in experimental pancreatic cancer. Carcinogenesis 2013, 34, 2361–2369. [Google Scholar] [CrossRef] [PubMed]
- Spratlin, J.; Sangha, R.; Glubrecht, D.; Dabbagh, L.; Young, J.D.; Dumontet, C. The absence of human equilibrative nucleoside transporter 1 is associated with reduced survival in patients with gemcitabine-treated pancreas adenocarcinoma. Clin. Cancer Res. 2004, 10, 6956–6961. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sinha, R. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Sun, Q.; Xu, W.; Ji, S.; Qin, Y.; Liu, W.; Hu, Q.; Zhang, Z.; Liu, M.; Yu, X.; Xu, X.; et al. Role of hepatocyte nuclear factor 4 alpha in cell proliferation and gemcitabine resistance in pancreatic adenocarcinoma. Cancer Cell Int. 2019, 19, 49. [Google Scholar] [CrossRef]
- Martinez-Useros, J.; Garcia-Foncillas, J. Can Molecular Biomarkers Change the Paradigm of Pancreatic Cancer Prognosis? Biomed. Res. Int. 2016, 2016, 4873089. [Google Scholar] [CrossRef]
- Human hg38 chr12:130329199-130381325 UCSC Genome Browser v396. Available online: https://genome.ucsc.edu/cgi-bin/hgTracks?db=hg38&lastVirtModeType=default&lastVirtModeExtraState=&virtModeType=default&virtMode=0&nonVirtPosition=&position=chr12%3A130329199%2D130381325&hgsid=823730117_z8PjbiTzqWd3ZdNvqbZjD0RPpY1I (accessed on 6 March 2020).
- Human hg38 chr8:22275316-22357568 UCSC Genome Browser v395. Available online: https://genome.ucsc.edu/cgi-bin/hgTracks?db=hg38&lastVirtModeType=default&lastVirtModeExtraState=&virtModeType=default&virtMode=0&nonVirtPosition=&position=chr8%3A22275316%2D22357568&hgsid=813260055_s97A3RqtBxTmk9YMuex2A8Ay0wgb (accessed on 6 March 2020).
- Sohn, E.J.; Jo, Y.R.; Park, H.T. Downregulation MIWI-piRNA regulates the migration of Schwann cells in peripheral nerve injury. Biochem. Biophys. Res. Commun. 2019, 519, 605–612. [Google Scholar] [CrossRef]
- Heyn, H.; Ferreira, H.J.; Bassas, L.; Bonache, S.; Sayols, S.; Sandoval, J. Epigenetic Disruption of the PIWI Pathway in Human Spermatogenic Disorders. PLoS ONE 2012, 7, e47892. [Google Scholar] [CrossRef]
- Abell, N.S.; Mercado, M.; Cañeque, T.; Rodriguez, R.; Xhemalce, B. Click Quantitative Mass Spectrometry Identifies PIWIL3 as a Mechanistic Target of RNA Interference Activator Enoxacin in Cancer Cells. J. Am. Chem. Soc. 2017, 139, 1400–1403. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, X.; Song, D.; Wei, J. Piwil2 modulates the invasion and metastasis of prostate cancer by regulating the expression of matrix metalloproteinase-9 and epithelial-mesenchymal transitions. Oncol. Lett. 2015, 10, 1735–1740. [Google Scholar] [CrossRef] [PubMed]
- Ling, W.; Zhigang, H.; Tian, H.; Bin, Z.; Xiaolin, X.; Hongxiu, Z. HPV 16 infection up-regulates Piwil2, which affects cell proliferation and invasion in cervical cancer by regulating MMP-9 via the MAPK pathway. Eur. J. Gynaecol. Oncol. 2015, 36, 647–654. [Google Scholar] [PubMed]
- Tan, L.; Mai, D.; Zhang, B.; Jiang, X.; Zhang, J.; Bai, R.; Zhao, Q.; Li, X.X.; Yang, J.; Li, D.X.; et al. PIWI-interacting RNA-36712 restrains breast cancer progression and chemoresistance by interaction with SEPW1 pseudogene SEPW1P RNA. Mol. Cancer 2019, 18, 9. [Google Scholar] [CrossRef]
- Cheng, J.; Deng, H.; Xiao, B.; Zhou, H.; Zhou, F.; Shen, Z. piR-823, a novel non-coding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells. Cancer Lett. 2012, 315, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Jiang, X.; Qi, W.; Ji, C.; Xie, X.; Zhang, D. piR-823 contributes to colorectal tumorigenesis by enhancing the transcriptional activity of HSF1. Cancer Sci. 2017, 108, 1746–1756. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Luo, Y.; Gao, Y.; Yang, Y.; Wang, Y.; Xu, Y.; Tan, S.; Zhang, Y.; Duan, J.; Yang, Y. piR-651 promotes tumor formation in non-small cell lung carcinoma through the upregulation of cyclin D1 and CDK4. Int. J. Mol. Med. 2016, 38, 927–936. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Neilson, E.G. Epithelial-mesenchymal transition and its implications for fibrosis. J. Clin. Investig. 2003, 112, 1776–1784. [Google Scholar] [CrossRef] [PubMed]
- López-Novoa, J.M.; Nieto, M.A. Inflammation and EMT: An alliance towards organ fibrosis and cancer progression. EMBO Mol. Med. 2009, 1, 303–314. [Google Scholar] [CrossRef]
- Joglekar, M.V.; Hardikar, A. Epithelial-to-mesenchymal transition in pancreatic islet β cells. Cell Cycle 2010, 9, 4077–4079. [Google Scholar] [CrossRef][Green Version]
- Klattenhoff, C.; Theurkauf, W. Biogenesis and germline functions of piRNAs. Development 2008, 135, 3–9. [Google Scholar] [CrossRef]
- Rojas-Ríos, P.; Simonelig, M. piRNAs and PIWI proteins: Regulators of gene expression in development and stem cells. Development 2018, 145, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Jung, C.; Javadian-Elyaderani, P.; Schweyer, S.; Schütte, D.; Shoukier, M.; Navernia, K. Pathways of Proliferation and Antiapoptosis Driven in Breast Cancer Stem Cells by Stem Cell Protein Piwil2. Cancer Res. 2010, 70, 4569–4579. [Google Scholar] [CrossRef] [PubMed]
- Rinkevich, Y.; Voskoboynik, A.; Rosner, A.; Rabinowitz, C.; Paz, G.; Oren, M. Repeated, long-term cycling of putative stem cells between niches in a basal chordate. Dev. Cell 2013, 24, 76–88. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Gao, Q.; Chen, K.; Xue, X.; Li, M.; Chen, Q.; Zhu, G.; Gao, Y. Hiwi facilitates chemoresistance as a cancer stem cell marker in cervical cancer. Oncol. Rep. 2014, 32, 1853–1860. [Google Scholar] [CrossRef]
- Wang, Y.; Gable, T.; Ma, M.Z.; Clark, D.; Zhao, J.; Zhang, Y. A piRNA-like Small RNA Induces Chemoresistance to Cisplatin-Based Therapy by Inhibiting Apoptosis in Lung Squamous Cell Carcinoma. Mol. Ther. Nucleic Acids 2017, 6, 269–278. [Google Scholar] [CrossRef]
- Wang, H.; Mao, C.; Li, N.; Sun, L.; Zheng, Y.; Xu, N. A case report of a dramatic response to olaparib in a patient with metastatic pancreatic cancer harboring a germline BRCA2 mutation. Medicine 2019, 98, e17443. [Google Scholar] [CrossRef]
- Wang, H.; Word, B.R.; Lyn-Cook, B.D. Enhanced Efficacy of Gemcitabine by Indole-3-carbinol in Pancreatic Cell Lines: The Role of Human Equilibrative Nucleoside Transporter 1. Anticancer. Res. 2011, 31, 3171–3180. [Google Scholar]
- Stoffel, M.; Duncan, S.A. The maturity-onset diabetes of the young (MODY1) transcription factor HNF4alpha regulates expression of genes required for glucose transport and metabolism. Proc. Natl. Acad. Sci. USA 1997, 94, 13209–13214. [Google Scholar] [CrossRef]
- Hamdan, F.H.; Zihlif, M.A. Gene expression alterations in chronic hypoxic MCF7 breast cancer cell line. Genomics 2014, 104, 477–481. [Google Scholar] [CrossRef]
- Kiselyuk, A.; Lee, S.-H.; Farber-Katz, S.; Zhang, M.; Athavankar, S.; Cohen, T. HNF4α antagonists discovered by a high-throughput screen for modulators of the human insulin promoter. Chem. Biol. 2012, 19, 806–818. [Google Scholar] [CrossRef]
- Tissue Expression of PIWIL3—Staining in Pancreas—The Human Protein Atlas version 19.3. Available online: https://www.proteinatlas.org/ENSG00000184571-PIWIL3/tissue/pancreas (accessed on 6 March 2020).
- Tissue Expression of PIWIL4—Staining in Pancreas—The Human Protein Atlas version 19.3. Available online: https://www.proteinatlas.org/ENSG00000134627-PIWIL4/tissue/pancreas (accessed on 6 March 2020).
- Egawa, S.; Takeda, K.; Fukuyama, S.; Motoi, F.; Sunamura, M.; Matsuno, S. Clinicopathological Aspects of Small Pancreatic Cancer. Pancreas 2004, 28, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Ariyama, J.; Suyama, M.; Satoh, K.; Sai, J. Imaging of Small Pancreatic Ductal Adenocarcinoma. Pancreas 1998, 16, 396–401. [Google Scholar] [CrossRef] [PubMed]

| Clinical Characteristics | N | % | Clinical Characteristics | N | % |
|---|---|---|---|---|---|
| Age | Neural invasion | ||||
| <65 years | 16 | 43 | No | 12 | 32 |
| >65 years | 21 | 57 | Yes | 25 | 68 |
| Gender | Lymph nodes involved | ||||
| Male | 21 | 57 | N0 | 14 | 38 |
| Female | 16 | 43 | N1 | 23 | 62 |
| Size | Adjuvant treatment | ||||
| <2 cm | 20 | 54 | No | 21 | 57 |
| >2 cm | 17 | 46 | Yes | 14 | 38 |
| Stage | N/A | 2 | 5 | ||
| I | 9 | 24 | pT | ||
| II | 28 | 76 | T1 | 6 | 16 |
| Grade | T2 | 5 | 14 | ||
| High | 30 | 81 | T3 | 26 | 70 |
| Low | 7 | 19 | N/A | 3 | 2 |
| Vascular invasion | |||||
| No | 12 | 32 | Total | 37 | 100 |
| Yes | 25 | 68 |
| Clinical Characteristics | N | % | Clinical Characteristics | N | % |
|---|---|---|---|---|---|
| Age | Grade | ||||
| <65 years | 25 | 20 | High | 19 | 15 |
| >65 years | 103 | 80 | Low | 105 | 82 |
| Gender | N/A | 4 | 3 | ||
| Male | 63 | 49 | Vascular invasion | ||
| Female | 65 | 51 | No | 75 | 59 |
| Diabetes Mellitus | Yes | 43 | 33 | ||
| No | 88 | 69 | N/A | 10 | 8 |
| Yes | 33 | 26 | Neural invasion | ||
| N/A | 7 | 5 | No | 47 | 37 |
| Adjuvant treatment | Yes | 71 | 55 | ||
| No | 75 | 58 | N/A | 10 | 8 |
| Yes | 24 | 19 | pT | ||
| N/A | 29 | 23 | T1 | 30 | 23 |
| Size | T2 | 44 | 35 | ||
| <2 cm | 31 | 24 | T3 | 51 | 40 |
| >2 cm | 69 | 54 | N/A | 3 | 2 |
| N/A | 28 | 22 | Lymph nodes involved | ||
| Stage | N0 | 70 | 55 | ||
| I | 46 | 36 | N1 | 51 | 40 |
| II | 74 | 58 | N/A | 7 | 5 |
| N/A | 8 | 6 | Total | 128 | 100 |
| Univariate PFS (95% CI) | Univariate OS (95% CI) | |||||||
|---|---|---|---|---|---|---|---|---|
| HR | Lower | Upper | p | HR | Lower | Upper | p | |
| Age (< 65 years vs. > 65 years) | 1.060 | 0.604 | 1.860 | 0.840 | 1.198 | 0.723 | 1.986 | 0.484 |
| Gender (Male vs. Female) | 1.494 | 0.920 | 2.425 | 0.104 | 1.182 | 0.785 | 1.778 | 0.423 |
| Diabetes Mellitus (No vs. Yes) | 1.070 | 0.614 | 1.864 | 0.811 | 1.113 | 0.694 | 1.784 | 0.658 |
| Adjuvant treatment (Yes vs. No) | 1.016 | 0.556 | 1.857 | 0.959 | 1.226 | 0.781 | 2.094 | 0.456 |
| Size (<2 cm vs. >2 cm) | 3.023 | 1.413 | 6.465 | 0.004 | 1.255 | 0.754 | 2.087 | 0.382 |
| pT (I / II vs. III) | 1.682 | 1.033 | 2.738 | 0.037 | 1.679 | 1.110 | 2.540 | 0.014 |
| Stage (I vs. II) | 1.866 | 1.105 | 3.151 | 0.020 | 1.795 | 1.148 | 2.807 | 0.010 |
| Grade (low vs. high) | 1.406 | 0.695 | 2.845 | 0.343 | 1.221 | 0.664 | 2.245 | 0.522 |
| Lymph nodes involved (No vs. Yes) | 1.548 | 0.943 | 2.540 | 0.084 | 1.573 | 1.025 | 2.414 | 0.038 |
| Vascular invasion (No vs. Yes) | 1.348 | 0.807 | 2.252 | 0.254 | 1.481 | 0.959 | 2.287 | 0.077 |
| Neural invasion (No vs. Yes) | 1.757 | 1.027 | 3.007 | 0.040 | 1.658 | 1.060 | 2.593 | 0.027 |
| PIWIL3 (high vs. low) | 1.380 | 0.819 | 2.327 | 0.227 | 1.237 | 0.798 | 1.917 | 0.342 |
| PIWIL4 (high vs. low) | 1.979 | 1.178 | 3.325 | 0.010 | 2.093 | 1.344 | 3.260 | 0.001 |
| Multivariate PFS (95% CI) | Multivariate OS (95% CI) | |||||||
| Size (<2 cm vs. > 2 cm) | 3.095 | 1.237 | 7.744 | 0.016 | ||||
| pT (I / II vs. III) | 1.339 | 0.609 | 2.944 | 0.467 | 1.178 | 0.608 | 2.284 | 0.627 |
| Stage (I vs. II) | 1.596 | 0.655 | 3.890 | 0.304 | 1.691 | 0.683 | 4.188 | 0.256 |
| Lymph nodes involved (No vs. Yes) | 1.084 | 0.549 | 2.141 | 0.817 | ||||
| Neural invasion | 1.232 | 0.620 | 2.449 | 0.551 | 1.229 | 0.761 | 1.985 | 0.398 |
| PIWIL4 (high vs. low) | 2.036 | 1.025 | 4.044 | 0.042 | 2.185 | 1.313 | 3.636 | 0.003 |
| PIWIL3 Low | PIWIL3 High | PIWIL4 Low | PIWIL4 High | |||
|---|---|---|---|---|---|---|
| Parameters | N (%) | N (%) | p-Value | N (%) | N (%) | p-Value |
| Gender | 0.946 | 0.050 | ||||
| Male | 43 (34%) | 20 (16%) | 34 (26%) | 29 (23%) | ||
| Female | 44 (34%) | 21 (16%) | 46 (36%) | 19 (15%) | ||
| Age | 0.630 | 0.227 | ||||
| <65 years | 18 (14%) | 7 (5%) | 13 (10%) | 12 (9%) | ||
| >65 years | 69 (54%) | 34 (27%) | 67 (53%) | 36 (28%) | ||
| Diabetes Mellitus | 0.724 | 0.939 | ||||
| No | 59 (49%) | 29 (24%) | 54 (45%) | 34 (28%) | ||
| Yes | 21 (17%) | 12 (10%) | 20 (16%) | 13 (11%) | ||
| Stage | 0.791 | 0.204 | ||||
| I | 30 (25%) | 16 (13%) | 26 (22%) | 20 (16%) | ||
| II | 50 (42%) | 24 (20%) | 49 (41%) | 25 (21%) | ||
| pT | 0.503 | 0.020 | ||||
| I/II | 48 (38%) | 26 (21%) | 40 (32%) | 34 (27%) | ||
| III | 36 (29%) | 15 (12%) | 38 (31%) | 13 (10%) | ||
| Adjuvant treatment | 0.704 | 0.085 | ||||
| No | 50 (51%) | 25 (25%) | 52 (53%) | 23 (23%) | ||
| Yes | 17 (17%) | 7 (7%) | 12 (12%) | 12 (12%) | ||
| Size | 0.264 | 0.705 | ||||
| <2 cm | 19 (19%) | 12 (12%) | 19 (19%) | 12 (12%) | ||
| >2 cm | 50 (50%) | 19 (19%) | 45 (45%) | 24 (24%) | ||
| Lymph nodes involved | 0.956 | 0.713 | ||||
| No | 47 (39%) | 23 (19%) | 43 (36%) | 27 (22%) | ||
| Yes | 34 (28%) | 17 (14%) | 33 (27%) | 18 (15%) | ||
| Vascular Invasion | 0.950 | 0.875 | ||||
| No | 51 (43%) | 24 (20%) | 46 (34%) | 29 (30%) | ||
| Yes | 29 (25%) | 14 (12%) | 27 (25%) | 16 (11%) | ||
| Neural Invasion | 0.050 | 0.019 | ||||
| No | 27 (23%) | 20 (17%) | 23 (20%) | 24 (20%) | ||
| Yes | 53 (45%) | 18 (15%) | 50 (42%) | 21 (18%) | ||
| Grade | 0.095 | 0.917 | ||||
| Low | 68 (55%) | 37 (30%) | 65 (52%) | 40 (32%) | ||
| High | 16 (13%) | 3 (2%) | 12 (10%) | 7 (6%) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Martinez-Useros, J.; Garcia-Carbonero, N.; Fernandez-Aceñero, M.J.; Orta, A.; Ortega-Medina, L.; Garcia-Botella, S.; Perez-Aguirre, E.; Diez-Valladares, L.; Celdran, A.; et al. The Clinical Significance of PIWIL3 and PIWIL4 Expression in Pancreatic Cancer. J. Clin. Med. 2020, 9, 1252. https://doi.org/10.3390/jcm9051252
Li W, Martinez-Useros J, Garcia-Carbonero N, Fernandez-Aceñero MJ, Orta A, Ortega-Medina L, Garcia-Botella S, Perez-Aguirre E, Diez-Valladares L, Celdran A, et al. The Clinical Significance of PIWIL3 and PIWIL4 Expression in Pancreatic Cancer. Journal of Clinical Medicine. 2020; 9(5):1252. https://doi.org/10.3390/jcm9051252
Chicago/Turabian StyleLi, Weiyao, Javier Martinez-Useros, Nuria Garcia-Carbonero, Maria J. Fernandez-Aceñero, Alberto Orta, Luis Ortega-Medina, Sandra Garcia-Botella, Elia Perez-Aguirre, Luis Diez-Valladares, Angel Celdran, and et al. 2020. "The Clinical Significance of PIWIL3 and PIWIL4 Expression in Pancreatic Cancer" Journal of Clinical Medicine 9, no. 5: 1252. https://doi.org/10.3390/jcm9051252
APA StyleLi, W., Martinez-Useros, J., Garcia-Carbonero, N., Fernandez-Aceñero, M. J., Orta, A., Ortega-Medina, L., Garcia-Botella, S., Perez-Aguirre, E., Diez-Valladares, L., Celdran, A., & García-Foncillas, J. (2020). The Clinical Significance of PIWIL3 and PIWIL4 Expression in Pancreatic Cancer. Journal of Clinical Medicine, 9(5), 1252. https://doi.org/10.3390/jcm9051252







