Looking for a Better Characterization of Triple-Negative Breast Cancer by Means of Circulating Tumor Cells
Abstract
1. Introduction
2. Experimental Section
2.1. Patient Inclusion and Sample Collection
2.2. CTC Immunoisolation
2.3. Gene Expression Analysis by RT-qPCR
2.4. Cell Lines
2.5. TIMP1 Knock-Down
2.6. Western Blot
2.7. Transwell Migration Assay
2.8. Proliferation Assay
2.9. Colony Formation Assay
2.10. Adhesion Assay
2.11. Zebrafish Care and Breeding
2.12. Zebrafish Xenograft Assays and Image Analysis
2.13. Mouse Xenograft
2.14. Statistical Analysis
3. Results
3.1. Patient Characteristics
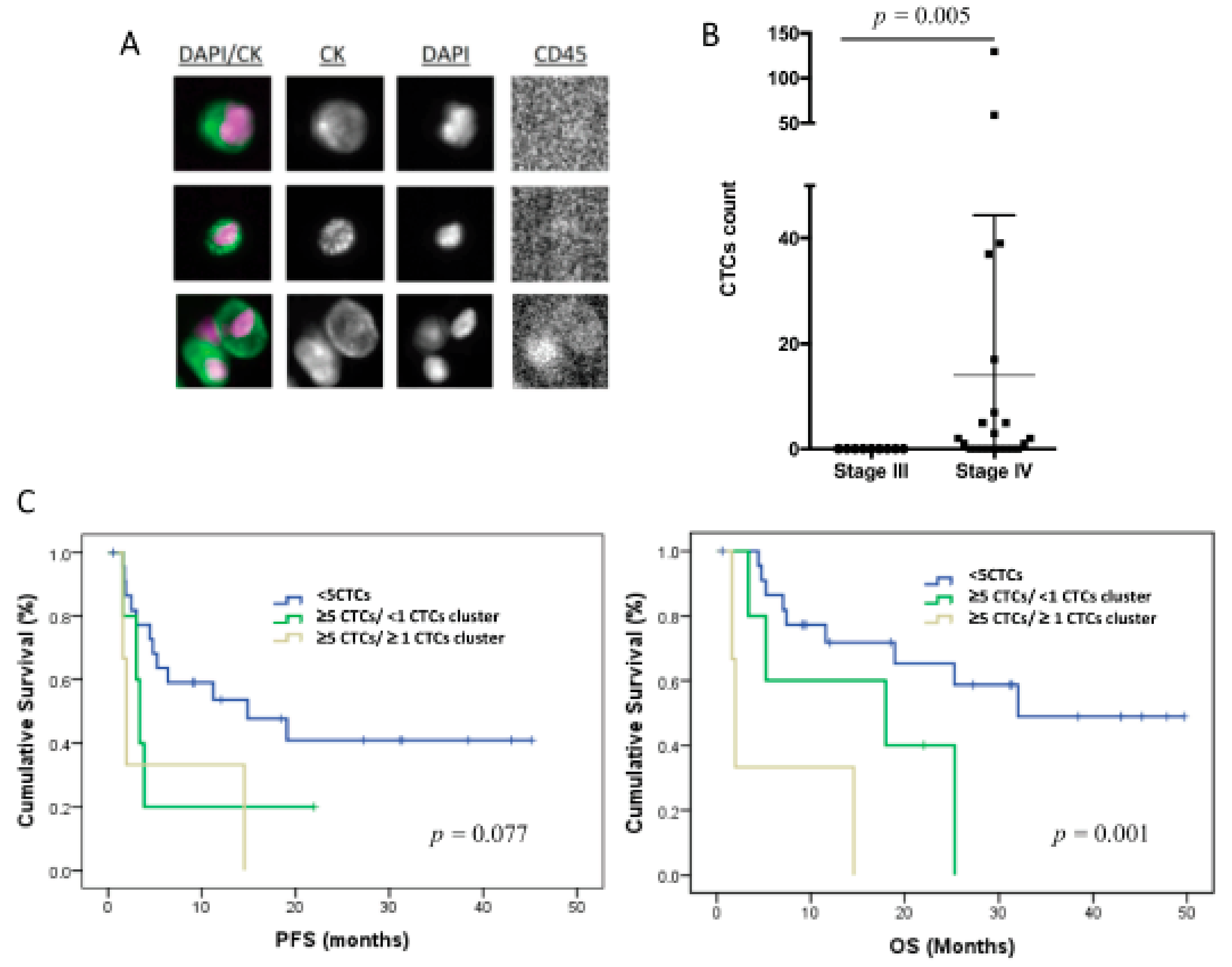
3.2. CTC Enumeration Correlates with a More Aggressive Disease
3.3. CTCs from TNBC Patients Are Characterized by High Cell Plasticity
3.4. The CTC Expression Signature Predicted the Patients’ Outcome
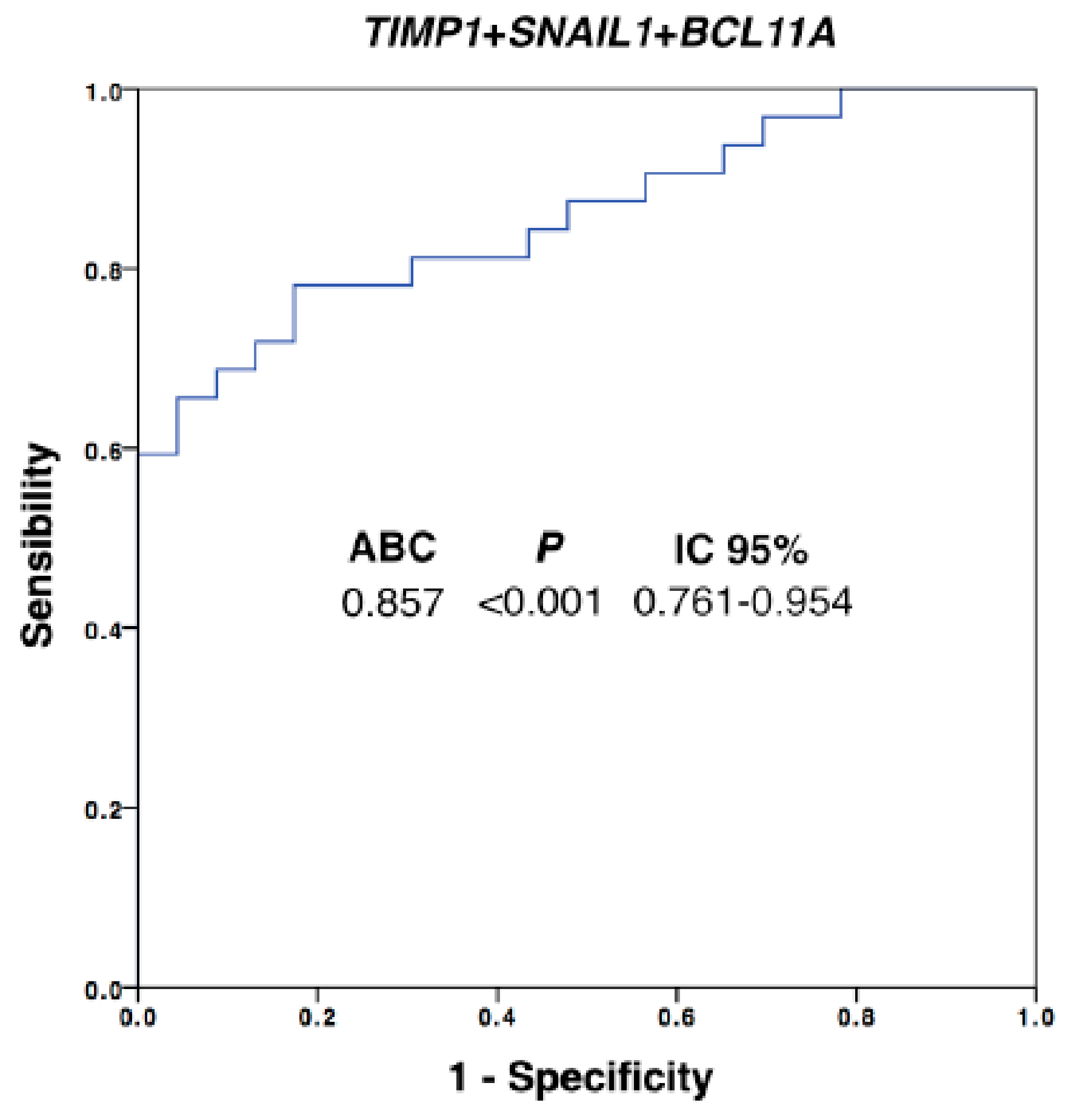
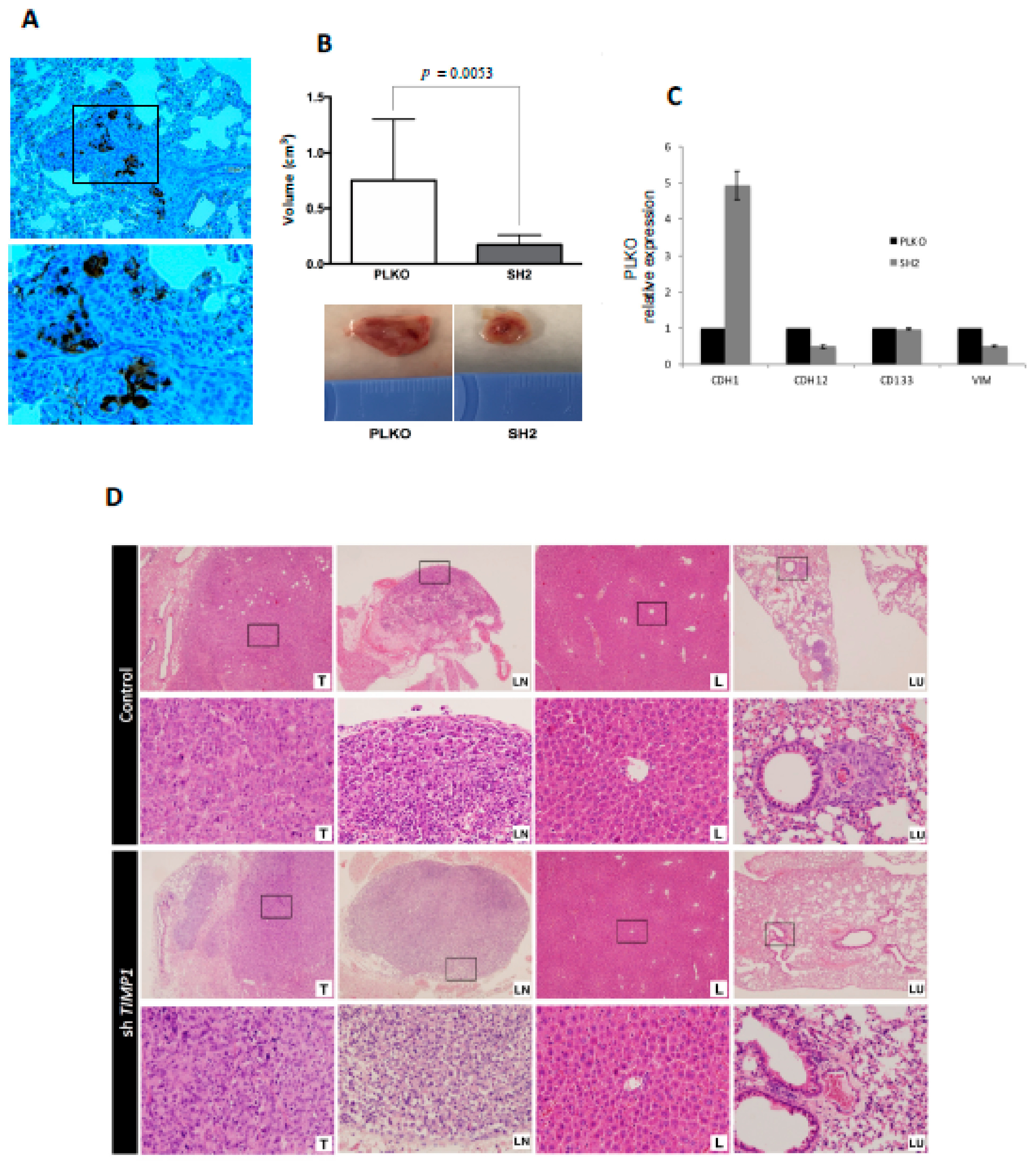
3.5. CTC Profiling Helps to Identify Therapeutic Targets such as AR and TIMP1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Senkus, E.; Kyriakides, S.; Ohno, S.; Penault, F.; Poortmans, P.; Rutgers, E.; Zackrisson, S.; Cardoso, F. Primary breast cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2015, 26, 8–30. [Google Scholar] [CrossRef] [PubMed]
- Bianchini, G.; Balko, J.; Mayer, I.; Sanders, M.; Gianni, L. Triple-negative breast cancer: Challenges and opportunities of a heterogeneous disease. Nat. Rev. Clin. Oncol. 2016, 11, 674–690. [Google Scholar] [CrossRef] [PubMed]
- Baccelli, I.; Schneeweiss, A.; Riethdorf, S.; Stenzinger, A.; Schillert, A.; Vogel, V.; Klein, C.; Saini, M.; Bauerle, T.; Wallwiener, M.; et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat. Biotechnol. 2013, 31, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Iwata, H.; Masuda, N.; Yamamoto, D.; Sagara, Y.; Sato, N.; Yamamoto, Y.; Saito, N.; Fujita, T.; Oura, S.; Watanabe, J.; et al. Circulating tumor cells as a prognostic marker for efficacy in the randomized phase III JO21095 trial in Japanese patients with HER2-negative metastatic breast cancer. Breast Cancer Res. Treat. 2017, 162, 501–510. [Google Scholar] [CrossRef]
- Lu, Y.; Wang, P.; Wang, X.; Peng, J.; Zhu, Y.; Shen, N. The significant prognostic value of circulating tumor cells in triple-negative breast cancer: A meta-analysis. Oncotarget 2016, 7, 37361–37369. [Google Scholar] [CrossRef]
- Riethdorf, S.; Muller, V.; Loibl, S.; Nekjudova, V.; Weber, K.; Huober, J.; Fehm, T.; Scharder, I.; Hilfrich, J.; Holms, F.; et al. Prognostic Impact of Circulating Tumor Cells for Breast Cancer Patients Treated in the Neoadjuvant ‘Geparquattro’ Trial. Clin. Cancer Res. 2017, 23, 5384–5393. [Google Scholar] [CrossRef]
- Zhang, Y.; Lv, Y.; Niu, Y.; Su, H.; Feng, A. Role of Circulating Tumor Cell (CTC) Monitoring in Evaluating Prognosis of Triple-Negative Breast Cancer Patients in China. Med. Sci. Monit. 2017, 23, 3071–3079. [Google Scholar] [CrossRef]
- Jansson, S.; Bendahl, P.; Larsson, A.; Aaltonen, K.; Rydén, L. Prognostic impact of circulating tumor cell apoptosis and clusters in serial blood samples from patients with metastatic breast cancer in a prospective observational cohort. BMC Cancer 2016, 16, 433. [Google Scholar] [CrossRef]
- Agelaki, S.; Dragolia, M.; Markonanolaki, H.; Alkahtani, S.; Stournaras, C.; Georgoulias, V.; Kallergi, G. Phenotypic characterization of circulating tumor cells in triple negative breast cancer patients. Oncotarget 2017, 8, 5309–5322. [Google Scholar] [CrossRef]
- Massagué, J.; Obenauf, A. Metastatic colonization by circulating tumour cells. Nature 2016, 529, 298–306. [Google Scholar] [CrossRef]
- Yu, M.; Bardia, A.; Wittner, B.; Stott, S.; Smas, M.; Ting, D.; Isakoff, S.; Ciciliano, J.; Wlls, M.; Shah, A.; et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013, 339, 580–584. [Google Scholar] [CrossRef] [PubMed]
- Santamaría, P.; Moreno-Bueno, G.; Cano, A. Contribution of Epithelial Plasticity to Therapy Resistance. J. Clin. Med. 2019, 8, 676. [Google Scholar] [CrossRef] [PubMed]
- Aktas, B.; Tewes, M.; Fehm, T.; Hauch, S.; Kimmig, R.; Kasimir-Bauer, S. Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res. 2009, 11, 46–52. [Google Scholar] [CrossRef]
- Kallergi, G.; Papadaki, M.; Politaki, E.; Mavroudis, D.; Georgoulias, V.; Agelaki, S. Epithelial to mesenchymal transition markers expressed in circulating tumour cells of early and metastatic breast cancer patients. Breast Cancer Res. 2011, 13, 59–64. [Google Scholar] [CrossRef]
- Kasimir-Bauer, S.; Bittner, A.; Konig, L.; Reiter, K.; Keller, T.; Kimming, R.; Hoffmann, O. Does primary neoadjuvant systemic therapy eradicate minimal residual disease? Analysis of disseminated and circulating tumor cells before and after therapy. Breast Cancer Res. 2016, 18, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Kasimir-Bauer, S.; Hoffmann, O.; Wallwiener, D.; Kimmig, R.; Fehm, T. Expression of stem cell and epithelial-mesenchymal transition markers in primary breast cancer patients with circulating tumor cells. Breast Cancer Res. 2012, 14, 1–15. [Google Scholar] [CrossRef]
- Papadaki, M.; Stoupis, G.; Theodoropoulos, P.; Mavroudis, D.; Georgoulias, V.; Agelaki, S. Circulating Tumor Cells with Stemness and Epithelial-to-Mesenchymal Transition Features Are Chemoresistant and Predictive of Poor Outcome in Metastatic Breast Cancer. Mol. Cancer Ther. 2019, 18, 437–447. [Google Scholar] [CrossRef]
- Barbazán, J.; Alonso-Alconada, L.; Muinelo-Romay, L.; Alonso-Nocelo, M.; León, L.; Candamio, S.; Gallardo, E.; Anido, U.; Doll, A.; Casares, M.A.; et al. A logistic model for the detection of circulating tumour cells in human metastatic colorectal cancer. J. Cell. Mol. Med. 2012, 16, 2342–2349. [Google Scholar] [CrossRef]
- Alonso-Alconada, L.; Muinelo-Romay, L.; Madissoo, K.; Diaz-Lopez, A.; Krakstad, C.; Trovik, J.; Wik, E.; Hapangama, D.; Coenegrachts, L.; Cano, A.; et al. Molecular profiling of circulating tumor cells links plasticity to the metastatic process in endometrial cancer. Mol. Cancer 2014, 14, 1–10. [Google Scholar] [CrossRef]
- Cheng, G.; Fan, X.; Hao, M.; Wang, J.; Zhou, X.; Sun, X. Higher levels of TIMP-1 expression are associated with a poor prognosis in triple-negative breast cancer. Mol. Cancer 2016, 15, 1–30. [Google Scholar] [CrossRef]
- Li, D.; Zhou, W.; Pang, J.; Tang, Q.; Zhong, B.; Shen, C.; Xiao, L.; Hou, T. A magic drug target: Androgen receptor. Med. Res. Rev. 2019, 5, 1485–1514. [Google Scholar] [CrossRef]
- Barton, V.; Gordon, M.; Richer, J.; Elias, J. Anti-androgen therapy in triple-negative breast cancer. Ther. Adv. Med. Oncol. 2016, 8, 305–308. [Google Scholar] [CrossRef] [PubMed]
- Bidard, F.; Cottu, P.; Park, J.; Loibl, S.; Latouche, A.; Pierga, J.; Pantel, K. Circulating Tumor Cells in Breast Cancer Patients Treated by Neoadjuvant Chemotherapy: A Meta-analysis. J. Natl. Cancer Inst. 2018, 110, 560–567. [Google Scholar] [CrossRef]
- Cristofanilli, M.; Hayes, D.; Budd, G.; Ellis, M.; Stopeck, A.; Reuben, J.; Doyle, G.; Matera, J.; Allard, W.; Miller, M.; et al. Circulating tumor cells: A novel prognostic factor for newly diagnosed metastatic breast cancer. J. Clin. Oncol. 2005, 7, 1420–1430. [Google Scholar] [CrossRef] [PubMed]
- Cristofanilli, M.; Zorcino, L.; Darrigues, L.; Reis-Filho, J.; Gerratana, L.; Michiels, S.; Bidard, F.; Pantel, K. The clinical use of circulating tumor cells (CTCs) enumeration for staging of metastatic breast cancer (MBC): International expert consensus paper. Crit. Rev. Oncol. Hematol. 2019, 134, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Pereira-Veiga, T.; Abreu, M.; Matias-Guiu, X.; Santacana, M.; Sanchez, L.; Cueva, J.; Palacios, P.; Abdulkader, I.; Lopez-Lopez, R.; Muinelo-Romay, L.; et al. CTCs-derived xenograft development in a Triple Negative breast cancer case. Int. J. Cancer 2018, 5, 485–501. [Google Scholar] [CrossRef] [PubMed]
- Mladinich, M.; Ruan, D.; Chan, C. Tackling Cancer Stem Cells via Inhibition of EMT Transcription Factors. Stem Cells Int. 2016, 2016. [Google Scholar] [CrossRef]
- Pastushenko, I.; Brisebarre, A.; Sifrim, A.; Fioramonti, M.; Revenco, T.; Boumahdi, S.; Keymeulen, A.; Brown, D.; Moers, V.; Lemaire, S.; et al. Identification of the tumour transition states occurring during EMT. Nature 2018, 556, 463–468. [Google Scholar] [CrossRef]
- Nieto, M.; Huang, R.; Jackson, R.; Thiery, J. EMT: 2016. Cell 2016, 166, 21–45. [Google Scholar] [CrossRef]
- Bao, H.; Burke, P.; Huang, J.; Chen, X.; Brohawn, P.; Yao, Y.; Lechleider, R.; Sikorski, R.; Buzoianu, M.; Zhang, J.; et al. Circulating tumor cells: Application as a biomarker for molecular characterization and predictor of survival in an all-comer solid tumor phase I clinical study. PLoS ONE 2013, 8, 58557. [Google Scholar] [CrossRef]
- Mitra, A.; Mishra, L.; Li, S. EMT, CTCs and CSCs in tumor relapse and drug-resistance. Oncotarget 2015, 13, 10697–10711. [Google Scholar] [CrossRef] [PubMed]
- Mego, M.; Giorgi, U.; Broglio, K.; Dawood, S.; Valero, V.; Andreopoulou, E.; Handy, B.; Reuben, J.; Cristofanilli, M. Circulating tumour cells are associated with increased risk of venous thromboembolism in metastatic breast cancer patients. Br. J. Cancer 2009, 101, 1813–1816. [Google Scholar] [CrossRef] [PubMed]
- Akalay, I.; Janji, B.; Hasmim, M.; Noman, M.; Andre, F.; Cremoux, P.; Berthearu, P.; Badoual, C.; Vielh, P.; Larsen, A.; et al. Epithelial-to-Mesenchymal Transition and Autophagy Induction in Breast Carcinoma Promote Escape from T-cell-Mediated Lysis. Cancer Res. 2013, 73, 2418–2427. [Google Scholar] [CrossRef] [PubMed]
- Collins, L.; Cole, K.; Marotti, J.; Hu, R.; Schnitt, S.; Tamimi, M. Androgen receptor expression in breast cancer in relation to molecular phenotype: Results from the Nurses’ Health Study. Mod. Pathol. 2011, 24, 924–931. [Google Scholar] [CrossRef]
- Mrklić, I.; Pogorelić, Z.; Capkun, V.; Tomić, S. Expression of androgen receptors in triple negative breast carcinomas. Acta Histochem. 2013, 115, 344–348. [Google Scholar] [CrossRef]
- Masuda, H.; Baggerly, K.; Wang, Y.; Zhang, Y.; Gonzalez-Angulo, A.; Meric-Bernstam, F.; Valero, V.; Lehmann, B.D.; Pietenpol, J.; Hortobagyi, G.; et al. Differential response to neoadjuvant chemotherapy among 7 triple-negative breast cancer molecular subtypes. Clin. Cancer Res. 2013, 19, 5533–5540. [Google Scholar] [CrossRef]
- Aceto, N.; Bardia, A.; Wittner, B.; Donaldson, M.; O’Keefe, R.; Engstrom, A.; Bersani, F.; Zheng, Y.; Comaills, V.; Niederhoffer, K.; et al. AR Expression in Breast Cancer CTCs Associates with Bone Metastases. Mol. Cancer Res. 2018, 16, 720–727. [Google Scholar] [CrossRef]
- Antonarakis, E.; Lu, C.; Luber, B.; Wang, H.; Chen, Y.; Zhu, Y.; Silberstein, J.; Taylor, M.; Maughan, B.; Denmeade, S.; et al. Clinical Significance of Androgen Receptor Splice Variant-7 mRNA Detection in Circulating Tumor Cells of Men With Metastatic Castration-Resistant Prostate Cancer Treated With First- and Second-Line Abiraterone and Enzalutamide. J. Clin. Oncol. 2017, 35, 2149–2156. [Google Scholar] [CrossRef]
- Dehm, S.; Montgomery, B.; Plymate, S. AR-Variant-Positive CTC: A Surrogate for a Surrogate for Taxane Therapy Outcome? Clin. Cancer Res. 2019, 25, 1696–1698. [Google Scholar] [CrossRef]
- Stetler-Stevenson, W. Tissue inhibitors of metalloproteinases in cell signaling: Metalloproteinase-independent biological activities. Sci. Signal. 2008, 27, 124–133. [Google Scholar] [CrossRef]
- Murphy, G.; Nagase, H. Progress in matrix metalloproteinase research. Mol. Aspects Med. 2008, 29, 290–308. [Google Scholar] [CrossRef] [PubMed]
- Oh, W.; Vargas, R.; Jacobus, S.; Leitzel, K.; Regan, M.; Hamer, P.; Pierce, K.; Brown-Shimer, S.; Carney, W.; Ali, S.; et al. Elevated plasma tissue inhibitor of metalloproteinase-1 levels predict decreased survival in castration-resistant prostate cancer patients. Cancer 2011, 117, 517–525. [Google Scholar] [CrossRef] [PubMed]
- Gouyer, V.; Conti, M.; Devos, P.; Zerimech, F.; Copin, M.; Crème, E.; Wurtz, A.; Porte, H.; Huet, G. Tissue inhibitor of metalloproteinase 1 is an independent predictor of prognosis in patients with nonsmall cell lung carcinoma who undergo resection with curative intent. Cancer 2005, 103, 1676–1684. [Google Scholar] [CrossRef] [PubMed]
- Kluger, H.; Bacchiocchi, A.; Mayer, T.; Kirsch, J.; Kluger, Y.; Sznol, M.; Ariyan, S.; Molinaro, A.; Halaban, R. Plasma markers for identifying patients with metastatic melanoma. Clin. Cancer Res. 2001, 17, 2417–2425. [Google Scholar] [CrossRef]
- Aaberg-Jessen, C.; Christensen, K.; Offenberg, H.; Bartels, A.; Breehsen, T.; Schroder, H.; Brunner, N.; Kristensen, B. Low expression of tissue inhibitor of metalloproteinases-1 (TIMP-1) in glioblastoma predicts longer patient survival. J. Neurooncol. 2009, 95, 117–128. [Google Scholar] [CrossRef]
- Ree, A.; Flørenes, V.; Berg, J.; Mælandsmo, G.; Nesland, J.; Fodstad, O. High levels of messenger RNAs for tissue inhibitors of metalloproteinases (TIMP-1 and TIMP-2) in primary breast carcinomas are associated with development of distant metastases. Clin. Cancer Res. 1997, 9, 1623–1628. [Google Scholar]
- Porter, J.; Shen, S.; Denhardt, D. Tissue inhibitor of metalloproteinase-1 stimulates proliferation of human cancer cells by inhibiting a metalloproteinase. Br. J. Cancer 2004, 90, 463–470. [Google Scholar] [CrossRef]


| Age (Years) | Mean (Range) | ||
|---|---|---|---|
| 58.5 (33–80) | |||
| Stage * | n (%) | Previous Surgery | n (%) |
| III | 9 (28.1%) | Yes | 23 (71.8%) |
| IV | 23 (71.9%) | No | 9 (28.2%) |
| Status * | Treatment * | ||
| First Diagnosis | 24 (75.0%) | Yes | 5 (15.6%) |
| Recurrence | 8 (25.0%) | No | 27 (84.4%) |
| Metastasis location | Ki67 levels | ||
| Visceral/bone | 4 (17.4%) | Low | 4 (12.5%) |
| Visceral | 17 (73.9%) | High | 27 (84.4%) |
| Unknown | 2 (8.7%) | Unknown | 1 (3.1%) |
| Histology | Disease evolution | ||
| Ductal | 29 (90.6%) | Progressions | 20 (62.5%) |
| Lobulillar | 1 (3.1%) | PFS (months) | median (range) |
| Metaplasic | 2 (6.3%) | 12.4 (0.5–45.2) | |
| Histology Grade | Survival | ||
| 3 | 22 (68.7%) | Deaths | 17 (53.1%) |
| 2 | 10 (31.3%) | OS (months) | median (range) |
| 18.4 (0.5–45.2) |
| Marker | n | PFS (Months) | OS (Months) | ||
|---|---|---|---|---|---|
| Mean (95% CI) | p | Mean (95% CI) | p | ||
| CTCs levels | |||||
| <5 CTCs | 23 | 22.7 (14.3–31.1) | 0.033 | 32.3 (23.7–40.8) | 0.006 |
| ≥5 CTCs | 8 | 6.5 (1.6–11.4) | 11.9 (4.8–19.1) | ||
| CTCs clusters | |||||
| No (<1CTC cluster) | 28 | 20.7 (13.2–28.3) | 0.071 | 29.4 (21.7–37.1) | 0.001 |
| Yes (≥1CTC cluster) | 3 | 6.04 (0.0–14.3) | 6.04 (0.0–14.3) | ||
| CTCs levels/clusters | |||||
| <5 CTCs | 23 | 22.7 (14.3–31.2) | 32.3 (23.7–40.8) | ||
| ≥5 CTCs and 0 CTCs cluster | 5 | 6.8 (0.8–13.4) | 0.077 | 15.5 (5.8–25.0) | 0.001 |
| ≥5 CTCs and ≥1CTCs cluster | 3 | 6.04 (0.0–14.3) | 6.04 (0–14.3) | ||
| Marker | AUROC | p | 95% CI |
|---|---|---|---|
| EPCAM | 0.697 | 0.013 | 0.560–0.834 |
| AR | 0.758 | 0.001 | 0.633–0.884 |
| TIMP1 | 0.764 | 0.001 | 0.636–0.891 |
| CRIPTO1 | 0.727 | 0.004 | 0.595–0.858 |
| CDH1 | 0.712 | 0.008 | 0.571–0.853 |
| VIM | 0.659 | 0.046 | 0.515–0.803 |
| CD49F | 0.755 | 0.001 | 0.624–0.887 |
| ALDH2 | 0.648 | 0.063 | 0.502–0.795 |
| CD44 | 0.696 | 0.014 | 0.554–0.837 |
| SNAIL1 | 0.793 | <0.001 | 0.673–0.912 |
| BCL11A | 0.766 | 0.001 | 0.637–0.894 |
| GAPDH | 0.665 | 0.027 | 0.520–0.811 |
| Marker | HR (95% CI) | p | |
|---|---|---|---|
| OS | CD49F (high vs. low) | 5.12 (1.61–15.25) | 0.006 |
| ALDH2 (high vs. low) | 3.12 (1.16–8.41) | 0.024 | |
| CD44 (high vs. low) | 3.70 (1.37–9.95) | 0.010 | |
| TIMP1 (high vs. low) | 5.12 (1.87–14.02) | 0.001 | |
| GAPDH (high vs. low) | 5.18 (1.49–15.29) | 0.004 | |
| PFS | CD49F (high vs. low) | 3.33 (1.23–9.01) | 0.018 |
| TIMP1 (high vs. low) | 3.86 (1.39–10.70) | 0.004 | |
| GAPDH (high vs. low) | 3.56 (1.34–10.17) | 0.014 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abreu, M.; Cabezas-Sainz, P.; Pereira-Veiga, T.; Falo, C.; Abalo, A.; Morilla, I.; Curiel, T.; Cueva, J.; Rodríguez, C.; Varela-Pose, V.; et al. Looking for a Better Characterization of Triple-Negative Breast Cancer by Means of Circulating Tumor Cells. J. Clin. Med. 2020, 9, 353. https://doi.org/10.3390/jcm9020353
Abreu M, Cabezas-Sainz P, Pereira-Veiga T, Falo C, Abalo A, Morilla I, Curiel T, Cueva J, Rodríguez C, Varela-Pose V, et al. Looking for a Better Characterization of Triple-Negative Breast Cancer by Means of Circulating Tumor Cells. Journal of Clinical Medicine. 2020; 9(2):353. https://doi.org/10.3390/jcm9020353
Chicago/Turabian StyleAbreu, Manuel, Pablo Cabezas-Sainz, Thais Pereira-Veiga, Catalina Falo, Alicia Abalo, Idoia Morilla, Teresa Curiel, Juan Cueva, Carmela Rodríguez, Vanesa Varela-Pose, and et al. 2020. "Looking for a Better Characterization of Triple-Negative Breast Cancer by Means of Circulating Tumor Cells" Journal of Clinical Medicine 9, no. 2: 353. https://doi.org/10.3390/jcm9020353
APA StyleAbreu, M., Cabezas-Sainz, P., Pereira-Veiga, T., Falo, C., Abalo, A., Morilla, I., Curiel, T., Cueva, J., Rodríguez, C., Varela-Pose, V., Lago-Lestón, R., Mondelo, P., Palacios, P., Moreno-Bueno, G., Cano, A., García-Caballero, T., Pujana, M. Á., Sánchez-Piñón, L., Costa, C., ... Muinelo-Romay, L. (2020). Looking for a Better Characterization of Triple-Negative Breast Cancer by Means of Circulating Tumor Cells. Journal of Clinical Medicine, 9(2), 353. https://doi.org/10.3390/jcm9020353






