Artificial Intelligence-Based Diagnosis of Cardiac and Related Diseases
Abstract
1. Introduction
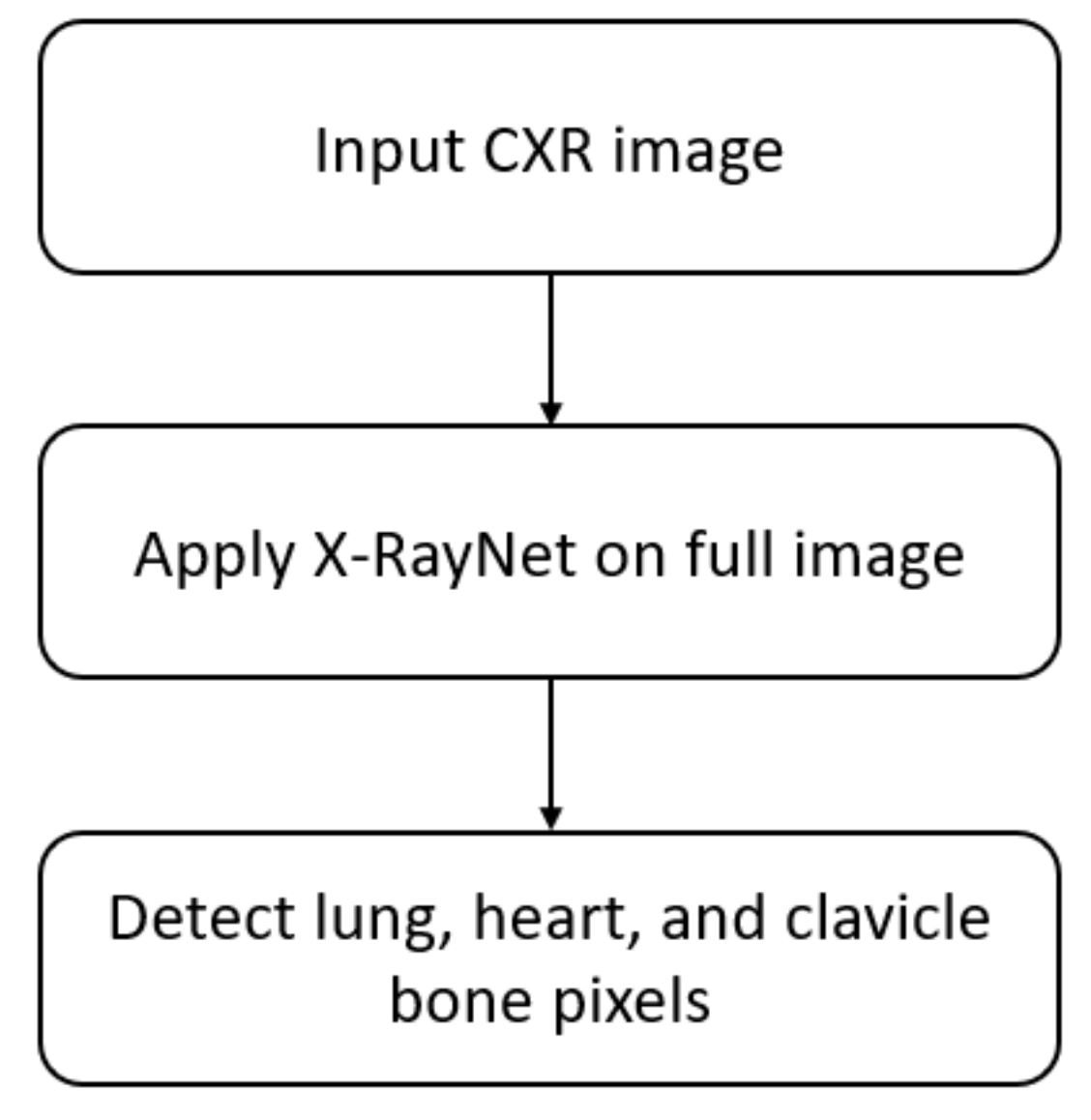
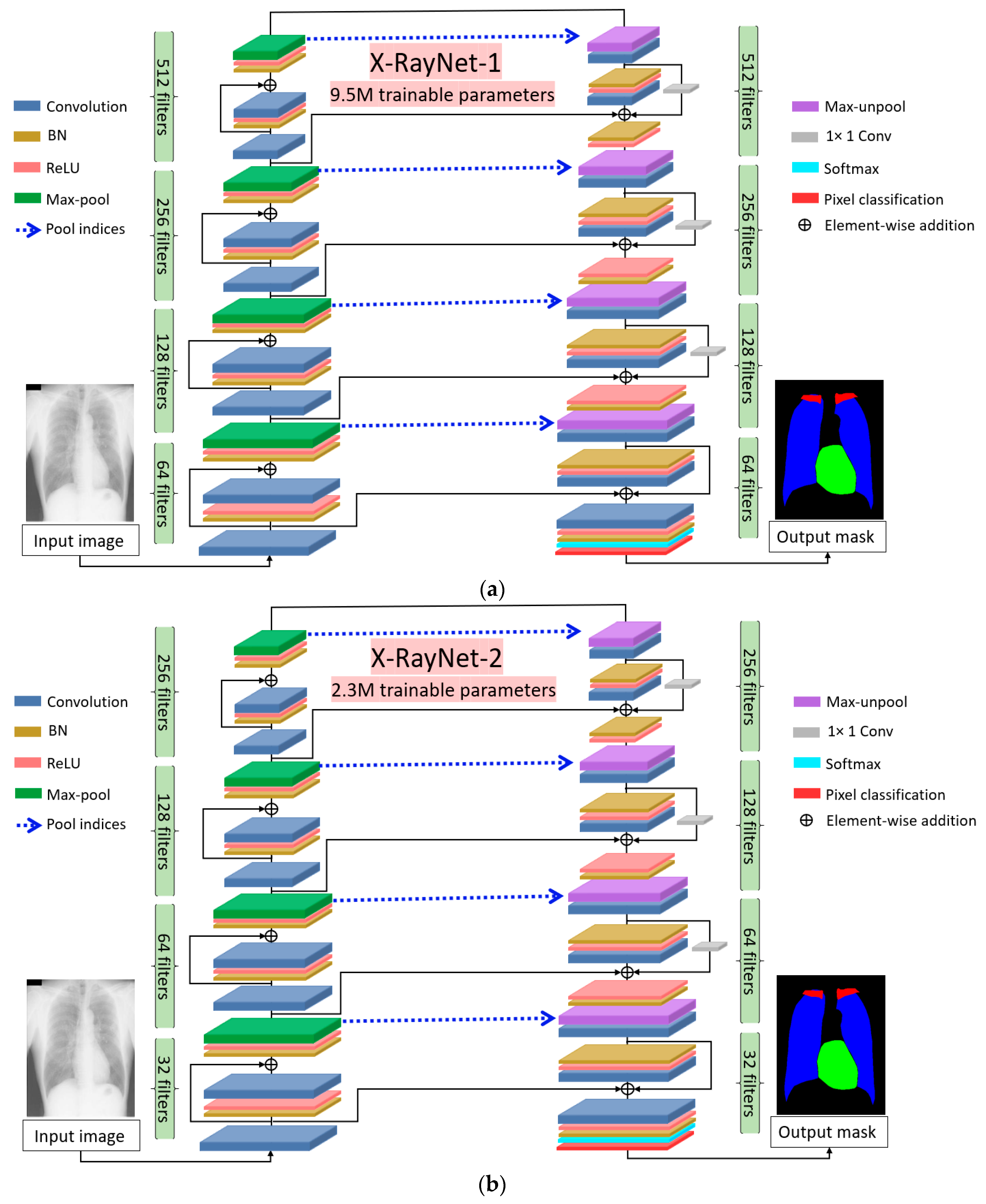
- X-RayNet does not require preprocessing for multiclass semantic segmentation to detect the lungs, heart, and clavicle bones at the same time. X-RayNet considers the importance of computational cost; therefore, X-RayNet-2 reduces the trainable parameters by 75% with a competitive performance.
- This study presents two separate identical semantic segmentation networks with a simple fully convolutional architecture.
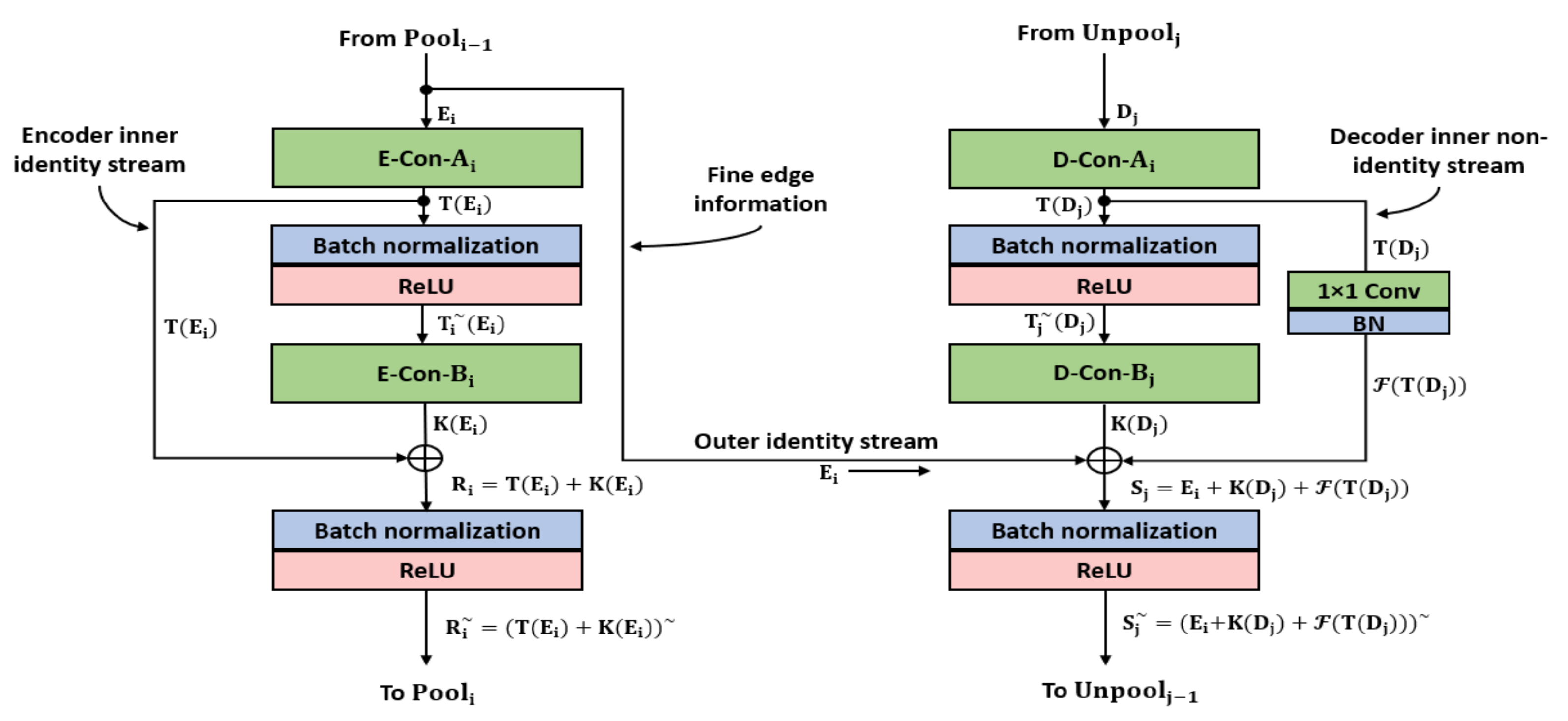
- X-RayNet utilizes a mesh of internal and external residual paths that transfers the enriched features from the preceding layers and at the end of the network. X-RayNet uses identity and nonidentity mappings for faster edge information transfer to ensure the residual mesh connects all the convolutional layers, including the first convolutional layer.
- For a fair comparison with other research results, the trained X-RayNet models and algorithms are made publicly available in [50].
2. Materials and Methods
2.1. Overview of Proposed Architecture
2.2. Chest Anatomy Segmentation Using X-RayNet
2.2.1. X-RayNet Encoder
2.2.2. X-RayNet Decoder
3. Results
3.1. Experimental Data and Environment
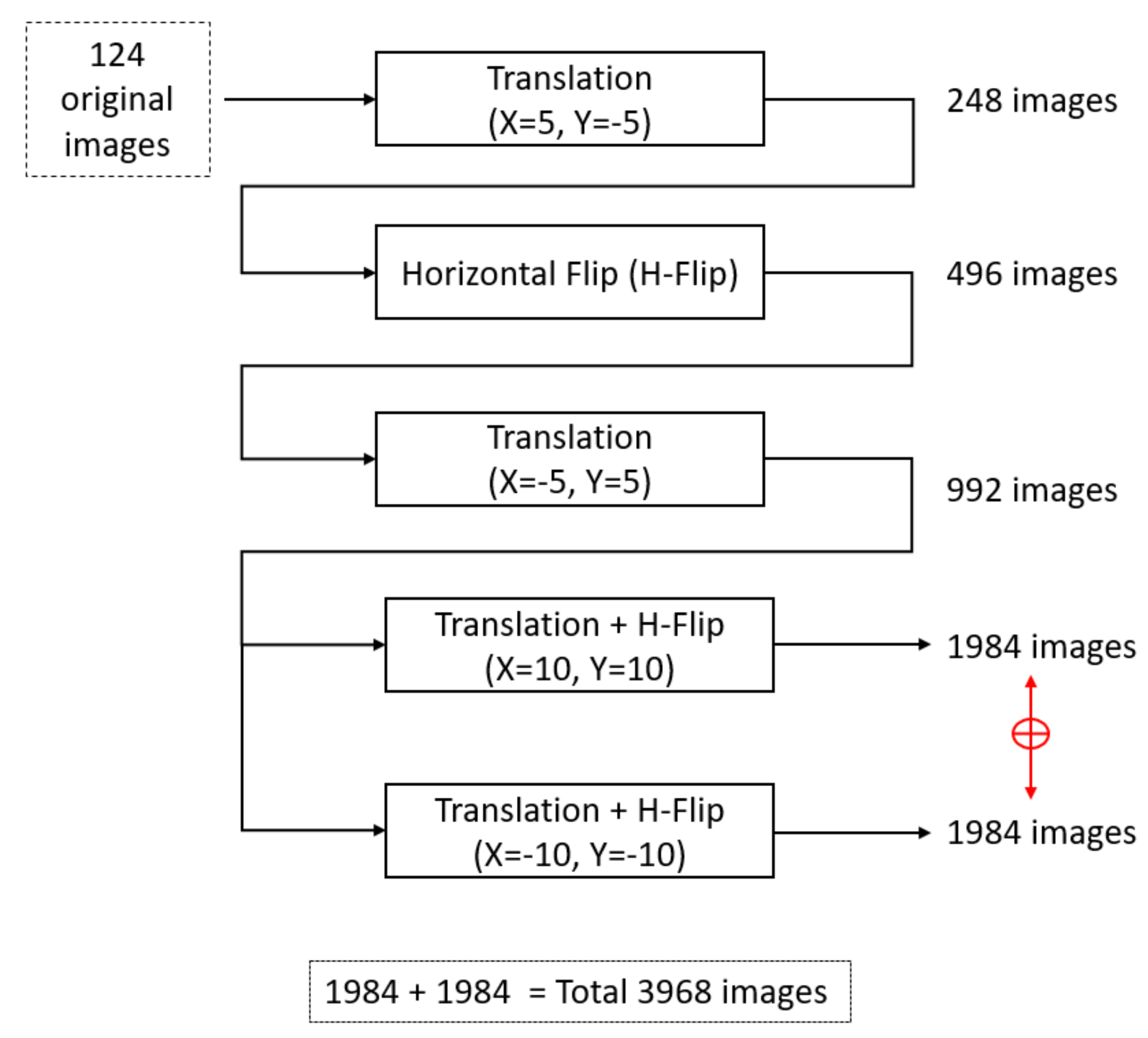
3.2. Data Augmentation
3.3. X-RayNet Training
3.4. Testing of the Proposed Method
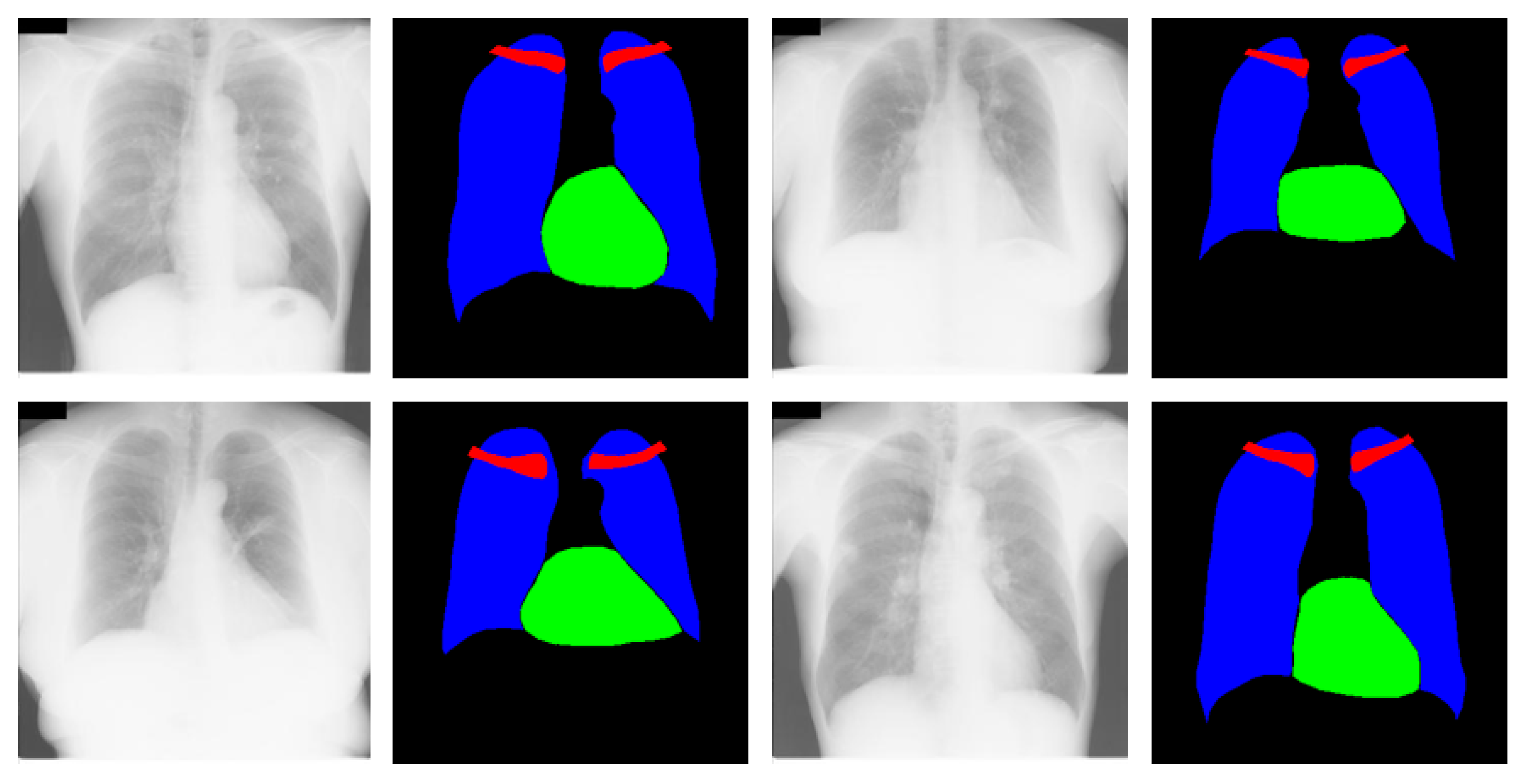
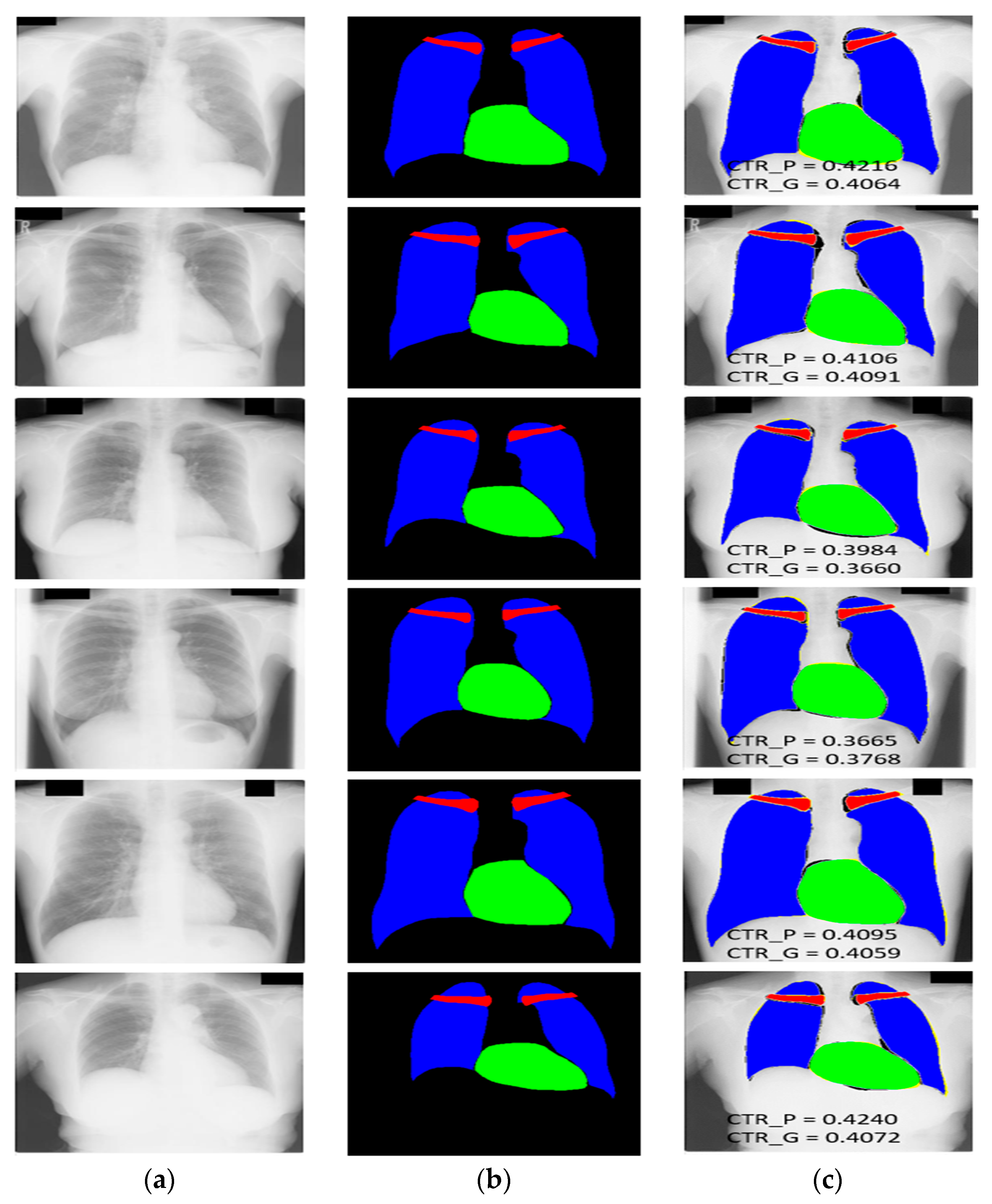
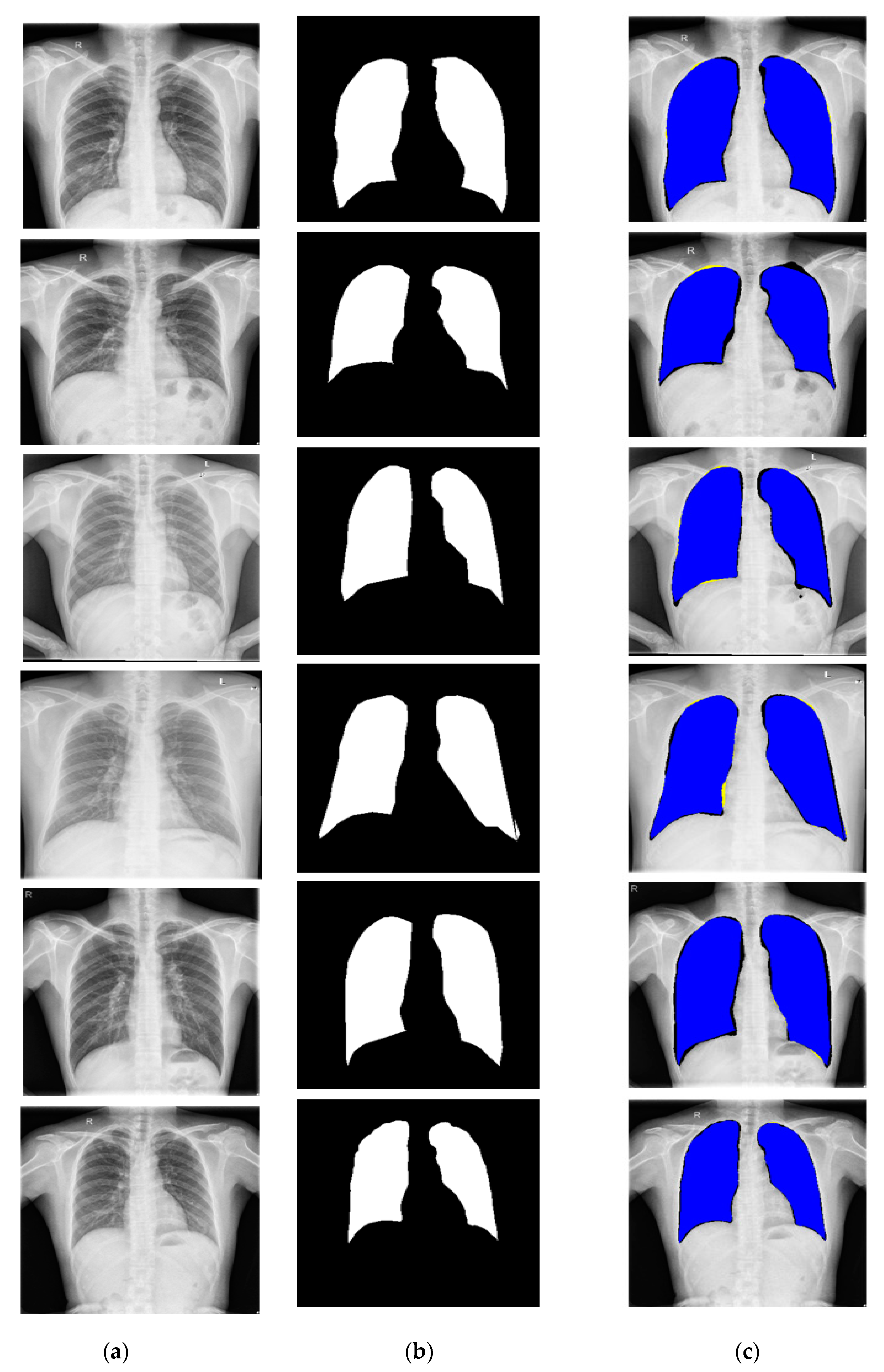
3.4.1. X-RayNet Testing for Chest Anatomy Segmentation
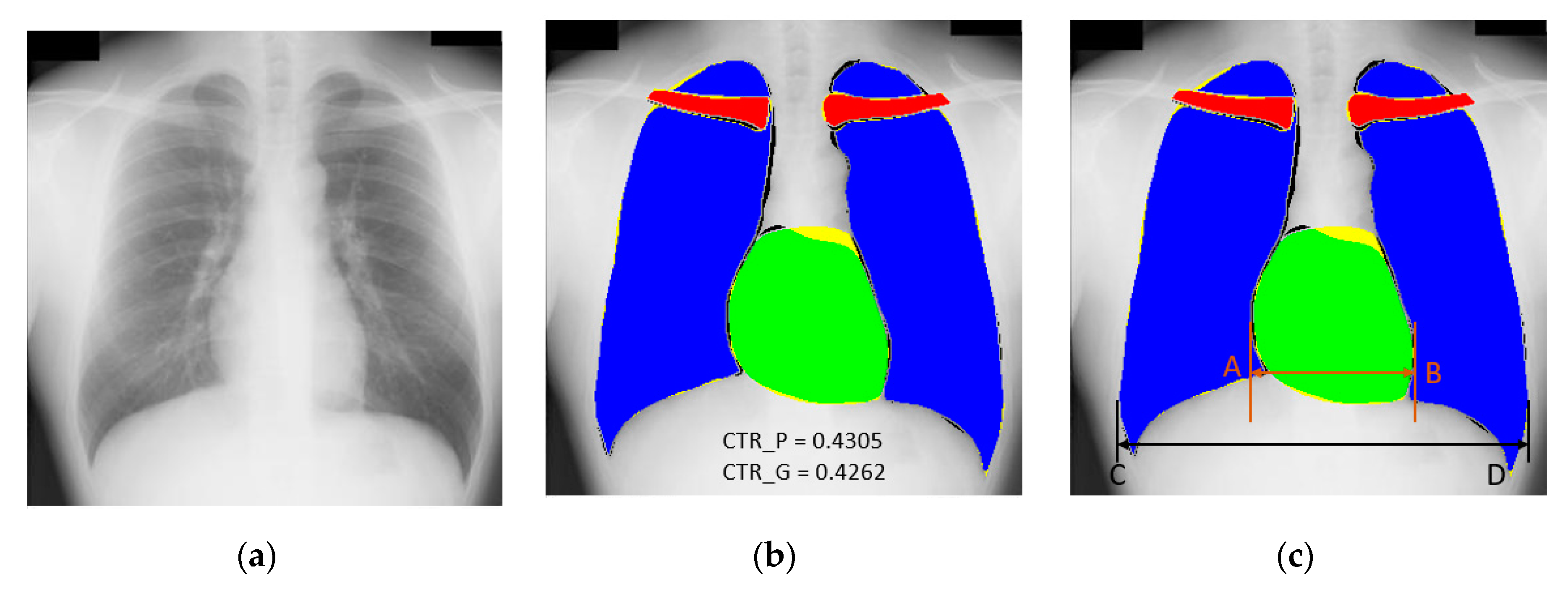
3.4.2. Chest Organ Segmentation Results by X-RayNet
3.4.3. Comparison of X-RayNet with Other Methods
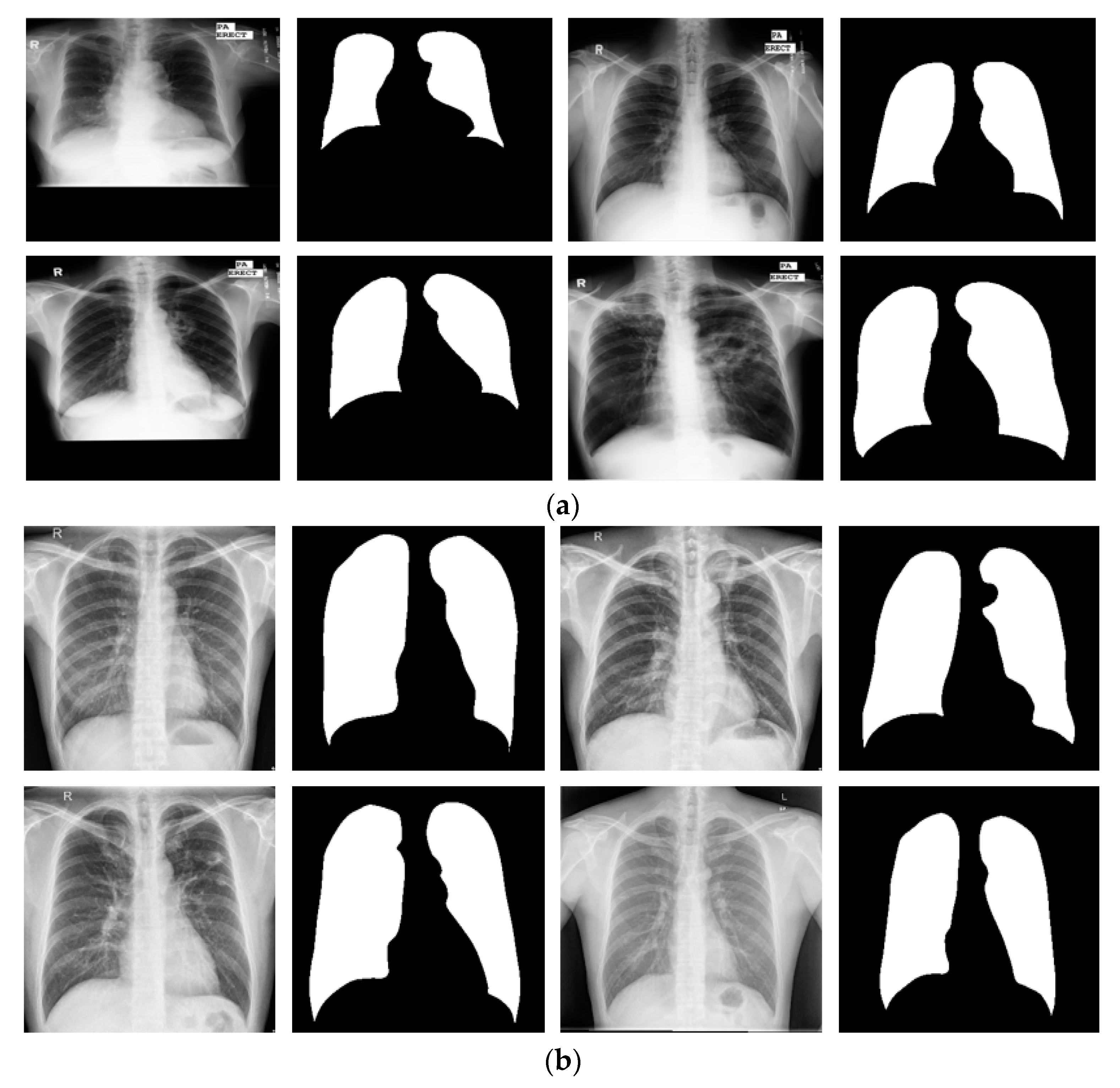
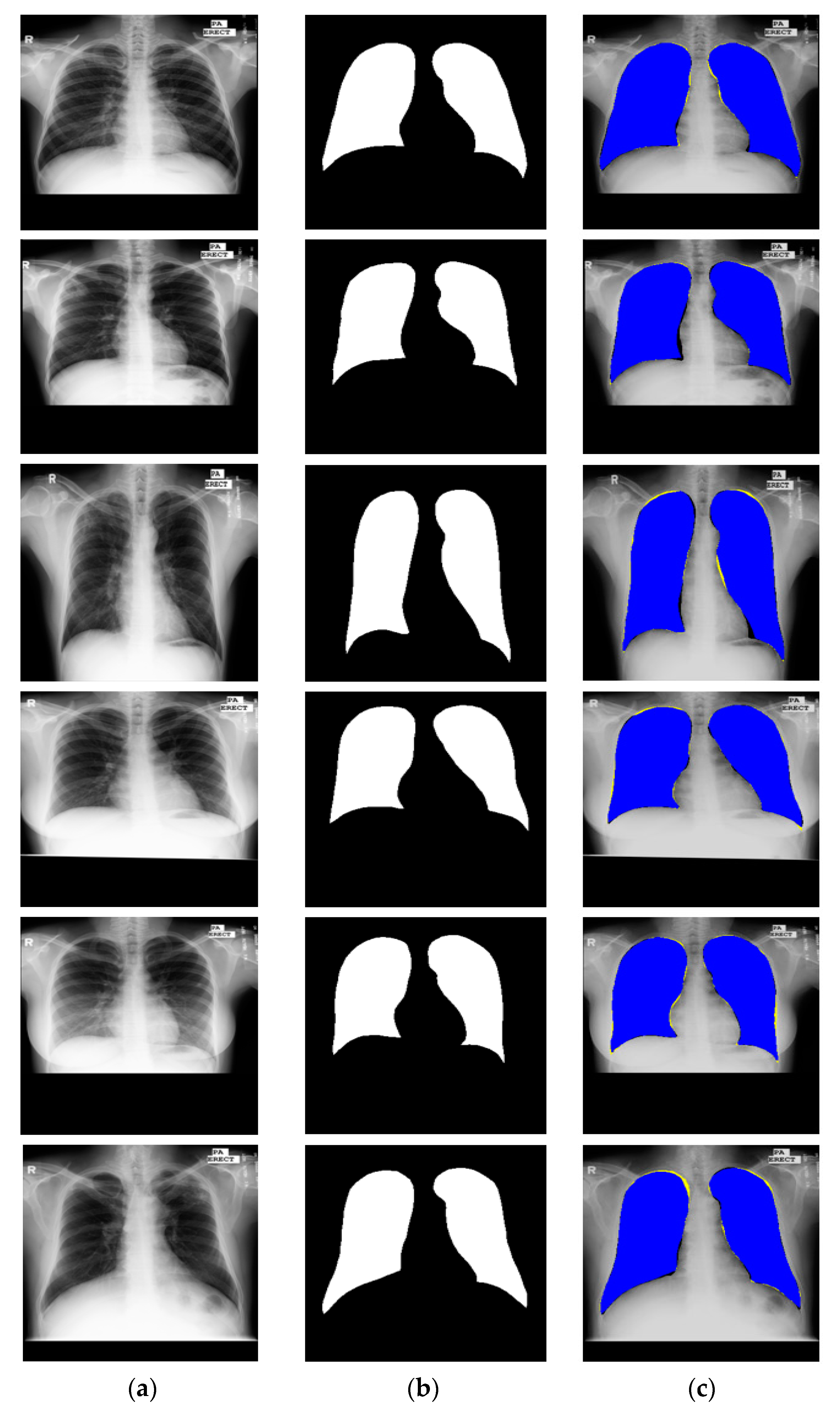
3.4.4. Lung Segmentation with Other Open Datasets Using X-RayNet
4. Discussion
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
Appendix A
| Block | Name/Size | Number of Filters | Output Feature Map Size (Width × Height × Number of Channels) | Number of Trainable Parameters (EC + BN) |
|---|---|---|---|---|
| EB-1 | EC-1_1 **/3 × 3 × 3 To decoder (OIS-1) and E-Add-1 | 64 | 350 × 350 × 64 | 1792 + 128 |
| EC-1_2 /3 × 3 × 64 | 64 | 36,928 | ||
| E-Add-1 (EC-1_1 + EC-1_2) using IIS | - | |||
| BN + ReLU | 128 | |||
| Pool-1 | Pool-1/2 × 2 To decoder (OIS-2) | - | 175 × 175 × 64 | - |
| EB-2 | EC-2_1 **/3 × 3 × 64 To E-Add-2 | 128 | 175 × 175 × 128 | 73,856 + 256 |
| EC-2_2 */3 × 3 × 128 | 128 | 147,584 | ||
| E-Add-2 (EC-2_1 + EC-2_2) using IIS | - | - | ||
| BN + ReLU | 256 | |||
| Pool-2 | * Pool-2/2 × 2 To decoder (OIS-3) | - | 87 × 87 × 128 | - |
| EB-3 | EC-3_1 **/3 × 3 × 128 To E-Add-3 | 256 | 87 × 87 × 256 | 295,168 + 512 |
| EC-3_2 /3 × 3 × 256 | 256 | 590,080 + 512 | ||
| E-Add-3 (EC-3_1 + EC-3_2) using IIS | - | - | ||
| BN + ReLU | ||||
| Pool-3 | * Pool-3/2 × 2 To decoder (OIS-4) | - | 43 × 43 × 256 | - |
| EB-4 | EC-4_1 **/3 × 3 × 256 To E-Add-4 | 512 | 43 × 43 × 512 | 1,180,160 + 1024 |
| EC-4_2 */3 × 3 × 512 | 512 | 2,359,808 | ||
| E-Add-4 (EC-4_1 + EC-4_2) using IIS | - | - | ||
| BN + ReLU | 1024 | |||
| Pool-4 | * Pool-4/2 × 2 | - | 21× 21 × 512 | - |
| Block | Name/Size | Number of Filters | Output Feature Map Size (Width × Height × Number of Channels) | Number of Trainable Parameters (DCon + BN) |
|---|---|---|---|---|
| Unpool-4 | Unpool-4 | - | 43 × 43 × 512 | - |
| DB-4 | DCon-4_2 **/3 × 3 × 512 | 512 | 2,359,808 + 1024 | |
| INIS-4 */1 × 1 × 512 | 256 | 43 × 43 × 256 | 131,328 + 512 | |
| DCon-4_1 */3 × 3 × 512 | 256 | 1,179,904 | ||
| Add-5 (DCon-4_2 + INIS-4 * + Pool-3^) | - | - | ||
| BN + ReLU | 512 | |||
| Unpool-3 | * Unpool-3 | - | 87 × 87 × 256 | - |
| DB-3 | DCon-3_2 **/3 × 3 × 256 | 256 | 590,080 + 512 | |
| INIS -3 */1 × 1 × 256 | 128 | 87 × 87 × 128 | 32,896 + 256 | |
| DCon-3_1 **/3 × 3 × 256 | 128 | 295,040 | ||
| Add-6 (DCon-3_2 + INIS-3 * + Pool-2^) | - | - | ||
| BN + ReLU | 256 | |||
| Unpool-2 | * Unpool-2 | - | 175 × 175 × 128 | - |
| DB-2 | DCon-2_2 **/3 × 3 × 128 | 128 | 147,584 + 256 | |
| INIS -2 */1 × 1 × 128 | 64 | 175 × 175 × 64 | 8256 + 128 | |
| DCon-2_1 **/3 × 3 × 128 | 64 | 73,792 | ||
| Add-7 (DCon-2_2 + INIS-2 * + Pool-1^) | - | - | ||
| BN + ReLU | 128 | |||
| Unpool-1 | * Unpool-1 | - | 350 × 350 × 64 | - |
| DB-1 | DConv-1_2 **/3 × 3 × 64 | 64 | 36,928 + 128 | |
| DConv-1_1 /3 × 3 × 64 | 2 | 36,928 | ||
| Add-8 (DCon-1_1 + DConv-1_2 + EC-1_1^) | - | |||
| MConv **/3 × 3 × 64 | 4 | 350 × 350 × 4 | 2308 | |
| BN + ReLU | 8 |
References
- Novikov, A.A.; Lenis, D.; Major, D.; Hladůvka, J.; Wimmer, M.; Bühler, K. Fully convolutional architectures for multiclass segmentation in chest radiographs. IEEE Trans. Med. Imaging 2018, 37, 1865–1876. [Google Scholar] [CrossRef] [PubMed]
- Peng, T.; Wang, Y.; Xu, T.C.; Chen, X. Segmentation of lung in chest radiographs using hull and closed polygonal line method. IEEE Access 2019, 7, 137794–137810. [Google Scholar] [CrossRef]
- Candemir, S.; Antani, S. A review on lung boundary detection in chest X-rays. Int. J. Comput. Assist. Radiol. Surg. 2019, 14, 563–576. [Google Scholar] [CrossRef]
- Coppini, G.; Miniati, M.; Monti, S.; Paterni, M.; Favilla, R.; Ferdeghini, E.M. A computer-aided diagnosis approach for emphysema recognition in chest radiography. Med. Eng. Phys. 2013, 35, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Miniati, M.; Coppini, G.; Monti, S.; Bottai, M.; Paterni, M.; Ferdeghini, E.M. Computer-aided recognition of emphysema on digital chest radiography. Eur. J. Radiol. 2011, 80, 169–175. [Google Scholar] [CrossRef]
- Coppini, G.; Miniati, M.; Paterni, M.; Monti, S.; Ferdeghini, E.M. Computer-aided diagnosis of emphysema in COPD patients: Neural-network-based analysis of lung shape in digital chest radiographs. Med. Eng. Phys. 2007, 29, 76–86. [Google Scholar] [CrossRef]
- Tavora, F.; Zhang, Y.; Zhang, M.; Li, L.; Ripple, M.; Fowler, D.; Burke, A. Cardiomegaly is a common arrhythmogenic substrate in adult sudden cardiac deaths, and is associated with obesity. Pathology (Phila.) 2012, 44, 187–191. [Google Scholar] [CrossRef]
- Candemir, S.; Jaeger, S.; Lin, W.; Xue, Z.; Antani, S.; Thoma, G. Automatic heart localization and radiographic index computation in chest x-rays. In Proceedings of the Medical Imaging 2016: Computer-Aided Diagnosis, San Diego, CA, USA, 28 February–2 March 2016; p. 978517. [Google Scholar]
- Hasan, M.A.; Lee, S.-L.; Kim, D.-H.; Lim, M.-K. Automatic evaluation of cardiac hypertrophy using cardiothoracic area ratio in chest radiograph images. Comput. Methods Programs Biomed. 2012, 105, 95–108. [Google Scholar] [CrossRef]
- Browne, R.F.J.; O’Reilly, G.; McInerney, D. Extraction of the two-dimensional cardiothoracic ratio from digital PA chest radiographs: Correlation with cardiac function and the traditional cardiothoracic ratio. J. Digit. Imaging 2004, 17, 120–123. [Google Scholar] [CrossRef][Green Version]
- Dong, N.; Kampffmeyer, M.; Liang, X.; Wang, Z.; Dai, W.; Xing, E.P. Unsupervised domain adaptation for automatic estimation of cardiothoracic ratio. arXiv 2018, arXiv:1807.03434. [Google Scholar]
- Solovyev, R.; Melekhov, I.; Lesonen, T.; Vaattovaara, E.; Tervonen, O.; Tiulpin, A. Bayesian feature pyramid networks for automatic multi-label segmentation of chest X-rays and assessment of cardio-thoratic ratio. In Proceedings of the International Conference on Advanced Concepts for Intelligent Vision Systems, Auckland, New Zealand, 10–14 February 2020; pp. 117–130. [Google Scholar]
- Brakohiapa, E.K.K.; Botwe, B.O.; Sarkodie, B.D.; Ofori, E.K.; Coleman, J. Radiographic determination of cardiomegaly using cardiothoracic ratio and transverse cardiac diameter: Can one size fit all? Part one. Pan Afr. Med. J. 2017, 27. [Google Scholar] [CrossRef] [PubMed]
- Owais, M.; Arsalan, M.; Choi, J.; Mahmood, T.; Park, K.R. Artificial intelligence-based classification of multiple gastrointestinal diseases using endoscopy videos for clinical diagnosis. J. Clin. Med. 2019, 8, 986. [Google Scholar] [CrossRef] [PubMed]
- Arsalan, M.; Owais, M.; Mahmood, T.; Cho, S.W.; Park, K.R. Aiding the diagnosis of diabetic and hypertensive retinopathy using artificial intelligence-based semantic segmentation. J. Clin. Med. 2019, 8, 1446. [Google Scholar] [CrossRef] [PubMed]
- Owais, M.; Arsalan, M.; Choi, J.; Park, K.R. Effective diagnosis and treatment through content-based medical image retrieval (CBMIR) by using artificial intelligence. J. Clin. Med. 2019, 8, 462. [Google Scholar] [CrossRef]
- Nguyen, D.T.; Pham, T.D.; Batchuluun, G.; Yoon, H.S.; Park, K.R. Artificial intelligence-based thyroid nodule classification using information from spatial and frequency domains. J. Clin. Med. 2019, 8, 1976. [Google Scholar] [CrossRef]
- Salk, J.J.; Loubet-Senear, K.; Maritschnegg, E.; Valentine, C.C.; Williams, L.N.; Higgins, J.E.; Horvat, R.; Vanderstichele, A.; Nachmanson, D.; Baker, K.T.; et al. Ultra-sensitive TP53 sequencing for cancer detection reveals progressive clonal selection in normal tissue over a century of human lifespan. Cell Rep. 2019, 28, 132–144. [Google Scholar] [CrossRef]
- Esteva, A.; Robicquet, A.; Ramsundar, B.; Kuleshov, V.; DePristo, M.; Chou, K.; Cui, C.; Corrado, G.; Thrun, S.; Dean, J. A guide to deep learning in healthcare. Nat. Med. 2019, 25, 24–29. [Google Scholar] [CrossRef]
- Hwang, E.J.; Nam, J.G.; Lim, W.H.; Park, S.J.; Jeong, Y.S.; Kang, J.H.; Hong, E.K.; Kim, T.M.; Goo, J.M.; Park, S.; et al. Deep learning for chest radiograph diagnosis in the emergency department. Radiology 2019. [Google Scholar] [CrossRef]
- Zhou, S.; Nie, D.; Adeli, E.; Yin, J.; Lian, J.; Shen, D. High-resolution encoder–decoder networks for low-contrast medical image segmentation. IEEE Trans. Image Process. 2020, 29, 461–475. [Google Scholar] [CrossRef]
- Pan, X.; Li, L.; Yang, D.; He, Y.; Liu, Z.; Yang, H. An accurate nuclei segmentation algorithm in pathological image based on deep semantic network. IEEE Access 2019, 7, 110674–110686. [Google Scholar] [CrossRef]
- Gordienko, Y.; Gang, P.; Hui, J.; Zeng, W.; Kochura, Y.; Alienin, O.; Rokovyi, O.; Stirenko, S. Deep learning with lung segmentation and bone shadow exclusion techniques for chest X-ray analysis of lung cancer. In Proceedings of the Advances in Computer Science for Engineering and Education, Kiev, Ukraine, 18–20 January 2019; pp. 638–647. [Google Scholar]
- Mittal, A.; Hooda, R.; Sofat, S. LF-SegNet: A fully convolutional encoder–decoder network for segmenting lung fields from chest radiographs. Wirel. Pers. Commun. 2018, 101, 511–529. [Google Scholar] [CrossRef]
- Candemir, S.; Jaeger, S.; Palaniappan, K.; Musco, J.P.; Singh, R.K.; Xue, Z.; Karargyris, A.; Antani, S.; Thoma, G.; McDonald, C.J. Lung segmentation in chest radiographs using anatomical atlases with nonrigid registration. IEEE Trans. Med. Imaging 2014, 33, 577–590. [Google Scholar] [CrossRef]
- Jaeger, S.; Karargyris, A.; Antani, S.; Thoma, G. Detecting tuberculosis in radiographs using combined lung masks. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, San Diego, CA, USA, 28 August–1 September 2012; pp. 4978–4981. [Google Scholar]
- Supanta, C.; Kemper, G.; del Carpio, C. An algorithm for feature extraction and detection of pulmonary nodules in digital radiographic images. In Proceedings of the IEEE International Conference on Automation/XXIII Congress of the Chilean Association of Automatic Control, Concepcion, Chile, 17–19 October 2018; pp. 1–5. [Google Scholar]
- Jangam, E.; Rao, A.C.S. Segmentation of lungs from chest X rays using firefly optimized fuzzy C-means and level set algorithm. In Proceedings of the Recent Trends in Image Processing and Pattern Recognition, Solapur, India, 21–22 December 2019; pp. 303–311. [Google Scholar]
- Vital, D.A.; Sais, B.T.; Moraes, M.C. Automatic pulmonary segmentation in chest radiography, using wavelet, morphology and active contours. In Proceedings of the XXVI Brazilian Congress on Biomedical Engineering, Armação de Buzios, RJ, Brazil, 21–25 October 2018; pp. 77–82. [Google Scholar]
- Wan Ahmad, W.S.H.M.; Zaki, W.M.D.; Ahmad Fauzi, M.F. Lung segmentation on standard and mobile chest radiographs using oriented Gaussian derivatives filter. Biomed. Eng. OnLine 2015, 14. [Google Scholar] [CrossRef] [PubMed]
- Iakovidis, D.K.; Papamichalis, G. Automatic segmentation of the lung fields in portable chest radiographs based on Bézier interpolation of salient control points. In Proceedings of the IEEE International Workshop on Imaging Systems and Techniques, Crete, Greece, 10–12 October 2008; pp. 82–87. [Google Scholar]
- Pattrapisetwong, P.; Chiracharit, W. Automatic lung segmentation in chest radiographs using shadow filter and multilevel thresholding. In Proceedings of the International Computer Science and Engineering Conference, Chiang Mai, Thailand, 14–17 December 2016; pp. 1–6. [Google Scholar]
- Li, X.; Chen, L.; Chen, J. A visual saliency-based method for automatic lung regions extraction in chest radiographs. In Proceedings of the 14th International Computer Conference on Wavelet Active Media Technology and Information Processing, Chengdu, China, 15–17 December 2017; pp. 162–165. [Google Scholar]
- Chen, P.-Y.; Lin, C.-H.; Kan, C.-D.; Pai, N.-S.; Chen, W.-L.; Li, C.-H. Smart pleural effusion drainage monitoring system establishment for rapid effusion volume estimation and safety confirmation. IEEE Access 2019, 7, 135192–135203. [Google Scholar] [CrossRef]
- Dawoud, A. Lung segmentation in chest radiographs by fusing shape information in iterative thresholding. IET Comput. Vis. 2011, 5, 185–190. [Google Scholar] [CrossRef]
- Saad, M.N.; Muda, Z.; Ashaari, N.S.; Hamid, H.A. Image segmentation for lung region in chest X-ray images using edge detection and morphology. In Proceedings of the IEEE International Conference on Control System, Computing and Engineering, Batu Ferringhi, Malaysia, 28–30 November 2014; pp. 46–51. [Google Scholar]
- Lee, W.-L.; Chang, K.; Hsieh, K.-S. Unsupervised segmentation of lung fields in chest radiographs using multiresolution fractal feature vector and deformable models. Med. Biol. Eng. Comput. 2016, 54, 1409–1422. [Google Scholar] [CrossRef]
- Chondro, P.; Yao, C.-Y.; Ruan, S.-J.; Chien, L.-C. Low order adaptive region growing for lung segmentation on plain chest radiographs. Neurocomputing 2018, 275, 1002–1011. [Google Scholar] [CrossRef]
- Chung, H.; Ko, H.; Jeon, S.J.; Yoon, K.-H.; Lee, J. Automatic lung segmentation with juxta-pleural nodule identification using active contour model and bayesian approach. IEEE J. Transl. Eng. Health Med. 2018, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Dong, N.; Wang, Z.; Liang, X.; Zhang, H.; Xing, E.P. SCAN: Structure correcting adversarial network for organ segmentation in chest X-rays. In Proceedings of the Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, Granada, Spain, 20 September 2018; pp. 263–273. [Google Scholar]
- Dong, N.; Kampffmeyer, M.; Liang, X.; Wang, Z.; Dai, W.; Xing, E. Unsupervised domain adaptation for automatic estimation of cardiothoracic ratio. In Proceedings of the Medical Image Computing and Computer Assisted Intervention, Granada, Spain, 16–20 September 2018; pp. 544–552. [Google Scholar]
- Tang, Y.-B.; Tang, Y.-X.; Xiao, J.; Summers, R.M. XLSor: A robust and accurate lung segmentor on chest X-rays using criss-cross attention and customized radiorealistic abnormalities generation. In Proceedings of the International Conference on Medical Imaging with Deep Learning, London, UK, 8–10 July 2019; pp. 457–467. [Google Scholar]
- Souza, J.C.; Bandeira Diniz, J.O.; Ferreira, J.L.; França da Silva, G.L.; Corrêa Silva, A.; de Paiva, A.C. An automatic method for lung segmentation and reconstruction in chest X-ray using deep neural networks. Comput. Methods Programs Biomed. 2019, 177, 285–296. [Google Scholar] [CrossRef] [PubMed]
- Venkataramani, R.; Ravishankar, H.; Anamandra, S. Towards continuous domain adaptation for medical imaging. In Proceedings of the IEEE 16th International Symposium on Biomedical Imaging, Venice, Italy, 8–11 April 2019; pp. 443–446. [Google Scholar]
- Oliveira, H.; dos Santos, J. Deep transfer learning for segmentation of anatomical structures in chest radiographs. In Proceedings of the 2018 31st SIBGRAPI Conference on Graphics, Patterns and Images, Parana, Brazil, 29 October–1 November 2018; pp. 204–211. [Google Scholar]
- Islam, J.; Zhang, Y. Towards robust lung segmentation in chest radiographs with deep learning. arXiv 2018, arXiv:1811.12638. [Google Scholar]
- Wang, J.; Li, Z.; Jiang, R.; Xie, Z. Instance segmentation of anatomical structures in chest radiographs. In Proceedings of the IEEE 32nd International Symposium on Computer-Based Medical Systems, Corodoba, Spain, 5–7 June 2019; pp. 441–446. [Google Scholar]
- Dong, N.; Xu, M.; Liang, X.; Jiang, Y.; Dai, W.; Xing, E. Neural architecture search for adversarial medical image segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention, Shenzhen, China, 13–17 October 2019; pp. 828–836. [Google Scholar]
- Jiang, F.; Grigorev, A.; Rho, S.; Tian, Z.; Fu, Y.; Jifara, W.; Adil, K.; Liu, S. Medical image semantic segmentation based on deep learning. Neural Comput. Appl. 2018, 29, 1257–1265. [Google Scholar] [CrossRef]
- X-RayNet Model with Algorithms. Available online: http://dm.dgu.edu/link.html (accessed on 16 January 2020).
- Yu, F.; Koltun, V.; Funkhouser, T. Dilated residual networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 636–644. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. SegNet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef] [PubMed]
- Arsalan, M.; Naqvi, R.A.; Kim, D.S.; Nguyen, P.H.; Owais, M.; Park, K.R. IrisDenseNet: Robust iris segmentation using densely connected fully convolutional networks in the images by visible light and near-infrared light camera sensors. Sensors 2018, 18, 1501. [Google Scholar] [CrossRef] [PubMed]
- Arsalan, M.; Kim, D.S.; Lee, M.B.; Owais, M.; Park, K.R. FRED-Net: Fully residual encoder–decoder network for accurate iris segmentation. Expert Syst. Appl. 2019, 122, 217–241. [Google Scholar] [CrossRef]
- Arsalan, M.; Kim, D.S.; Owais, M.; Park, K.R. OR-Skip-Net: Outer residual skip network for skin segmentation in non-ideal situations. Expert Syst. Appl. 2020, 141. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Shiraishi, J.; Katsuragawa, S.; Ikezoe, J.; Matsumoto, T.; Kobayashi, T.; Komatsu, K.; Matsui, M.; Fujita, H.; Kodera, Y.; Doi, K. Development of a digital image database for chest radiographs with and without a lung nodule. Am. J. Roentgenol. 2000, 174, 71–74. [Google Scholar] [CrossRef]
- Van Ginneken, B.; Stegmann, M.B.; Loog, M. Segmentation of anatomical structures in chest radiographs using supervised methods: A comparative study on a public database. Med. Image Anal. 2006, 10, 19–40. [Google Scholar] [CrossRef]
- GeForce GTX TITAN X Graphics Processing Unit. Available online: https://www.geforce.com/hardware/desktop-gpus/geforce-gtx-titan-x/specifications (accessed on 4 December 2019).
- MATLAB R2019a. Available online: https://ch.mathworks.com/products/new_products/latest_features.html (accessed on 4 December 2019).
- Kingma, D.P.; Ba, J.L. Adam: A method for stochastic optimization. In Proceedings of the International Conference for Learning Representations, San Diego, CA, USA, 7–9 May 2015; pp. 1–15. [Google Scholar]
- Jaeger, S.; Candemir, S.; Antani, S.; Wáng, Y.-X.J.; Lu, P.-X.; Thoma, G. Two public chest X-ray datasets for computer-aided screening of pulmonary diseases. Quant. Imaging Med. Surg. 2014, 4, 475–477. [Google Scholar]
- Vajda, S.; Karargyris, A.; Jaeger, S.; Santosh, K.C.; Candemir, S.; Xue, Z.; Antani, S.; Thoma, G. Feature selection for automatic tuberculosis screening in frontal chest radiographs. J. Med. Syst. 2018, 42. [Google Scholar] [CrossRef]
- Santosh, K.C.; Antani, S. Automated chest X-ray screening: Can lung region symmetry help detect pulmonary abnormalities? IEEE Trans. Med. Imaging 2018, 37, 1168–1177. [Google Scholar] [CrossRef]

| Type | Methods | Strength | Weakness |
|---|---|---|---|
| Using handcrafted local features * | Lung segmentation using Hull-CPLM [2] | Selects the ROI for lung detection | Preprocessing is required |
| Nongrid registration lung segmentation [25] | Sift-flow modeling for registration provides an advantage | Boundary refinement is required | |
| Probabilistic lung shape model [26,32,35] | Probabilistic shape model mask helps in shape segmentation | Single threshold creates the segmentation error | |
| Otsu thresholding [27] | Excludes the noise area for lung nodule segmentation | Gamma correction is required | |
| Fuzzy c-means clustering [28,30,37] | Better performance compared to K-means | The lower value of β requires more iterations | |
| Active contour and morphology [29,39] | Active contour can estimate the real lung boundary | The iterative method takes many iterations | |
| Salient point-based lung segmentation [31,33] | Interpolation of salient points approximate lung boundary well | Results are affected by overlapped regions | |
| Harris corner detector [34,36] | Convolutional mask refines the contour | Edge detection is affected by noise | |
| Region growing [38] | Region growing methods are good towards the real boundary | ROI is required | |
| Using features based on machine learning or deep learning | Structural correcting adversarial network [40,49] | Adversarial training is good for a small number of training images | Critic network requires fully connected layer and consumes a lot of parameters |
| Domain adaptation [41,44] | Domain adaption is good to enhance segmentation performance | FCN-based segmentation consumes many parameters | |
| Lung segmentation by criss-cross attention [42] | Image-to-image translation is used for augmentation | Three separate deep models of ResNet101, UNet, and MUNIT are used | |
| Similar structure as AlexNet [43] | Semantic segmentation is close to real boundary | Patch-based deep learning scheme is computationally expensive | |
| FCN, U-Net, and SegNet for CXR segmentation [45] | Semantic segmentation provides good results for multiclass segmentation | FCN consumes many trainable parameters owing to fully connected layer | |
| U-Net [46] | U-Net is popular for medical image segmentation | Preprocessing is required | |
| Mask-RCNN [47] | Multiclass efficient segmentation is performed | Region proposals are also required with pixel-wise annotation | |
| ResNet [49] | Dropping 5th convolutional block from VGG-16 reduces the number of parameters | Clavicle bone segmentation is not considered | |
| X-RayNet (proposed) | 12 residual mesh streams enhance features to provide good segmentation performance | Data augmentation is required to artificially increase the amount of data |
| Method | Other Architectures | X-RayNet |
|---|---|---|
| ResNet [52] | Only adjacent convolutional layers have residual skip paths | Both adjacent and nonadjacent layers have residual skip connections. There are paths between the encoder and decoder. |
| 1 × 1 convolution is employed as bottleneck layer in all ResNet variants | 1 × 1 convolution is used to connect three blocks of the decoder based on nonidentity mapping | |
| Max-pooling layers are without indices information | Max-pool to max-unpool indices information is shared between the corresponding encoder and decoder block | |
| All variants use fully connected layers for classification purposes | The fully connected layers are not used to make the network a fully convolutional network (FCN) for semantic segmentation | |
| Average pooling is employed at the end of the network | Max-pooling layers and max-unpooling layers are used in each encoder and decoder block | |
| IrisDenseNet [54] | Encoder and decoder consist of 13 convolutional layers each, resulting in a total of 26 convolutional layers | Encoder and decoder consist of eight and nine (3 × 3) convolutional layers, respectively |
| Uses dense connectivity in encoder with depth-wise concatenation | Residual connectivity between encoder and decoder by elementwise addition | |
| First two blocks have two convolutional layers and the rest of the blocks have three convolutional layers in the encoder and decoder | Two convolutional layers in each encoder and decoder convolutional block, where one convolutional layer is at the end of the network to produce respective class masks | |
| The decoder is the same as the VGG-16 network without feature reuse by dense connectivity | Both encoder and decoder use the residual mesh connectivity for feature reuse | |
| FRED-Net [55] | Only uses residual skip connections between adjacent convolutional layers of same block | Uses residual skip connections for adjacent convolutional layers and between encoder and decoder externally |
| There is no skip connection between encoder and decoder | Inner and outer residual connections for spatial information flow | |
| The overall network has six skip paths | The overall network has 12 residual skip paths that create the residual mesh | |
| Overall network is based on nonidentity mapping | Among the 12 residual paths that create a residual mesh, nine are with identity mapping and three are with nonidentity mapping | |
| The ReLU is used after the elementwise addition that represents the postactivation only | The network is based on pre- and post-activation | |
| SegNet [53] | 26 convolutional layers | 17 convolutional layers |
| No residual connectivity that causes vanishing gradient problem | Vanishing gradient problem is handled by residual mesh | |
| Each block has a different number of convolutional layers | All the blocks have the same two convolutional layers | |
| 512-depth block used twice to increase the depth of the network | Used 512-depth block once for X-RayNet-1 and 512-depth block is not used in X-RayNet-2 | |
| OR-Skip-Net [56] | There is no internal connectivity between the convolutional layers in the encoder and decoder | Both encoder and decoder convolutional layers are connected with residual mesh for feature empowerment |
| The outer skip connections are with nonidentity mapping | The encoder-to-decoder connections are with identity mapping | |
| Only pre-activation is used as ReLU exists before elementwise addition | The network is based on pre- and post-activation | |
| Four residual connections are used | 12 residual skip connections are used | |
| Vess-Net [15] | 16 convolutional layers are used | 16 convolutions are used with an extra convolution in the decoder for fine edge connectivity |
| The first convolutional layer has no internal or external residual connection | The features from the first convolutional layer are important for edge information for the minor class, like the clavicle bones; therefore, it is internally and externally connected | |
| All the convolutional layers are internally connected with each other inside the encoder and decoder with nonidentity mapping | Most of the internal layers of the encoder and decoder are connected using identity mapping | |
| 10 residual paths | 12 residual paths | |
| U-Net [57] | 23 convolutional layers are used | 17 convolution layers are used |
| Up convolutions are used in the expansive part for upsampling | The unpool layer in combination with normal convolution is used for upsampling | |
| 1 × 1 convolution is used at the end of the network | 1 × 1 convolution is only used in the decoder internal residual connections | |
| Feature concatenation is utilized for empowerment | Feature elementwise addition is utilized for feature empowerment | |
| Cropping is required owing to border pixel loss during convolution | The feature map size is controlled by indices information transfer between pooling and unpooling layers |
| Type | Method | Lungs | Heart | Clavicle Bones | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Acc | J | D | Acc | J | D | Acc | J | D | ||
| Local feature-based methods | Peng et al. [22] | 97.0 | 93.6 | 96.7 | - | - | - | - | - | - |
| Candemir et al. [25] | - | 95.4 | 96.7 | - | - | - | - | - | - | |
| Jangam et al. [28] | - | 95.6 | 97.4 | - | - | - | - | - | - | |
| Wan Ahmed et al. [30] | 95.77 | - | - | - | - | - | - | - | - | |
| Vital et al. [29] | - | - | 95.9 | - | - | - | - | - | - | |
| Iakovidis et al. [31] | - | - | 91.66 | - | - | - | - | - | - | |
| Chondro et al. [38] | - | 96.3 | - | - | - | - | - | - | - | |
| Hybrid voting [59] | - | 94.9 | - | - | 86.0 | - | - | 73.6 | - | |
| PC post-processed [59] | - | 94.5 | - | - | 82.4 | - | - | 61.5 | - | |
| Human Observer [59] | - | 94.6 | - | - | 87.8 | - | - | 89.6 | - | |
| PC [59] | - | 93.8 | - | - | 81.1 | - | - | 61.8 | - | |
| Hybrid ASM/PC [59] | - | 93.4 | - | - | 83.6 | - | - | 66.3 | - | |
| Hybrid AAM/PC [59] | - | 93.3 | - | - | 82.7 | - | - | 61.3 | - | |
| ASM tuned [59] | - | 92.7 | - | - | 81.4 | - | - | 73.4 | - | |
| AAM whiskers BFGS [59] | - | 92.2 | - | - | 83.4 | - | - | 64.2 | - | |
| ASM default [59] | - | 90.3 | - | - | 79.3 | - | - | 69.0 | - | |
| AAM whiskers [59] | - | 91.3 | - | - | 81.3 | - | - | 62.5 | - | |
| AAM default [59] | - | 84.7 | - | - | 77.5 | - | - | 50.5 | - | |
| Mean shape [59] | - | 71.3 | - | - | 64.3 | - | - | 30.3 | - | |
| Dawoud [35] | - | 94.0 | - | - | - | - | - | - | - | |
| Coppini et al. [4] | - | 92.7 | 95.5 | - | - | - | - | - | - | |
| Deep feature-based methods | Dai et al. FCN [40] | - | 92.9 | 96.3 | - | 86.5 | 92.7 | - | - | - |
| Dong et al. [41] | 95.5 | - | 90.2 | |||||||
| Mittal et al. [24] | 98.73 | 95.10 | - | - | - | - | - | - | - | |
| Oliveira et al. FCN [45] | 95.05 | 97.45 | 89.25 | 94.24 | 75.52 | 85.90 | ||||
| Oliveira et al. U-Net [45] | 96.02 | 97.96 | 89.21 | 94.16 | 86.54 | 92.58 | ||||
| Oliveira et al. SegNet [45] | 95.54 | 97.71 | 89.64 | 94.44 | 87.30 | 93.08 | ||||
| Novikov et al. InvertedNet [1] | 94.9 | 97.4 | 88.8 | 94.1 | 83.3 | 91.0 | ||||
| ContextNet-1 [44] | 95.8 | - | - | - | - | - | - | - | ||
| ContextNet-2 [44] | - | 96.5 | - | - | - | - | - | |||
| ResNet50 (512, C = 4) ~* [47] | 93.9 | 96.8 | 88.3 | 93.7 | 79.4 | 88.3 | ||||
| ResNet50 (512, C = 4) * [47] | 95.3 | 97.6 | 89.4 | 94.3 | 84.9 | 91.8 | ||||
| ResNet50 (512, C = 6) * [47] | 94.5 | 97.2 | 89.3 | 94.3 | 84.3 | 91.5 | ||||
| ResNet50 (512, C = 8) * [47] | 94.9 | 97.4 | 89.7 | 94.5 | 84.7 | 91.6 | ||||
| ResNet101 (512, C = 4) * [47] | 95.3 | 97.6 | 90.4 | 94.9 | 85.2 | 92.0 | ||||
| ResNet50 (256, C = 4) * [47] | 95.0 | 97.4 | 89.8 | 94.6 | 82.3 | 90.2 | ||||
| ResNet101 (256, C = 4) * [47] | 94.9 | 97.4 | 90.1 | 94.7 | 79.6 | 88.5 | ||||
| BFPN [12] | - | 87.0 | 93.0 | - | 82.0 | 91.0 | - | - | - | |
| OR-Skip-Net [56] | 98.92 | 96.14 | 98.02 | 98.94 | 88.8 | 94.01 | 99.7 | 83.79 | 91.07 | |
| X-RayNet-1 (proposed method) | 99.06 | 96.65 | 98.29 | 99.16 | 90.99 | 95.22 | 99.8 | 88.72 | 93.94 | |
| X-RayNet-2 (proposed method) | 98.93 | 96.14 | 98.02 | 98.96 | 89.30 | 94.25 | 99.8 | 86.65 | 92.73 | |
| Type | Method | Acc | J | D |
|---|---|---|---|---|
| Handcrafted local feature-based methods | Candemir et al. [25] | - | 94.1 | 96.0 |
| Peng et al. [2] | 97.0 | - | - | |
| Vajda et al. [64] * | 69.0 | - | - | |
| Learned/deep feature-based methods | Souza et al. [43] | 96.97 | 88.07 | 96.97 |
| Feature selection with BN [65] * | 77.0 | - | - | |
| Feature selection with MLP [65] * | 79.0 | - | - | |
| Feature selection with RF [65] * | 81.0 | - | - | |
| Feature selection and Vote [65] * | 83.0 | - | - | |
| Bayesian feature pyramid network [12] | - | 87.0 | 93.0 | |
| X-RayNet-1 (proposed method) | 99.11 | 96.36 | 98.14 | |
| X-Ray-Net-2 (proposed method) | 98.72 | 94.96 | 97.40 |
| Type | Method | Acc | J | D |
|---|---|---|---|---|
| Handcrafted local feature-based methods | Peng et al. [2] | 97.0 | - | - |
| Vajda et al. [64] * | 92.0 | - | - | |
| Learned/deep feature-based methods | Feature selection with BN [65] * | 81.0 | - | - |
| Feature selection with MLP [65] * | 88.0 | - | - | |
| Feature selection with RF [65] * | 89.0 | - | - | |
| Feature selection and Vote [65] * | 91.0 | - | - | |
| Bayesian feature pyramid network [12] | - | 87.0 | 93.0 | |
| X-RayNet-1 (proposed method) | 97.70 | 91.82 | 95.64 | |
| X-Ray-Net-2 (proposed method) | 97.32 | 90.56 | 95.0 |
| Method | Train | Test | Acc | J | D |
|---|---|---|---|---|---|
| X-RayNet-1 | MC | SC | 96.27 | 87.74 | 93.24 |
| X-RayNet-1 | SC | MC | 98.10 | 92.52 | 96.06 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arsalan, M.; Owais, M.; Mahmood, T.; Choi, J.; Park, K.R. Artificial Intelligence-Based Diagnosis of Cardiac and Related Diseases. J. Clin. Med. 2020, 9, 871. https://doi.org/10.3390/jcm9030871
Arsalan M, Owais M, Mahmood T, Choi J, Park KR. Artificial Intelligence-Based Diagnosis of Cardiac and Related Diseases. Journal of Clinical Medicine. 2020; 9(3):871. https://doi.org/10.3390/jcm9030871
Chicago/Turabian StyleArsalan, Muhammad, Muhammad Owais, Tahir Mahmood, Jiho Choi, and Kang Ryoung Park. 2020. "Artificial Intelligence-Based Diagnosis of Cardiac and Related Diseases" Journal of Clinical Medicine 9, no. 3: 871. https://doi.org/10.3390/jcm9030871
APA StyleArsalan, M., Owais, M., Mahmood, T., Choi, J., & Park, K. R. (2020). Artificial Intelligence-Based Diagnosis of Cardiac and Related Diseases. Journal of Clinical Medicine, 9(3), 871. https://doi.org/10.3390/jcm9030871






