Molecular Testing in CML between Old and New Methods: Are We at a Turning Point?
Abstract
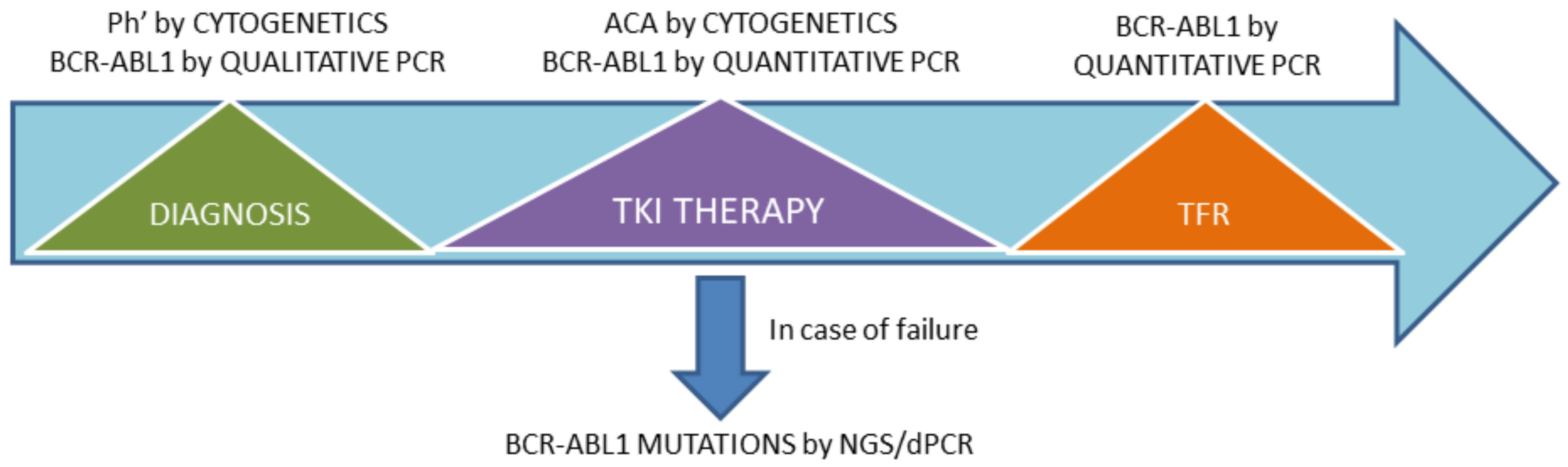
1. Introduction
2. MR Monitoring
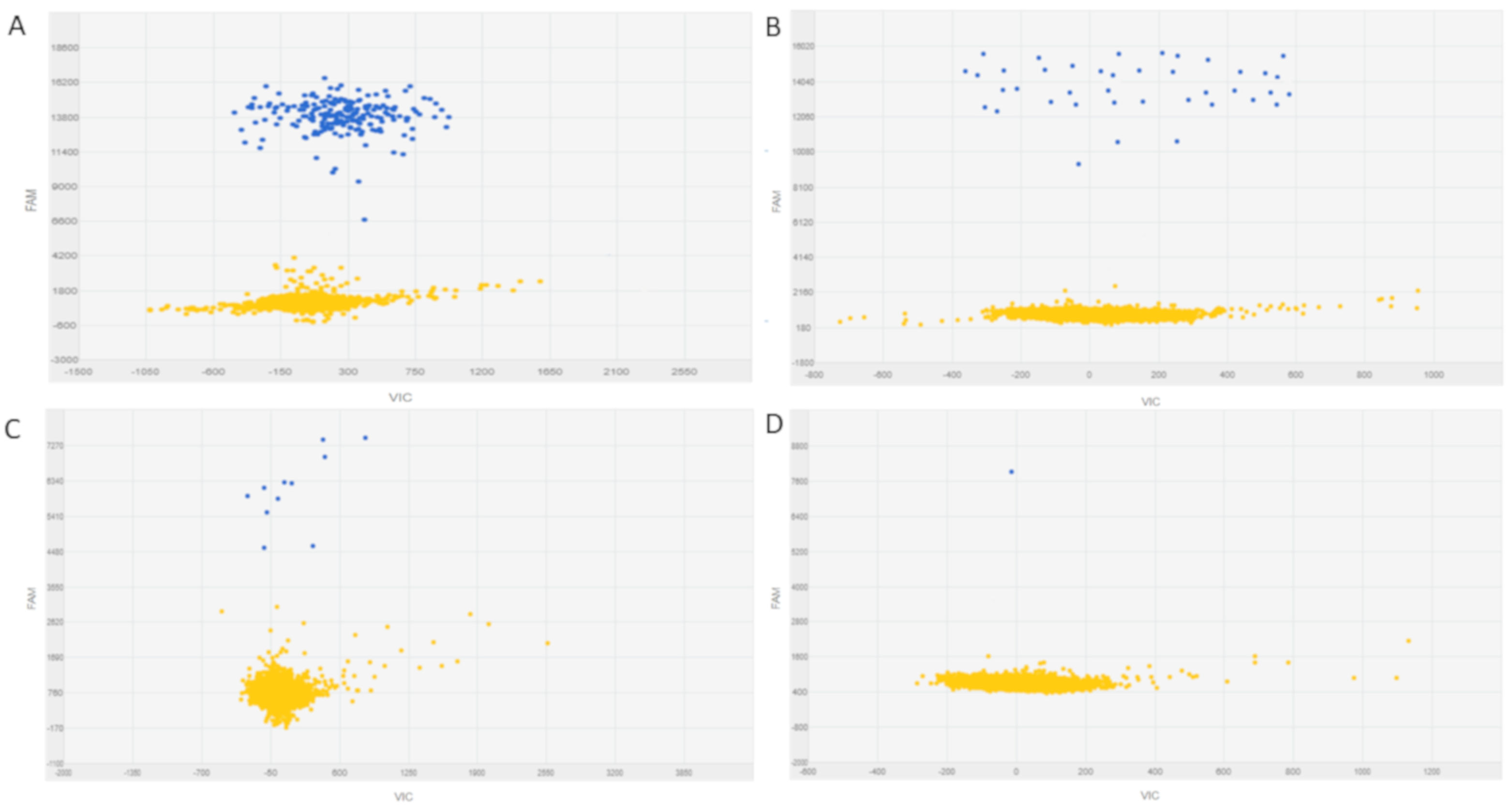
3. BCR-ABL1 Mutation Testing
4. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Soverini, S.; Mancini, M.; Bavaro, L.; Cavo, M.; Martinelli, G. Chronic myeloid leukemia: The paradigm of targeting oncogenic tyrosine kinase signaling and counteracting resistance for successful cancer therapy. Mol. Cancer 2018, 17, 1–15. [Google Scholar] [CrossRef]
- Hochhaus, A.; Baccarani, M.; Silver, R.T.; Schiffer, C.; Apperley, J.F.; Cervantes, F.; Clark, R.E.; Cortes, J.E.; Deininger, M.W.; Guilhot, F.; et al. European LeukemiaNet 2020 recommendations for treating chronic myeloid leukemia. Leukemia 2020, 34, 966–984. [Google Scholar] [CrossRef]
- O’Brien, S.G.; Guilhot, F.; Larson, R.A.; Gathmann, I.; Baccarani, M.; Cervantes, F.; Cornelissen, J.J.; Fischer, T.; Hochhaus, A.; Hughes, T.; et al. Imatinib Compared with Interferon and Low-Dose Cytarabine for Newly Diagnosed Chronic-Phase Chronic Myeloid Leukemia. N. Engl. J. Med. 2003, 348, 994–1004. [Google Scholar] [CrossRef] [PubMed]
- Baccarani, M.; Abruzzese, E.; Accurso, V.; Albano, F.; Annunziata, M.; Barulli, S.; Beltrami, G.; Bergamaschi, M.; Binotto, G.; Bocchia, M.; et al. Managing chronic myeloid leukemia for treatment-free remission: A proposal from the GIMEMA CML WP. Blood Adv. 2019, 3, 4280–4290. [Google Scholar] [CrossRef]
- Soverini, S.; Abruzzese, E.; Bocchia, M.; Bonifacio, M.; Galimberti, S.; Gozzini, A.; Iurlo, A.; Luciano, L.; Pregno, P.; Rosti, G.; et al. Next-generation sequencing for BCR-ABL1 kinase domain mutation testing in patients with chronic myeloid leukemia: A position paper. J. Hematol. Oncol. 2019, 12, 131. [Google Scholar] [CrossRef] [PubMed]
- Soverini, S.; Hochhaus, A.; Nicolini, F.E.; Gruber, F.; Lange, T.; Saglio, G.; Pane, F.; Müller, M.C.; Ernst, T.; Rosti, G.; et al. BCR-ABL kinase domain mutation analysis in chronic myeloid leukemia patients treated with tyrosine kinase inhibitors: Recommendations from an expert panel on behalf of European LeukemiaNet. Blood 2011, 118, 1208–1215. [Google Scholar] [CrossRef] [PubMed]
- Zabriskie, M.S.; Eide, C.A.; Tantravahi, S.K.; Vellore, N.A.; Estrada, J.; Nicolini, F.E.; Khoury, H.J.; Larson, R.A.; Konopleva, M.; Cortes, J.E.; et al. BCR-ABL1 Compound Mutations Combining Key Kinase Domain Positions Confer Clinical Resistance to Ponatinib in Ph Chromosome-Positive Leukemia. Cancer Cell 2014, 26, 428–442. [Google Scholar] [CrossRef] [PubMed]
- Dominy, K.; Mokretar, K.; Reid, A.G.; Khorashad, J.S. Molecular Monitoring of Chronic Myeloid Leukemia. Methods Mol. Biol. 2020, 2065, 153–173. [Google Scholar] [PubMed]
- Izzo, B.; Gottardi, E.M.; Errichiello, S.; Daraio, F.; Baratè, C.; Galimberti, S. Monitoring Chronic Myeloid Leukemia: How Molecular Tools May Drive Therapeutic Approaches. Front. Oncol. 2019, 9, 833. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, H.; Tabe, Y.; Ai, T.; Tsuchiya, K.; Yuri, M.; Misawa, S.; Horii, T.; Kawaguchi, A.; Ohsaka, A.; Kimura, S. A new highly sensitive real-time quantitative-PCR method for detection of BCR-ABL1 to monitor minimal residual disease in chronic myeloid leukemia after discontinuation of imatinib. PLoS ONE 2019, 14, e0207170. [Google Scholar] [CrossRef]
- Yoshida, C.; Nakamae, H.; Fletcher, L.; Koga, D.; Sogabe, T.; Matsumura, I.; Kanakura, Y.; Branford, S.; Naoe, T. Validation of a rapid one-step high sensitivity real-time quantitative PCR system for detecting major BCR-ABL1 mRNA on an International Scale. Springerplus 2016, 5, 569. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cumbo, C.; Anelli, L.; Specchia, G.; Albano, F. Monitoring of minimal residual disease (Mrd) in chronic myeloid leukemia: Recent advances. Cancer Manag. Res. 2020, 12, 3175–3189. [Google Scholar] [CrossRef] [PubMed]
- Cilloni, D.; Petiti, J.; Rosso, V.; Andreani, G.; Dragani, M.; Fava, C.; Saglio, G. Digital PCR in myeloid malignancies: Ready to replace quantitative PCR? Int. J. Mol. Sci. 2019, 20, 2249. [Google Scholar] [CrossRef] [PubMed]
- Franke, G.N.; Maier, J.; Wildenberger, K.; Cross, M.; Giles, F.J.; Müller, M.C.; Hochhaus, A.; Niederwieser, D.; Lange, T. Comparison of Real-Time Quantitative PCR and Digital Droplet PCR for BCR-ABL1 Monitoring in Patients with Chronic Myeloid Leukemia. J. Mol. Diagn. 2020, 22, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Coccaro, N.; Tota, G.; Anelli, L.; Zagaria, A.; Specchia, G.; Albano, F. Digital PCR: A Reliable Tool for Analyzing and Monitoring Hematologic Malignancies. Int. J. Mol. Sci. 2020, 21, 3141. [Google Scholar] [CrossRef] [PubMed]
- Kanagal-Shamanna, R. Digital PCR: Principles and applications. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2016; Volume 1392, pp. 43–50. [Google Scholar]
- Quan, P.-L.; Sauzade, M.; Brouzes, E. dPCR: A Technology Review. Sensors 2018, 18, 1271. [Google Scholar] [CrossRef]
- Guerrini, F.; Paolicchi, M.; Ghio, F.; Ciabatti, E.; Grassi, S.; Salehzadeh, S.; Ercolano, G.; Metelli, M.R.; del Re, M.; Iovino, L.; et al. The Droplet Digital PCR: A New Valid Molecular Approach for the Assessment of B-RAF V600E Mutation in Hairy Cell Leukemia. Front. Pharmacol. 2016, 7, 363. [Google Scholar] [CrossRef]
- Fontanelli, G.; Baratè, C.; Ciabatti, E.; Guerrini, F.; Grassi, S.; del Re, M.; Morganti, R.; Petrini, I.; Arici, R.; Barsotti, S.; et al. Real-Time PCR and Droplet Digital PCR: Two techniques for detection of the JAK2V617F mutation in Philadelphia-negative chronic myeloproliferative neoplasms. Int. J. Lab. Hematol. 2015, 37, 766–773. [Google Scholar] [CrossRef]
- Della Starza, I.; de Novi, L.A.; Santoro, A.; Salemi, D.; Tam, W.; Cavalli, M.; Menale, L.; Soscia, R.; Apicella, V.; Ilari, C.; et al. Digital droplet PCR and next-generation sequencing refine minimal residual disease monitoring in acute lymphoblastic leukemia. Leuk. Lymphoma 2019, 60, 2838–2840. [Google Scholar] [CrossRef]
- Cavalli, M.; de Novi, L.A.; della Starza, I.; Cappelli, L.V.; Nunes, V.; Pulsoni, A.; del Giudice, I.; Guarini, A.; Foà, R. Comparative analysis between RQ-PCR and digital droplet PCR of BCL2/IGH gene rearrangement in the peripheral blood and bone marrow of early stage follicular lymphoma. Br. J. Haematol. 2017, 177, 588–596. [Google Scholar] [CrossRef]
- Cross, N.C.P.; White, H.E.; Colomer, D.; Ehrencrona, H.; Foroni, L.; Gottardi, E.; Lange, T.; Lion, T.; Machova Polakova, K.; Dulucq, S.; et al. Laboratory recommendations for scoring deep molecular responses following treatment for chronic myeloid leukemia. Leukemia 2015, 29, 999–1003. [Google Scholar] [CrossRef] [PubMed]
- Mori, S.; Vagge, E.; Le Coutre, P.; Abruzzese, E.; Martino, B.; Pungolino, E.; Elena, C.; Pierri, I.; Assouline, S.; D’Emilio, A.; et al. Age and dPCR can predict relapse in CML patients who discontinued imatinib: The ISAV study. Am. J. Hematol. 2015, 90, 910–914. [Google Scholar] [CrossRef] [PubMed]
- Berdeja, J.G.; Heinrich, M.C.; Dakhil, S.R.; Goldberg, S.L.; Wadleigh, M.; Kuriakose, P.; Cortes, J.; Radich, J.; Helton, B.; Rizzieri, D.; et al. Rates of deep molecular response by digital and conventional PCR with frontline nilotinib in newly diagnosed chronic myeloid leukemia: A landmark analysis. Leuk. Lymphoma 2019, 60, 2384–2393. [Google Scholar] [CrossRef] [PubMed]
- Goh, H.G.; Lin, M.; Fukushima, T.; Saglio, G.; Kim, D.; Choi, S.Y.; Kim, S.H.; Lee, J.; Lee, Y.S.; Oh, S.M.; et al. Sensitive quantitation of minimal residual disease in chronic myeloid leukemia using nanofluidic digital polymerase chain reaction assay. Leuk. Lymphoma 2011, 52, 896–904. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-J.; Zheng, C.-F.; Liu, Z.; Tan, Y.-H.; Chen, X.-H.; Zhao, B.-L.; Li, G.-X.; Xu, Z.-F.; Ren, F.-G.; Zhang, Y.-F.; et al. Droplet digital PCR for BCR/ABL(P210) detection of chronic myeloid leukemia: A high sensitive method of the minimal residual disease and disease progression. Eur. J. Haematol. 2018, 101, 291–296. [Google Scholar] [CrossRef]
- Dello Sbarba, P.; Rovida, E.; Marzi, I.; Cipolleschi, M.G. One more stem cell niche: How the sensitivity of chronic myeloid leukemia cells to imatinib mesylate is modulated within a “hypoxic” environment. Hypoxia 2014, 214, 1. [Google Scholar] [CrossRef][Green Version]
- Krumbholz, M.; Karl, M.; Tauer, J.T.; Thiede, C.; Rascher, W.; Suttorp, M.; Metzler, M. Genomic BCR-ABL1 breakpoints in pediatric chronic myeloid leukemia. Genes Chromosomes Cancer 2012, 51, 1045–1053. [Google Scholar] [CrossRef]
- Krumbholz, M.; Goerlitz, K.; Albert, C.; Lawlor, J.; Suttorp, M.; Metzler, M. Large amplicon droplet digital PCR for DNA-based monitoring of pediatric chronic myeloid leukaemia. J. Cell. Mol. Med. 2019, 23, 4955–4961. [Google Scholar] [CrossRef]
- Lund, H.L.; Hughesman, C.B.; McNeil, K.; Clemens, S.; Hocken, K.; Pettersson, R.; Karsan, A.; Foster, L.J.; Haynes, C. Initial diagnosis of chronic myelogenous leukemia based on quantification of M-BCR status using droplet digital PCR. Anal. Bioanal. Chem. 2016, 408, 1079–1094. [Google Scholar] [CrossRef]
- Cumbo, C.; Impera, L.; Minervini, C.F.; Orsini, P.; Anelli, L.; Zagaria, A.; Coccaro, N.; Tota, G.; Minervini, A.; Casieri, P.; et al. Genomic BCR-ABL1 breakpoint characterization by a multistrategy approach for “personalized monitoring” of residual disease in chronic myeloid leukemia patients. Oncotarget 2018, 9, 10978–10986. [Google Scholar] [CrossRef]
- Bernardi, S.; Foroni, C.; Zanaglio, C.; Re, F.; Polverelli, N.; Turra, A.; Morello, E.; Farina, M.; Cattina, F.; Gandolfi, L.; et al. Feasibility of tumor-derived exosome enrichment in the onco-hematology leukemic model of chronic myeloid leukemia. Int. J. Mol. Med. 2019, 44, 2133–2144. [Google Scholar] [CrossRef] [PubMed]
- Bernardi, S.; Malagola, M.; Polverelli, N.; Russo, D. Exosomes in Chronic Myeloid Leukemia: Are We Reading a New Reliable Message? Acta Haematol. 2020, 143, 509–510. [Google Scholar] [CrossRef]
- Corrado, C.; Saieva, L.; Raimondo, S.; Santoro, A.; de Leo, G.; Alessandro, R. Chronic myelogenous leukaemia exosomes modulate bone marrow microenvironment through activation of epidermal growth factor receptor. J. Cell. Mol. Med. 2016, 20, 1829–1839. [Google Scholar] [CrossRef] [PubMed]
- Bernardi, S.; Ruggieri, G.; Malagola, M.; Cancelli, V.; Cattina, F.; Polverelli, N.; Zanaglio, C.; Perucca, S.; Re, F.; Montanelli, A.; et al. Digital PCR (Dpcr) a Step Forward to Detection and Quantification of Minimal Residual Disease (MRD) in Ph+/BCR-ABL1 Chronic Myeloid Leukemia (CML). J. Mol. Biomark. Diagn. 2017, 8, 1–3. [Google Scholar] [CrossRef]
- Bernardi, S.; Malagola, M.; Zanaglio, C.; Polverelli, N.; Dereli Eke, E.; D’Adda, M.; Farina, M.; Bucelli, C.; Scaffidi, L.; Toffoletti, E.; et al. Digital PCR improves the quantitation of DMR and the selection of CML candidates to TKIs discontinuation. Cancer Med. 2019, 8, 2041–2055. [Google Scholar] [CrossRef]
- Nicolini, F.E.; Dulucq, S.; Boureau, L.; Cony-Makhoul, P.; Charbonnier, A.; Escoffre-Barbe, M.; Rigal-Huguet, F.; Coiteux, V.; Varet, B.; Dubruille, V.; et al. Evaluation of residual disease and TKI duration are predictive factors for molecular recurrence after stopping Imatinib first-line in chronic phase CML Patients. Clin. Cancer Res. 2019, 25, 6606–6613. [Google Scholar] [CrossRef]
- Abruzzese, E.; Bocchia, M.; Bernardi, S.; Trawinska, M.M.; Raspadori, D.; Zanaglio, C.; Bondanini, F.; Sicuranza, A.; Pacelli, P.; Malagola, M.; et al. Minimal Residual Disease Detection at RNA and Leukemic Stem Cell (LSC) Level. Comparison of Qpcr, d-PCR and CD26 Stem Cell Measurements in Chronic Myeloid Leukemia (CML) Patients in Deep Molecular Response (DMR). Blood 2018, 132, 4244. [Google Scholar] [CrossRef]
- Dueck, M.E.; Lin, R.; Zayac, A.; Gallagher, S.; Chao, A.K.; Jiang, L.; Datwani, S.S.; Hung, P.; Stieglitz, E. Precision cancer monitoring using a novel, fully integrated, microfluidic array partitioning digital PCR platform. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef]
- Baccarani, M.; Castagnetti, F.; Gugliotta, G.; Rosti, G.; Soverini, S.; Albeer, A.; Pfirrmann, M. The proportion of different BCR-ABL1 transcript types in chronic myeloid leukemia. An international overview. Leukemia 2019, 33, 1173–1183. [Google Scholar] [CrossRef]
- Kjaer, L.; Skov, V.; Andersen, M.T.; Aggerholm, A.; Clair, P.; Gniot, M.; Soeby, K.; Udby, L.; Dorff, M.H.; Hasselbalch, H.; et al. Variant-specific discrepancy when quantitating BCR-ABL1 e13a2 and e14a2 transcripts using the Europe Against Cancer qPCR assay. Eur. J. Haematol. 2019, 103, 26–34. [Google Scholar] [CrossRef]
- Bernardi, S.; Bonifacio, M.; Iurlo, A.; Zanaglio, C.; Tiribelli, M.; Binotto, G.; Abruzzese, E.; Russo, D. “Variant-specific discrepancy when quantitating BCR-ABL1 e13a2 and e14a2 transcripts using the Europe Against Cancer qPCR assay.” Is dPCR the key? Eur. J. Haematol. 2019, 103, 272–273. [Google Scholar] [CrossRef]
- Chung, H.J.; Hur, M.; Yoon, S.; Hwang, K.; Lim, H.S.; Kim, H.; Moon, H.W.; Yun, Y.M. Performance evaluation of the QXdX BCR-ABL %Is droplet digital PCR assay. Ann. Lab. Med. 2020, 40, 72–75. [Google Scholar] [CrossRef]
- Maier, J.; Lange, T.; Cross, M.; Wildenberger, K.; Niederwieser, D.; Franke, G.N. Optimized Digital Droplet PCR for BCR-ABL. J. Mol. Diagn. 2019, 21, 27–37. [Google Scholar] [CrossRef]
- Fava, C.; Bernardi, S.; Gottardi, E.M.; Daraio, F.; Giugliano, E.; Lorenzatti, R.; Varotto, M.; Barberio, D.; Galeotti, L.; Ceccherini, F.; et al. Standardization of Two Dpcr Platforms for Detection of BCR/ABL1—Minimal Residual Disease (MRD) in Ph+ Chronic Myeloid Leukemia (CML). Blood 2017, 130, 2867. [Google Scholar]
- Asghari, H.; Talati, C.; Achille, A.; Powers, J.J.; Sahakian, E.; Chan, O.; Shelton, D.N.; Isenalumhe, L.; Nodzon, L.; Sweet, K.L.; et al. Conventional Real Time Quantitative Polymerase Chain Reaction Method Yields Similar Level of Sensitivity to Digital Droplet Polymerase Chain Reaction for Detection of BCR-ABL p210 Transcripts in Patients with Chronic Phase Chronic Myeloid Leukemia. Blood 2019, 134, 3382. [Google Scholar] [CrossRef]
- Brown, J.T.; Beldorth, I.J.; Laosinchai-Wolf, W.; Fahey, M.E.; Jefferson, K.L.; Ruskin, A.K.; Roth, J.J.; Cai, L.; Watt, C.D.; Press, R.D.; et al. Analytical Validation of a Highly Sensitive, Multiplexed Chronic Myeloid Leukemia Monitoring System Targeting BCR-ABL1 RNA. J. Mol. Diagn. 2019, 21, 718–733. [Google Scholar] [CrossRef]
- Scott, S.; Cartwright, A.; Tapley, A.; Boeckx, N.; Cayuela, J.M.; Corner, A.; Dulucq, S.; Galimberti, S.; Lauricella, C.; Rose, S.; et al. Digital PCR For The Measurement of BCR-ABL1 In CML: A New Dawn? EHA Library 2020, 294652. [Google Scholar]
- Spiess, B.; Rinaldetti, S.; Naumann, N.; Galuschek, N.; Kossak-Roth, U.; Wuchter, P.; Tarnopolscaia, I.; Rose, D.; Voskanyan, A.; Fabarius, A.; et al. Diagnostic performance of the molecular BCR-ABL1 monitoring system may impact on inclusion of CML patients in stopping trials. PLoS ONE 2019, 14, e0214305. [Google Scholar] [CrossRef]
- Zanaglio, C.; Bernardi, S.; Gandolfi, L.; Farina, M.; Re, F.; Polverelli, N.; Zollner, T.; Turra, A.; Morello, E.; Malagola, M.; et al. RT-qPCR versus Digital PCR: How Do They Impact Differently on Clinical Management of Chronic Myeloid Leukemia Patients? Case Rep. Oncol. 2020, 13, 1263–1269. [Google Scholar] [CrossRef]
- Russo, D.; Malagola, M.; Skert, C.; Cancelli, V.; Turri, D.; Pregno, P.; Bergamaschi, M.; Fogli, M.; Testoni, N.; de Vivo, A.; et al. Managing chronic myeloid leukaemia in the elderly with intermittent imatinib treatment. Blood Cancer J. 2015, 5, e347. [Google Scholar] [CrossRef]
- Machova Polakova, K.; Kulvait, V.; Benesova, A.; Linhartova, J.; Klamova, H.; Jaruskova, M.; de Benedittis, C.; Haferlach, T.; Baccarani, M.; Martinelli, G.; et al. Next-generation deep sequencing improves detection of BCR-ABL1 kinase domain mutations emerging under tyrosine kinase inhibitor treatment of chronic myeloid leukemia patients in chronic phase. J. Cancer Res. Clin. Oncol. 2015, 141, 887–899. [Google Scholar] [CrossRef]
- Baer, C.; Kern, W.; Koch, S.; Nadarajah, N.; Schindela, S.; Meggendorfer, M.; Haferlach, C.; Haferlach, T. Ultra-deep sequencing leads to earlier and more sensitive detection of the tyrosine kinase inhibitor resistance mutation T315I in chronic myeloid leukemia. Haematologica 2016, 101, 830–838. [Google Scholar] [CrossRef]
- Soverini, S.; de Benedittis, C.; Castagnetti, F.; Gugliotta, G.; Mancini, M.; Bavaro, L.; Machova Polakova, K.; Linhartova, J.; Iurlo, A.; Russo, D.; et al. In chronic myeloid leukemia patients on second-line tyrosine kinase inhibitor therapy, deep sequencing of BCR-ABL1 at the time of warning may allow sensitive detection of emerging drug-resistant mutants. BMC Cancer 2016, 16, 572. [Google Scholar] [CrossRef]
- Soverini, S.; de Benedittis, C.; Polakova, K.M.; Brouckova, A.; Horner, D.; Iacono, M.; Castagnetti, F.; Gugliotta, G.; Palandri, F.; Papayannidis, C.; et al. Unraveling the complexity of tyrosine kinase inhibitor-resistant populations by ultra-deep sequencing of the BCR-ABL kinase domain. Blood 2013, 122, 1634–1648. [Google Scholar] [CrossRef]
- Soverini, S.; de Benedittis, C.; Polakova, K.M.; Linhartova, J.; Castagnetti, F.; Gugliotta, G.; Papayannidis, C.; Mancini, M.; Klamova, H.; Salvucci, M.; et al. Next-generation sequencing for sensitive detection of BCR-ABL1 mutations relevant to tyrosine kinase inhibitor choice in imatinib-resistant patients. Oncotarget 2016, 7, 21982–21990. [Google Scholar] [CrossRef]
- Kizilors, A.; Crisà, E.; Lea, N.; Passera, R.; Mian, S.; Anwar, J.; Best, S.; Nicolini, F.E.; Ireland, R.; Aldouri, M.; et al. Effect of low-level BCR-ABL1 kinase domain mutations identified by next-generation sequencing in patients with chronic myeloid leukaemia: A population-based study. Lancet Haematol. 2019, 6, e276–e284. [Google Scholar] [CrossRef]
- Soverini, S.; Bavaro, L.; de Benedittis, C.; Martelli, M.; Iurlo, A.; Orofino, N.; Sica, S.; Sorà, F.; Lunghi, F.; Ciceri, F.; et al. Prospective assessment of NGS-detectable mutations in CML patients with nonoptimal response: The NEXT-in-CML study. Blood 2020, 135, 534–541. [Google Scholar] [CrossRef]
- Baccarani, M.; Deininger, M.W.; Rosti, G.; Hochhaus, A.; Soverini, S.; Apperley, J.F.; Cervantes, F.; Clark, R.E.; Cortes, J.E.; Guilhot, F.; et al. European LeukemiaNet recommendations for the management of chronic myeloid leukemia: 2013. Blood 2013, 122, 872–884. [Google Scholar] [CrossRef]
- Cayuela, J.M.; Chomel, J.C.; Coiteux, V.; Dulucq, S.; Escoffre-Barbe, M.; Etancelin, P.; Etienne, G.; Hayette, S.; Millot, F.; Nibourel, O.; et al. Recommendations from the French CML Study Group (Fi-LMC) for BCR-ABL1 kinase domain mutation analysis in chronic myeloid leukemia. Bull. Cancer 2020, 107, 113–128. [Google Scholar] [CrossRef]
- Schmitt, M.W.; Pritchard, J.R.; Leighow, S.M.; Aminov, B.I.; Beppu, L.; Kim, D.S.; Hodgson, J.G.; Rivera, V.M.; Loeb, L.A.; Radich, J.P. Single-molecule sequencing reveals patterns of preexisting drug resistance that suggest treatment strategies in Philadelphia-positive leukemias. Clin. Cancer Res. 2018, 24, 5321–5334. [Google Scholar] [CrossRef]
- Minervini, C.F.; Cumbo, C.; Orsini, P.; Anelli, L.; Zagaria, A.; Impera, L.; Coccaro, N.; Brunetti, C.; Minervini, A.; Casieri, P.; et al. Mutational analysis in BCR-ABL1 positive leukemia by deep sequencing based on nanopore MinION technology. Exp. Mol. Pathol. 2017, 103, 33–37. [Google Scholar] [CrossRef]
- Petiti, J.; Rosso, V.; Croce, E.; Franceschi, V.; Andreani, G.; Dragani, M.; de Gobbi, M.; Lunghi, M.; Saglio, G.; Fava, C.; et al. Highly Sensitive Detection of IDH2 Mutations in Acute Myeloid Leukemia. J. Clin. Med. 2020, 9, 271. [Google Scholar] [CrossRef]
- Grassi, S.; Guerrini, F.; Ciabatti, E.; Puccetti, R.; Salehzadeh, S.; Metelli, M.R.; Di Vita, A.; Domenichini, C.; Caracciolo, F.; Orciuolo, E.; et al. Digital Droplet PCR is a Specific and Sensitive Tool for Detecting IDH2 Mutations in Acute Myeloid LeuKemia Patients. Cancers 2020, 12, 1738. [Google Scholar] [CrossRef]
- Drandi, D.; Genuardi, E.; Dogliotti, I.; Ferrante, M.; Jiménez, C.; Guerrini, F.; Lo Schirico, M.; Mantoan, B.; Muccio, V.; Lia, G.; et al. Highly sensitive MYD88 l265p mutation detection by droplet digital polymerase chain reaction in waldenström macroglobulinemia. Haematologica 2018, 103, 1029–1037. [Google Scholar] [CrossRef]
- Zocco, D.; Bernardi, S.; Novelli, M.; Astrua, C.; Fava, P.; Zarovni, N.; Carpi, F.M.; Bianciardi, L.; Malavenda, O.; Quaglino, P.; et al. Isolation of extracellular vesicles improves the detection of mutant DNA from plasma of metastatic melanoma patients. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Tan, Y.; Liu, Z.; Wang, W.; Zhu, G.; Guo, J.; Chen, X.; Zheng, C.; Xu, Z.; Chang, J.; Ren, F.; et al. Monitoring of clonal evolution of double C-KIT exon 17 mutations by Droplet Digital PCR in patients with core-binding factor acute myeloid leukemia. Leuk. Res. 2018, 69, 89–93. [Google Scholar] [CrossRef]
- Akahoshi, Y.; Nakasone, H.; Kawamura, K.; Kusuda, M.; Kawamura, S.; Takeshita, J.; Yoshino, N.; Misaki, Y.; Yoshimura, K.; Gomyo, A.; et al. Detection of T315I using digital polymerase chain reaction in allogeneic transplant recipients with Ph-positive acute lymphoblastic anemia in the dasatinib era. Exp. Hematol. 2020, 81, 60–67. [Google Scholar] [CrossRef]
- Soverini, S.; Martelli, M.; Bavaro, L.; de Benedittis, C.; Iurlo, A.; Galimberti, S.; Pregno, P.; Bonifacio, M.; Lunghi, F.; Castagnetti, F.; et al. Detection of Actionable BCR-ABL1 Kinase Domain (KD) Mutations in Chronic Myeloid Leukemia (CML) Patients with Failure and Warning Response to Tyrosine Kinase Inhibitors (TKIs): Potential Impact of Next-Generation Sequencing (NGS) and Droplet Digital PCR (ddPCR) on Clinical Decision Making. Blood 2019, 134, 661. [Google Scholar]
- Vannuffel, P.; Bavaro, L.; Nollet, F.; Aynaci, A.; Martelli, M.; Devos, H.; de Rop, C.; Soverini, S. Droplet Digital PCR Phasing (DROP-PHASE): A Novel Method for Straightforward Detection of BCR-ABL1 Compound Mutations in Tyrosine Kinase Inhibitors Resistant Chronic Myeloid Leukemia (CML) and Acute Lymphoblastic Leukemia (ALL). Blood 2019, 134, 4660. [Google Scholar] [CrossRef]
- Galimberti, S.; Guerrini, F.; Grassi, S.; Bocchia, M.; Bavaro, L.; Ciabatti, E.; Dragani, M.; Gottardi, E.M.; Izzo, B.; Lunghi, F.; et al. Digital Droplet PCR is a fast and effective tool for detecting T315I mutation in Chronic Myeloid Leukemia. EHA Library 2020, 294653. [Google Scholar]
- Daraio, F.; Giugliano, E.; Fava, C.; Lorenzatti, R.; Varotto, M.; Barberio, D.; Bernardi, S.; Izzo, B.; Errichiello, S.; Bochicchio, M.T.; et al. An Italian Multicentre Study Using Different Digital PCR Instruments on BCR-ABL1 Positive Patients at Different Levels of CML Disease. Blood 2017, 130, 4176. [Google Scholar]
- Branford, S.; Wang, P.; Yeung, D.T.; Thomson, D.; Purins, A.; Wadham, C.; Shahrin, N.H.; Marum, J.E.; Nataren, N.; Parker, W.T.; et al. Integrative genomic analysis reveals cancer-associated mutations at diagnosis of CML in patients with high-risk disease. Blood 2018, 132, 948–961. [Google Scholar] [CrossRef]
- Branford, S.; Kim, D.D.H.; Apperley, J.F.; Eide, C.A.; Mustjoki, S.; Ong, S.T.; Nteliopoulos, G.; Ernst, T.; Chuah, C.; Gambacorti-Passerini, C.; et al. Laying the foundation for genomically-based risk assessment in chronic myeloid leukemia. Leukemia 2019, 33, 1835–1850. [Google Scholar] [CrossRef]
- Morello, E.; Malagola, M.; Bernardi, S.; Pristipino, C.; Russo, D. The role of allogeneic hematopoietic stem cell transplantation in the four P medicine era. Blood Res. 2018, 53, 3–6. [Google Scholar] [CrossRef]
- Giustacchini, A.; Thongjuea, S.; Barkas, N.; Woll, P.S.; Povinelli, B.J.; Booth, C.A.G.; Sopp, P.; Norfo, R.; Rodriguez-Meira, A.; Ashley, N.; et al. Single-cell transcriptomics uncovers distinct molecular signatures of stem cells in chronic myeloid leukemia. Nat. Med. 2017, 23, 692–702. [Google Scholar] [CrossRef]
- Srivastava, A.; Joshi, B.D.; Tandon, P.; Ayala, A.P.; Bansal, A.K.; Grillo, D. Study of polymorphism in imatinib mesylate: A quantum chemical approach using electronic and vibrational spectra. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2013, 103, 325–332. [Google Scholar] [CrossRef]


| Method | Pros | Cons |
|---|---|---|
| RT-qPCR | Widely available Internationally standardized | Poor sensitivity and low precision at low levels of target Poor robustness Standard curve required |
| dPCR | More sensitive and accurate Enables the detection of as little as 1 copy of BCR-ABL1 transcript Performs an absolute quantification of the target without the need for a standard curve | Not yet widely available Not yet standardized |
| Method | Pros | Cons |
|---|---|---|
| Sanger sequencing | Widely available Easy to use | Poor sensitivity |
| NGS | More sensitive than Sanger sequencing Enables to scan the entire KD for any mutation Enables clonal analysis in case of multiple mutations falling within the same sequence reads (discrimination between compound and polyclonal mutations) | Not yet widely available Requires pooling of a minimum of 8–10 samples to be cost-effective Labor-intensive Not yet standardized RT-PCR and sequencing errors generate background “noise” at lower levels of sensitivity Chemistries and instruments still evolving |
| dPCR | Cheap, fast, and simple Has the greatest sensitivity | Can be implemented only for a limited number of mutations Not yet standardized May confirm the presence of compound mutations only if the mutation partners are already known and, hence, specific probes can be designed and used |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soverini, S.; Bernardi, S.; Galimberti, S. Molecular Testing in CML between Old and New Methods: Are We at a Turning Point? J. Clin. Med. 2020, 9, 3865. https://doi.org/10.3390/jcm9123865
Soverini S, Bernardi S, Galimberti S. Molecular Testing in CML between Old and New Methods: Are We at a Turning Point? Journal of Clinical Medicine. 2020; 9(12):3865. https://doi.org/10.3390/jcm9123865
Chicago/Turabian StyleSoverini, Simona, Simona Bernardi, and Sara Galimberti. 2020. "Molecular Testing in CML between Old and New Methods: Are We at a Turning Point?" Journal of Clinical Medicine 9, no. 12: 3865. https://doi.org/10.3390/jcm9123865
APA StyleSoverini, S., Bernardi, S., & Galimberti, S. (2020). Molecular Testing in CML between Old and New Methods: Are We at a Turning Point? Journal of Clinical Medicine, 9(12), 3865. https://doi.org/10.3390/jcm9123865







