Porphyromonas gingivalis Mfa1 Induces Chemokine and Cell Adhesion Molecules in Mouse Gingival Fibroblasts via Toll-Like Receptors
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Purification of Mfa1 Fimbriae
2.3. RT2 Profiler PCR Array Analysis
2.4. Real-Time PCR
2.5. siRNA Transfection
2.6. Flow Cytometry
2.7. Statistical Analysis
3. Results
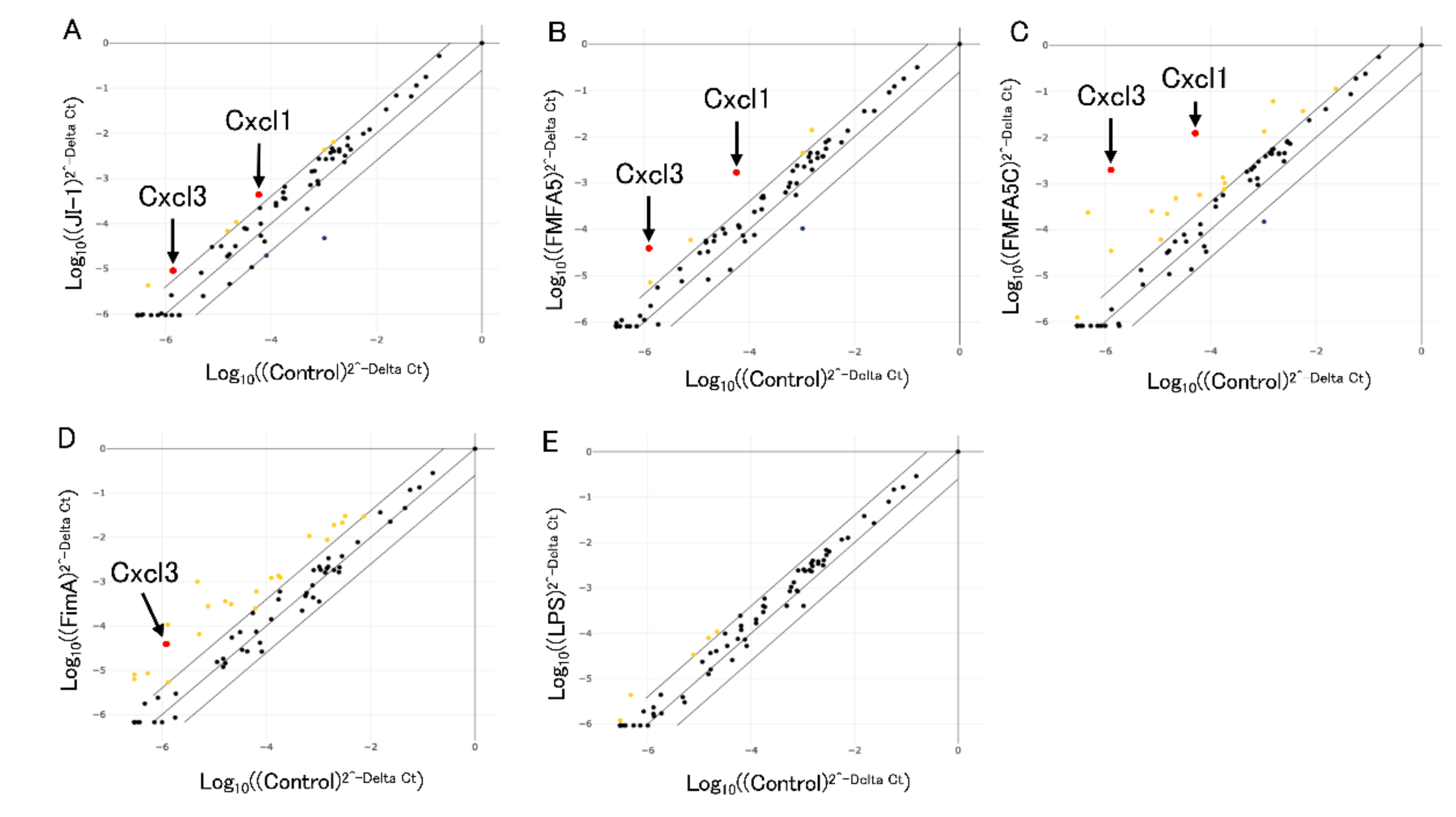
3.1. Analysis of Antibacterial Response-Associated Genes in Gingival Fibroblasts to Various Fimbriae
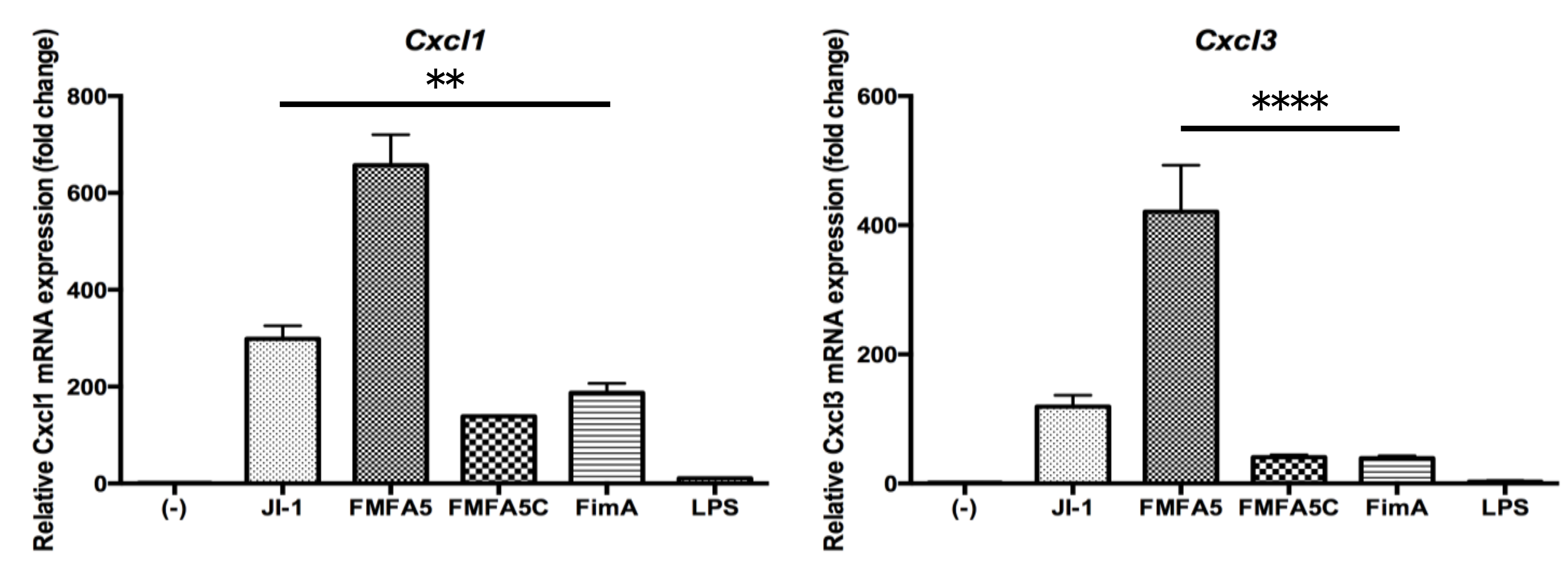
3.2. Confirmation of PCR Array Data for Selected Antibacterial Response-Associated Genes by Quantitative Real Time-PCR
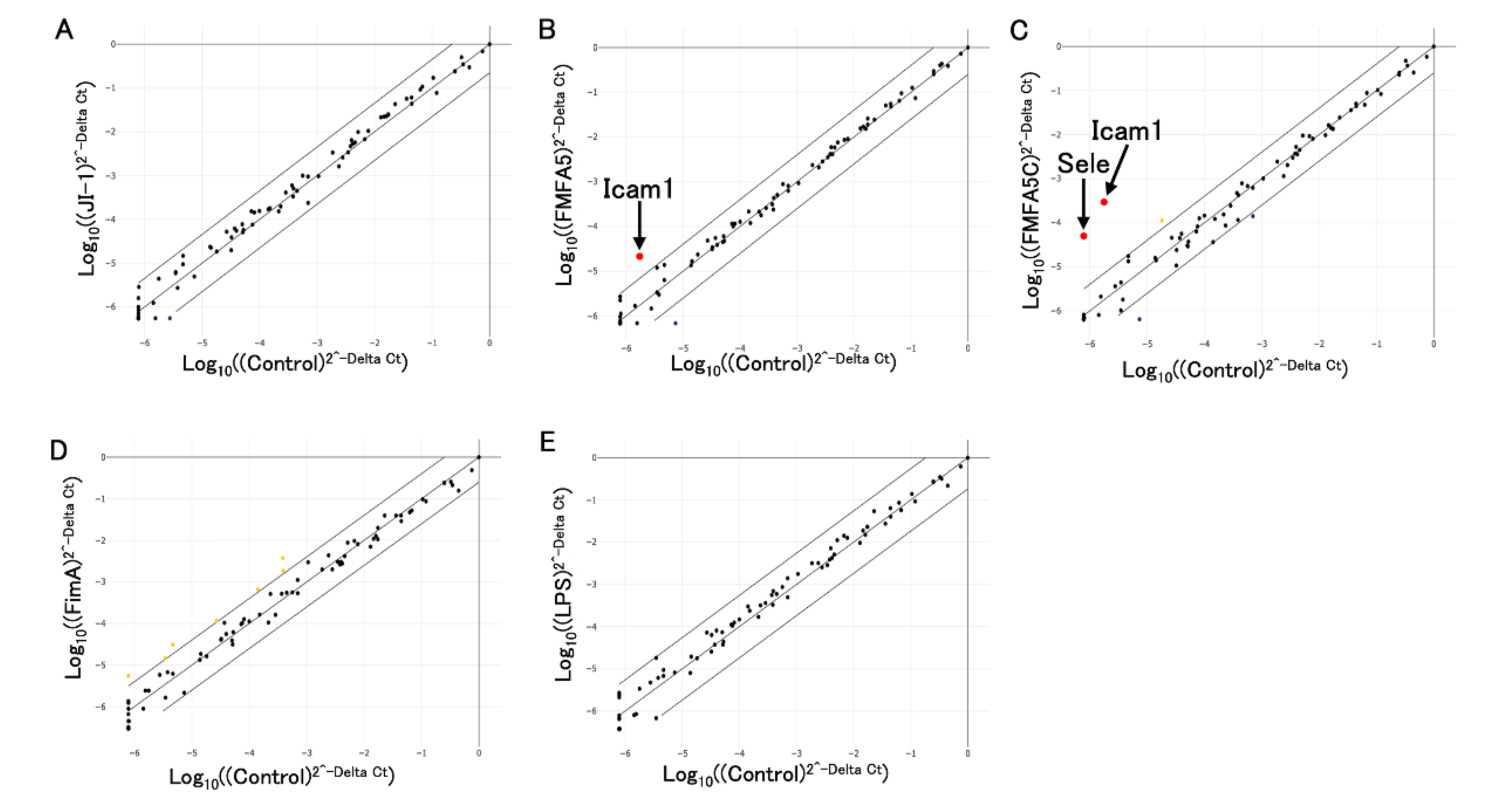
3.3. Analysis of Extracellular Matrix and Adhesion Molecule-Associated Genes in Gingival Fibroblasts in Response to Various Fimbriae
3.4. Confirmation of PCR Array Data for Selected Extracellular Matrix and Adhesion Molecule-Associated Genes by Quantitative RT-PCR
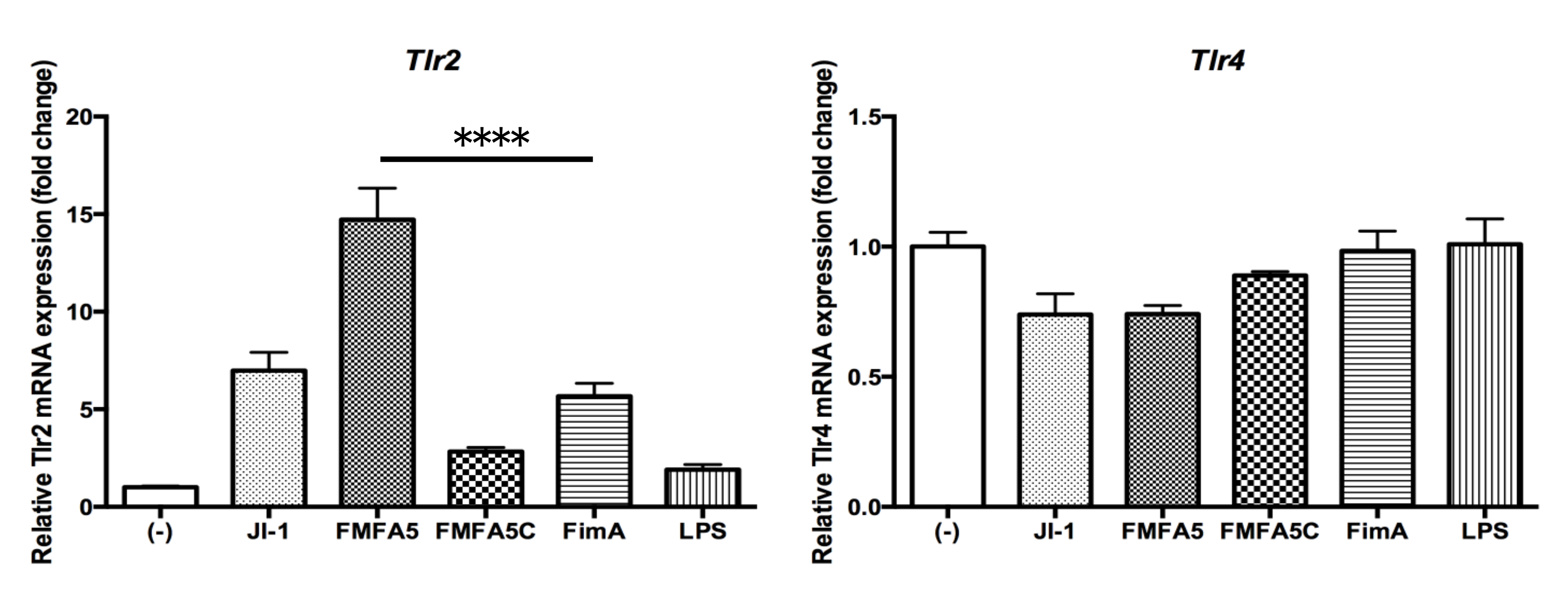
3.5. Induction of Tlr2 and Tlr4 Gene Expression by Various Fimbriae and LPS Stimulation in Gingival Fibroblasts
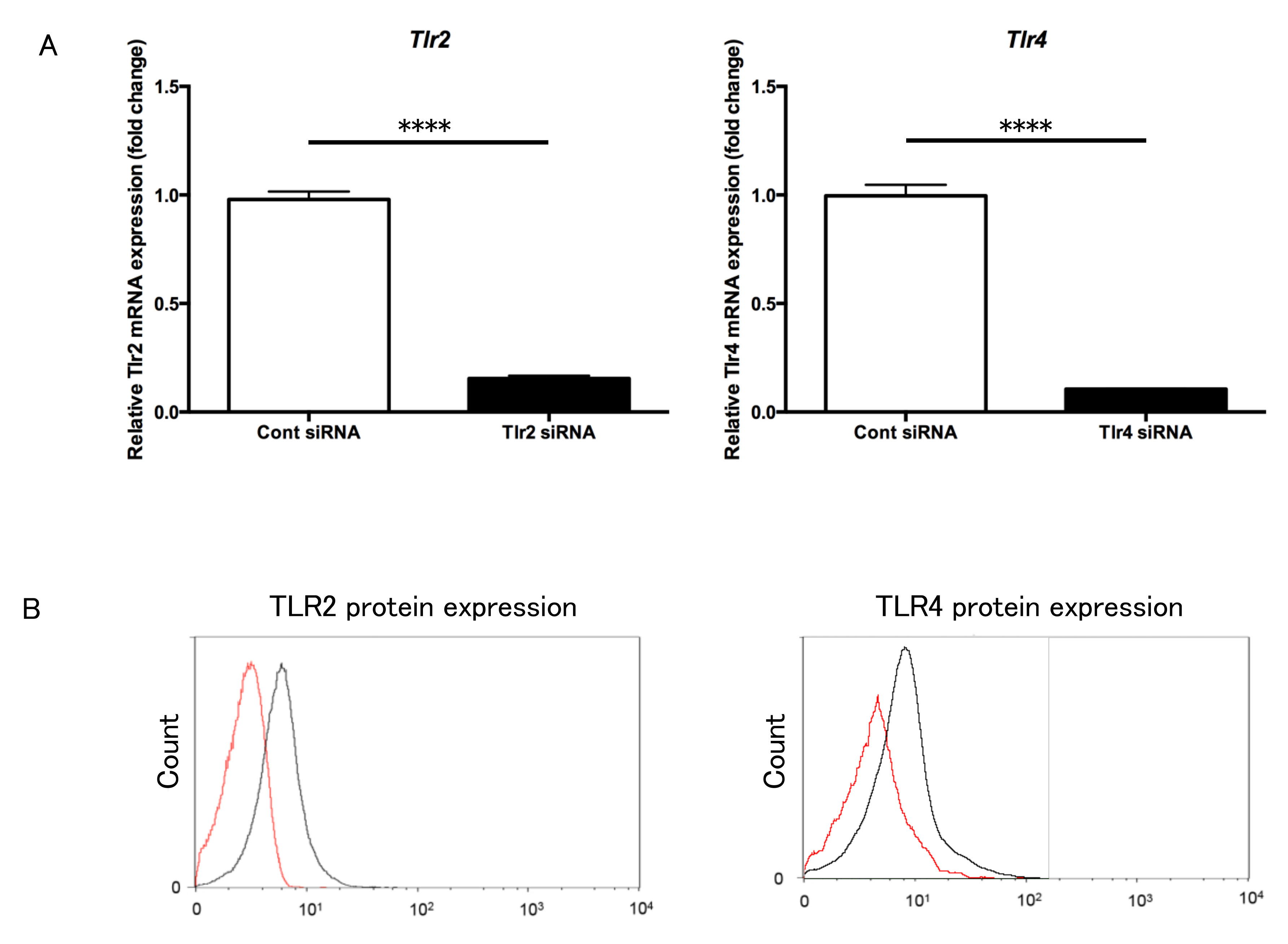
3.6. Transfection of Tlr2 and Tlr4 siRNA into Gingival Fibroblasts
3.7. Expression of Selected Genes in Tlr2 and Tlr4 siRNA-Transfected Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pihlstrom, B.L.; Michalowicz, B.S.; Johnson, N.W. Periodontal diseases. Lancet 2005, 366, 1809–1820. [Google Scholar] [CrossRef]
- Socransky, S.S.; Haffajee, A.D.; Cugini, M.A.; Smith, C.; Kent, R.L., Jr. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 1998, 25, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Riviere, G.R.; Smith, K.S.; Tzagaroulaki, E.; Kay, S.L.; Zhu, X.; DeRouen, T.A.; Adams, D.F. Periodontal status and detection frequency of bacteria at sites of periodontal health and gingivitis. J. Periodontol. 1996, 67, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, H.; Yoshimura, A.; Kaneko, T.; Golenbock, D.T.; Hara, Y. Analysis of the activity to induce toll-like receptor (TLR)2- and TLR4-mediated stimulation of supragingival plaque. J. Periodontol. 2008, 79, 920–928. [Google Scholar] [CrossRef]
- Darveau, R.P.; Hajishengallis, G.; Curtis, M.A. Porphyromonas gingivalis as a potential community activist for disease. J. Dent. Res. 2012, 91, 816–820. [Google Scholar] [CrossRef]
- Lamont, R.J.; Jenkinson, H.F. Life below the gum line: Pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 1998, 62, 1244–1263. [Google Scholar] [CrossRef]
- Hospenthal, M.K.; Costa, T.R.D.; Waksman, G. A comprehensive guide to pilus biogenesis in Gram-negative bacteria. Nat. Rev. Microbiol. 2017, 15, 365–379. [Google Scholar] [CrossRef]
- Lamont, R.J.; Jenkinson, H.F. Subgingival colonization by Porphyromonas gingivalis. Oral Microbiol. Immunol. 2000, 15, 341–349. [Google Scholar] [CrossRef]
- Yoshimura, F.; Murakami, Y.; Nishikawa, K.; Hasegawa, Y.; Kawaminami, S. Surface components of Porphyromonas gingivalis. J. Periodontal. Res. 2009, 44, 1–12. [Google Scholar] [CrossRef]
- Enersen, M.; Nakano, K.; Amano, A. Porphyromonas gingivalis fimbriae. J. Oral Microbiol. 2013, 5, 20265. [Google Scholar] [CrossRef]
- Ikai, R.; Hasegawa, Y.; Izumigawa, M.; Nagano, K.; Yoshida, Y.; Kitai, N.; Lamont, R.J.; Yoshimura, F.; Murakami, Y. Mfa4, an Accessory Protein of Mfa1 Fimbriae, Modulates Fimbrial Biogenesis, Cell Auto-Aggregation, and Biofilm Formation in Porphyromonas gingivalis. PLoS ONE 2015, 10, e0139454. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.; Hasegawa, Y.; Yoshimura, F.; Persson, K. Structural and functional characterization of shaft, anchor, and tip proteins of the Mfa1 fimbria from the periodontal pathogen Porphyromonas gingivalis. Sci. Rep. 2018, 8, 1793. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, O.; Watanabe, K.; Lamont, R.J. Involvement of integrins in fimbriae-mediated binding and invasion by Porphyromonas gingivalis. Cell Microbiol. 2002, 4, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Amano, A.; Nakagawa, I.; Okahashi, N.; Hamada, N. Variations of Porphyromonas gingivalis fimbriae in relation to microbial pathogenesis. J. Periodontol. Res. 2004, 39, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Hajishengallis, G.; Wang, M.; Liang, S.; Shakhatreh, M.A.; James, D.; Nishiyama, S.; Yoshimura, F.; Demuth, D.R. Subversion of innate immunity by periodontopathic bacteria via exploitation of complement receptor-3. Adv. Exp. Med. Biol. 2008, 632, 203–219. [Google Scholar] [CrossRef] [PubMed]
- Amano, A. Host-parasite interactions in periodontitis: Subgingival infection and host sensing. Periodontology 2000 2010, 52, 7–11. [Google Scholar] [CrossRef] [PubMed]
- Amano, A. Host-parasite interactions in periodontitis: Microbial pathogenicity and innate immunity. Periodontology 2000 2010, 54, 9–14. [Google Scholar] [CrossRef]
- Kuboniwa, M.; Amano, A.; Hashino, E.; Yamamoto, Y.; Inaba, H.; Hamada, N.; Nakayama, K.; Tribble, G.D.; Lamont, R.J.; Shizukuishi, S. Distinct roles of long/short fimbriae and gingipains in homotypic biofilm development by Porphyromonas gingivalis. BMC Microbiol. 2009, 9, 105. [Google Scholar] [CrossRef]
- Umemoto, T.; Hamada, N. Characterization of biologically active cell surface components of a periodontal pathogen. The roles of major and minor fimbriae of Porphyromonas gingivalis. J. Periodontol. 2003, 74, 119–122. [Google Scholar] [CrossRef]
- Hasegawa, Y.; Nagano, K.; Ikai, R.; Izumigawa, M.; Yoshida, Y.; Kitai, N.; Lamont, R.J.; Murakami, Y.; Yoshimura, F. Localization and function of the accessory protein Mfa3 in Porphyromonas gingivalis Mfa1 fimbriae. Mol. Oral Microbiol. 2013, 28, 467–480. [Google Scholar] [CrossRef]
- Hasegawa, Y.; Nagano, K.; Murakami, Y.; Lamont, R.J. Purification of Native Mfa1 Fimbriae from Porphyromonas gingivalis. Methods Mol. Biol. 2021, 2210, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, Y.; Iijima, Y.; Persson, K.; Nagano, K.; Yoshida, Y.; Lamont, R.J.; Kikuchi, T.; Mitani, A.; Yoshimura, F. Role of Mfa5 in Expression of Mfa1 Fimbriae in Porphyromonas gingivalis. J. Dent. Res. 2016, 95, 1291–1297. [Google Scholar] [CrossRef] [PubMed]
- Rath-Deschner, B.; Memmert, S.; Damanaki, A.; Nokhbehsaim, M.; Eick, S.; Cirelli, J.A.; Gotz, W.; Deschner, J.; Jager, A.; Nogueira, A.V.B. CXCL1, CCL2, and CCL5 modulation by microbial and biomechanical signals in periodontal cells and tissues-in vitro and in vivo studies. Clin. Oral Investig. 2020, 24, 3661–3670. [Google Scholar] [CrossRef] [PubMed]
- Sakai, A.; Ohshima, M.; Sugano, N.; Otsuka, K.; Ito, K. Profiling the cytokines in gingival crevicular fluid using a cytokine antibody array. J. Periodontol. 2006, 77, 856–864. [Google Scholar] [CrossRef] [PubMed]
- Buskermolen, J.K.; Roffel, S.; Gibbs, S. Stimulation of oral fibroblast chemokine receptors identifies CCR3 and CCR4 as potential wound healing targets. J. Cell. Physiol. 2017, 232, 2996–3005. [Google Scholar] [CrossRef] [PubMed]
- Miyauchi, M.; Kitagawa, S.; Hiraoka, M.; Saito, A.; Sato, S.; Kudo, Y.; Ogawa, I.; Takata, T. Immunolocalization of CXC chemokine and recruitment of polymorphonuclear leukocytes in the rat molar periodontal tissue after topical application of lipopolysaccharide. Histochem. Cell. Biol. 2004, 121, 291–297. [Google Scholar] [CrossRef]
- Hanazawa, S.; Murakami, Y.; Takeshita, A.; Kitami, H.; Ohta, K.; Amano, S.; Kitano, S. Porphyromonas gingivalis fimbriae induce expression of the neutrophil chemotactic factor KC gene of mouse peritoneal macrophages: Role of protein kinase C. Infect. Immun. 1992, 60, 1544–1549. [Google Scholar] [CrossRef]
- Baker, P.J.; DuFour, L.; Dixon, M.; Roopenian, D.C. Adhesion molecule deficiencies increase Porphyromonas gingivalis-induced alveolar bone loss in mice. Infect. Immun. 2000, 68, 3103–3107. [Google Scholar] [CrossRef]
- Niederman, R.; Westernoff, T.; Lee, C.; Mark, L.L.; Kawashima, N.; Ullman-Culler, M.; Dewhirst, F.E.; Paster, B.J.; Wagner, D.D.; Mayadas, T.; et al. Infection-mediated early-onset periodontal disease in P/E-selectin-deficient mice. J. Clin. Periodontol. 2001, 28, 569–575. [Google Scholar] [CrossRef]
- Takahashi, Y.; Davey, M.; Yumoto, H.; Gibson, F.C., 3rd; Genco, C.A. Fimbria-dependent activation of pro-inflammatory molecules in Porphyromonas gingivalis infected human aortic endothelial cells. Cell Microbiol. 2006, 8, 738–757. [Google Scholar] [CrossRef]
- Akira, S.; Takeda, K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004, 4, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Hajishengallis, G.; Lambris, J.D. Microbial manipulation of receptor crosstalk in innate immunity. Nat. Rev. Immunol. 2011, 11, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Burns, E.; Bachrach, G.; Shapira, L.; Nussbaum, G. Cutting Edge: TLR2 is required for the innate response to Porphyromonas gingivalis: Activation leads to bacterial persistence and TLR2 deficiency attenuates induced alveolar bone resorption. J. Immunol. 2006, 177, 8296–8300. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, N.; Yokoe, S.; Ogata, Y.; Sato, S.; Imai, K. Exposure to Porphyromonas gingivalis Induces Production of Proinflammatory Cytokine via TLR2 from Human Respiratory Epithelial Cells. J. Clin. Med. 2020, 9, 3433. [Google Scholar] [CrossRef]
- Darveau, R.P.; Pham, T.T.; Lemley, K.; Reife, R.A.; Bainbridge, B.W.; Coats, S.R.; Howald, W.N.; Way, S.S.; Hajjar, A.M. Porphyromonas gingivalis lipopolysaccharide contains multiple lipid A species that functionally interact with both toll-like receptors 2 and 4. Infect. Immun. 2004, 72, 5041–5051. [Google Scholar] [CrossRef]
- Olsen, I.; Singhrao, S.K. Importance of heterogeneity in Porhyromonas gingivalis lipopolysaccharide lipid A in tissue specific inflammatory signalling. J. Oral Microbiol. 2018, 10, 1440128. [Google Scholar] [CrossRef]
- Herath, T.D.K.; Darveau, R.P.; Seneviratne, C.J.; Wang, C.Y.; Wang, Y.; Jin, L. Heterogeneous Porphyromonas gingivalis LPS modulates immuno-inflammatory response, antioxidant defense and cytoskeletal dynamics in human gingival fibroblasts. Sci Rep. 2016, 6, 29829. [Google Scholar] [CrossRef]
- Andrukhov, O.; Ertlschweiger, S.; Moritz, A.; Bantleon, H.P.; Rausch-Fan, X. Different effects of P. gingivalis LPS and E. coli LPS on the expression of interleukin-6 in human gingival fibroblasts. Acta Odontol. Scand. 2014, 72, 337–345. [Google Scholar] [CrossRef]
- Herath, T.D.; Darveau, R.P.; Seneviratne, C.J.; Wang, C.Y.; Wang, Y.; Jin, L. Tetra- and penta-acylated lipid A structures of Porphyromonas gingivalis LPS differentially activate TLR4-mediated NF-kappaB signal transduction cascade and immuno-inflammatory response in human gingival fibroblasts. PLoS ONE 2013, 8, e58496. [Google Scholar] [CrossRef]
- Ogawa, T.; Asai, Y.; Hashimoto, M.; Uchida, H. Bacterial fimbriae activate human peripheral blood monocytes utilizing TLR2, CD14 and CD11a/CD18 as cellular receptors. Eur. J. Immunol. 2002, 32, 2543–2550. [Google Scholar] [CrossRef]
- Harokopakis, E.; Hajishengallis, G. Integrin activation by bacterial fimbriae through a pathway involving CD14, Toll-like receptor 2, and phosphatidylinositol-3-kinase. Eur. J. Immunol. 2005, 35, 1201–1210. [Google Scholar] [CrossRef]
- Tabeta, K.; Yamazaki, K.; Akashi, S.; Miyake, K.; Kumada, H.; Umemoto, T.; Yoshie, H. Toll-like receptors confer responsiveness to lipopolysaccharide from Porphyromonas gingivalis in human gingival fibroblasts. Infect. Immun. 2000, 68, 3731–3735. [Google Scholar] [CrossRef] [PubMed]
- Hiramine, H.; Watanabe, K.; Hamada, N.; Umemoto, T. Porphyromonas gingivalis 67-kDa fimbriae induced cytokine production and osteoclast differentiation utilizing TLR2. FEMS Microbiol. Lett. 2003, 229, 49–55. [Google Scholar] [CrossRef]
- Hajishengallis, G.; Martin, M.; Sojar, H.T.; Sharma, A.; Schifferle, R.E.; DeNardin, E.; Russell, M.W.; Genco, R.J. Dependence of bacterial protein adhesins on toll-like receptors for proinflammatory cytokine induction. Clin. Diagn. Lab. Immunol. 2002, 9, 403–411. [Google Scholar] [CrossRef]
- Aoki, Y.; Tabeta, K.; Murakami, Y.; Yoshimura, F.; Yamazaki, K. Analysis of immunostimulatory activity of Porphyromonas gingivalis fimbriae conferred by Toll-like receptor 2. Biochem. Biophys. Res. Commun. 2010, 398, 86–91. [Google Scholar] [CrossRef]
- Hashimoto, M.; Asai, Y.; Ogawa, T. Separation and structural analysis of lipoprotein in a lipopolysaccharide preparation from Porphyromonas gingivalis. Int. Immunol. 2004, 16, 1431–1437. [Google Scholar] [CrossRef]
- Asai, Y.; Hashimoto, M.; Fletcher, H.M.; Miyake, K.; Akira, S.; Ogawa, T. Lipopolysaccharide preparation extracted from Porphyromonas gingivalis lipoprotein-deficient mutant shows a marked decrease in toll-like receptor 2-mediated signaling. Infect. Immun. 2005, 73, 2157–2163. [Google Scholar] [CrossRef]
- Cai, J.; Chen, J.; Guo, H.; Pan, Y.; Zhang, Y.; Zhao, W.; Li, X.; Li, Y. Recombinant fimbriae protein of Porphyromonas gingivalis induces an inflammatory response via the TLR4/NFkappaB signaling pathway in human peripheral blood mononuclear cells. Int. J. Mol. Med. 2019, 43, 1430–1440. [Google Scholar] [CrossRef] [PubMed]
- Reife, R.A.; Coats, S.R.; Al-Qutub, M.; Dixon, D.M.; Braham, P.A.; Billharz, R.J.; Howald, W.N.; Darveau, R.P. Porphyromonas gingivalis lipopolysaccharide lipid A heterogeneity: Differential activities of tetra- and penta-acylated lipid A structures on E-selectin expression and TLR4 recognition. Cell Microbiol. 2006, 8, 857–868. [Google Scholar] [CrossRef] [PubMed]
- Ziauddin, S.M.; Montenegro Raudales, J.L.; Sato, K.; Yoshioka, H.; Ozaki, Y.; Kaneko, T.; Yoshimura, A.; Hara, Y. Analysis of Subgingival Plaque Ability to Stimulate Toll-Like Receptor 2 and 4. J. Periodontol. 2016, 87, 1083–1091. [Google Scholar] [CrossRef]
- El-Awady, A.R.; Miles, B.; Scisci, E.; Kurago, Z.B.; Palani, C.D.; Arce, R.M.; Waller, J.L.; Genco, C.A.; Slocum, C.; Manning, M.; et al. Porphyromonas gingivalis evasion of autophagy and intracellular killing by human myeloid dendritic cells involves DC-SIGN-TLR2 crosstalk. PLoS Pathog. 2015, 10, e1004647. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takayanagi, Y.; Kikuchi, T.; Hasegawa, Y.; Naiki, Y.; Goto, H.; Okada, K.; Okabe, I.; Kamiya, Y.; Suzuki, Y.; Sawada, N.; et al. Porphyromonas gingivalis Mfa1 Induces Chemokine and Cell Adhesion Molecules in Mouse Gingival Fibroblasts via Toll-Like Receptors. J. Clin. Med. 2020, 9, 4004. https://doi.org/10.3390/jcm9124004
Takayanagi Y, Kikuchi T, Hasegawa Y, Naiki Y, Goto H, Okada K, Okabe I, Kamiya Y, Suzuki Y, Sawada N, et al. Porphyromonas gingivalis Mfa1 Induces Chemokine and Cell Adhesion Molecules in Mouse Gingival Fibroblasts via Toll-Like Receptors. Journal of Clinical Medicine. 2020; 9(12):4004. https://doi.org/10.3390/jcm9124004
Chicago/Turabian StyleTakayanagi, Yuhei, Takeshi Kikuchi, Yoshiaki Hasegawa, Yoshikazu Naiki, Hisashi Goto, Kousuke Okada, Iichiro Okabe, Yosuke Kamiya, Yuki Suzuki, Noritaka Sawada, and et al. 2020. "Porphyromonas gingivalis Mfa1 Induces Chemokine and Cell Adhesion Molecules in Mouse Gingival Fibroblasts via Toll-Like Receptors" Journal of Clinical Medicine 9, no. 12: 4004. https://doi.org/10.3390/jcm9124004
APA StyleTakayanagi, Y., Kikuchi, T., Hasegawa, Y., Naiki, Y., Goto, H., Okada, K., Okabe, I., Kamiya, Y., Suzuki, Y., Sawada, N., Okabe, T., Suzuki, Y., Kondo, S., Ohno, T., Hayashi, J.-I., & Mitani, A. (2020). Porphyromonas gingivalis Mfa1 Induces Chemokine and Cell Adhesion Molecules in Mouse Gingival Fibroblasts via Toll-Like Receptors. Journal of Clinical Medicine, 9(12), 4004. https://doi.org/10.3390/jcm9124004




