Bacterial Colonization in Patients with Chronic Lymphocytic Leukemia and Factors Associated with Infections and Colonization
Abstract
1. Introduction
2. Experimental Section
2.1. Patients
2.2. Microbiological Procedures
2.3. Blood Sampling
2.4. Detection of Basic Peripheral Blood Lymphocyte Subsets
2.5. Analysis of CD38 and ZAP-70 Expression in CLL Cells
2.6. DNA Isolation and Calculation of EBV Load
2.7. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Linet, M.S.; Schubauer-Berigan, M.K.; Weisenburger, D.D.; Richardson, D.B.; Landgren, O.; Blair, A.; Silver, S.; Field, R.W.; Caldwell, G.; Hatch, M.; et al. Chronic lymphocytic leukaemia: An overview of aetiology in light of recent developments in classification and pathogenesis. Br. J. Haematol. 2007, 139, 672–686. [Google Scholar] [CrossRef] [PubMed]
- Oscier, D.; Fegan, C.; Hillmen, P.; Illidge, T.; Johnson, S.; Maguire, P.; Matutes, E.; Milligan, D. Guidelines working group of the UK CLL Forum. British committee for standards in haematology. Guidelines on the diagnosis and management of chronic lymphocytic leukaemia. Br. J. Haematol. 2004, 125, 294–317. [Google Scholar] [CrossRef] [PubMed]
- Morrison, V.A. Infectious complications in patients with chronic lymphocytic leukemia: Pathogenesis, spectrum of infection, and approaches to prophylaxis. Clin. Lymphoma Myeloma 2009, 9, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Wadhwa, P.; Morrison, V. Infectious Complications of Chronic Lymphocytic Leukemia. Semin. Oncol. 2006, 33, 240–249. [Google Scholar] [CrossRef] [PubMed]
- Morrison, V.A. Infectious complications of chronic lymphocytic leukaemia: pathogenesis, spectrum of infection, preventive approaches. Best Pr. Res. Clin. Haematol. 2010, 23, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Korona-Glowniak, I.; Grywalska, E.; Chudzik, B.; Bojarska-Junak, A.; Malm, A.; Roliński, J. Upper respiratory tract colonization by gram-negative rods in patients with chronic lymphocytic leukemia: Analysis of risk factors. Sci. World J. 2012, 2012, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Hallek, M. Chronic lymphocytic leukemia: 2013 update on diagnosis, risk stratification and treatment. Am. J. Hematol. 2013, 88, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Hallek, M.; Cheson, B.D.; Catovsky, D.; Caligaris-Cappio, F.; Dighiero, G.; Döhner, H.; Hillmen, P.; Keating, M.J.; Montserrat, E.; Rai, K.R.; et al. International workshop on chronic lymphocytic leukemia. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: A report from the international workshop on chronic lymphocytic leukemia updating the national cancer institute-Working group 1996 guidelines. Blood 2008, 111, 5446–5456. [Google Scholar] [PubMed]
- Grywalska, E.; Roliński, J.; Pasiarski, M.; Korona-Glowniak, I.; Maj, M.; Surdacka, A.; Grafka, A.; Stelmach-Gołdyś, A.; Zgurski, M.; Góźdź, S.; et al. High viral loads of epstein-barr virus DNA in Peripheral blood of patients with chronic lymphocytic leukemia associated with unfavorable prognosis. PLoS ONE 2015, 10, e0140178. [Google Scholar] [CrossRef] [PubMed]
- Zgodzinski, W.; Grywalska, E.; Zinkiewicz, K.; Surdacka, A.; Majewski, M.; Zakoscielny, A.; Bury, P.; Rolinski, J.; Wallner, G.T. Peripheral blood T lymphocytes are downregulated by the PD-1/PD-L1 axis in advanced gastric cancer. Arch. Med. Sci. 2018, 15, 774. [Google Scholar] [CrossRef] [PubMed]
- Hus, I.; Podhorecka, M.; Bojarska-Junak, A.; Roliński, J.; Schmitt, M.; Sieklucka, M.; Wasik-Szczepanek, E.; Dmoszynska, A. The clinical significance of ZAP-70 and CD38 expression in B-cell chronic lymphocytic leukaemia. Ann. Oncol. 2006, 17, 683–690. [Google Scholar] [CrossRef] [PubMed]
- Itala, M.; Helenius, H.; Nikoskelainen, J.; Remes, K. Infections and serum IgG levels in patients with chronic lymphocytic leukemia. Eur. J. Haematol. 1992, 48, 266–270. [Google Scholar] [CrossRef] [PubMed]
- Chapel, H.; Dicato, M.; Gamm, H.; Brennan, V.; Ries, F.; Bunch, C.; Lee, M. Immunoglobulin replacement in patients with chronic lymphocytic leukaemia: A comparison of two dose regimens. Br. J. Haematol. 1994, 88, 209–212. [Google Scholar] [CrossRef] [PubMed]
- Pasiarski, M.; Roliński, J.; Grywalska, E.; Stelmach-Gołdys, A.; Korona-Glowniak, I.; Gozdz, S.; Hus, I.; Malm, A. Antibody and plasmablast response to 13-valent pneumococcal conjugate vaccine in chronic lymphocytic leukemia patients—Preliminary report. PLoS ONE 2014, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Teh, B.W.; Tam, C.S.; Handunnetti, S.; Worth, L.J.; Slavin, M.A. Infections in patients with chronic lymphocytic leukaemia: Mitigating risk in the era of targeted therapies. Blood Rev. 2018, 32, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Skrupky, L.P.; Kerby, P.W.; Hotchkiss, R.S. Advances in the management of sepsis and in the understanding of key immunologic defects of the disorder anesthesiology. Anesthesiology 2011, 115, 1349–1362. [Google Scholar] [PubMed]
- Rabe, H.; Nordström, I.; Andersson, K.; Lundell, A.C.; Rudin, A. Staphylococcus aureus convert neonatal conventional CD4+ T cells into FOXP3+ CD25+ CD127low T cells via the PD-1/PD-L1 axis. Immunology 2014, 141, 467–481. [Google Scholar] [CrossRef]
- McKay, J.T.; Egan, R.P.; Yammani, R.D.; Chen, L.; Shin, T.; Yagita, H.; Haas, K.M. PD-1 suppresses protective immunity to Streptococcus pneumoniae through a B cell-intrinsic mechanism. J. Immunol. 2015, 194, 2289–2299. [Google Scholar] [CrossRef]
- Okazaki, T.; Chikuma, S.; Iwai, Y.; Fagarasan, S.; Honjo, T. A rheostat for immune responses: The unique properties of PD-1 and their advantages for clinical application. Nat. Immunol. 2013, 14, 1212–1218. [Google Scholar] [CrossRef]
- Wang, S.; Chen, L. Immunobiology of cancer therapies targeting CD137 and B7-H1/PD-1 cosignal pathways. Curr. Top. Microbiol. Immunol. 2011, 344, 245–267. [Google Scholar]
- Brown, K.E.; Freeman, G.J.; Wherry, E.J.; Sharpe, A.H. Role of PD-1 in regulating acute infections. Curr. Opin. Immunol. 2010, 22, 397–401. [Google Scholar] [CrossRef]
- Yao, S.; Wang, S.; Zhu, Y.; Luo, L.; Zhu, G.; Flies, S.; Xu, H.; Ruff, W.; Broadwater, M.; Choi, I.H.; et al. PD-1 on dendritic cells impedes innate immunity against bacterial infection. Blood 2009, 113, 5811–5818. [Google Scholar] [CrossRef] [PubMed]
- Lazar-Molnar, E.; Chen, B.; Sweeney, K.A.; Wang, E.J.; Liu, W.; Lin, J.; Porcelli, S.A.; Almo, S.C.; Nathenson, S.G.; Jacobs, W.R. Programmed death-1 (PD-1)-deficient mice are extraordinarily sensitive to tuberculosis. Proc. Natl. Acad. Sci. USA 2010, 107, 13402–13407. [Google Scholar] [CrossRef] [PubMed]
- Barber, D.L.; Mayer-Barber, K.D.; Feng, C.G.; Sharpe, A.H.; Sher, A. CD4 T cells promote rather than control tuberculosis in the absence of PD-1-mediated inhibition. J. Immunol. 2011, 186, 1598–1607. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Venet, F.; Wang, Y.L.; Lepape, A.; Yuan, Z.; Chen, Y.; Swan, R.; Kherouf, H.; Monneret, G.; Chung, C.S.; et al. PD-1 expression by macrophages plays a pathologic role in altering microbial clearance and the innate inflammatory response to sepsis. Proc. Natl. Acad. Sci. USA 2009, 106, 6303–6308. [Google Scholar] [CrossRef] [PubMed]
- Den Heijer, C.D.; van Bijnen, E.M.; Paget, W.J.; Pringle, M.; Goossens, H.; Bruggeman, C.A.; Schellevis, F.G.; Stobberingh, E.E.; APRES Study Team. Prevalence and resistance of commensal Staphylococcus aureus, including meticillin-resistant S. aureus, in nine European countries: A cross-sectional study. Lancet Infect. Dis. 2013, 13, 409–415. [Google Scholar] [CrossRef]
- Brown, A.F.; Leech, J.M.; Rogers, T.R.; McLoughlin, R.M. Staphylococcus aureus colonization: Modulation of host immune response and impact on human vaccine design. Front. Immunol. 2013, 4, 507. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.; Huggins, R.H.; Lertsburapa, T.; Bauer, K.; Rademaker, A.; Gerami, P.; Guitart, J. Cutaneous T-cell lymphoma and Staphylococcus aureus colonization. J. Am. Acad. Dermatol. 2008, 59, 949–952. [Google Scholar] [CrossRef]
- Niederman, M.S. Bacterial adherence as a mechanism of airway colonization. Eur. J. Microbiol. Infect. Dis. 1989, 8, 15–20. [Google Scholar] [CrossRef]
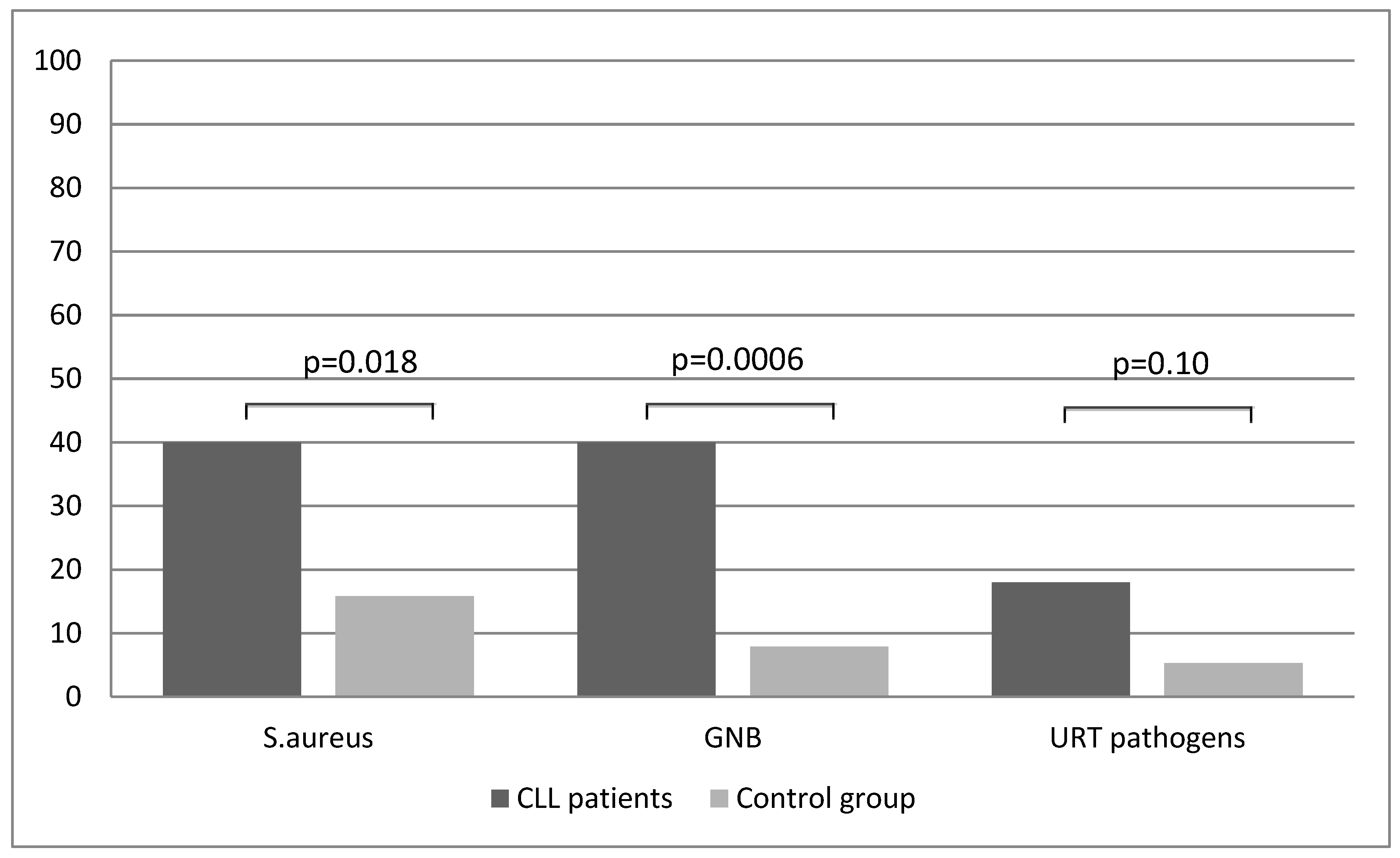
| CLL Patients (n = 50) | Control (n = 38) | p Value | |||
|---|---|---|---|---|---|
| Mean ± SD | Median (Range) | Mean ± SD | Median (Range) | ||
| Age | 63.1 ± 10.7 | 63.0 (38–80) | 62.6 ± 9.9 | 62.5 (41–83) | 0.85 |
| Male gender, n (%) | 19 (38.0) | 15 (39.5) | 1.0 | ||
| No of infections per year | 4.4 ± 1.7 | 4.0 (1–8) | 1.1 ± 0.91 | 1.0 (0–3) | <0.0001 |
| Type of infections | |||||
| URTIs, n (%) | 43 (86.0) | 34 (89.5) | 0.75 | ||
| LRTIs, n (%) | 34 (68.0) | 1 (2.6) | <0.0001 | ||
| UTIs, n (%) | 11 (22.0) | 0 (0) | 0.002 | ||
| Skin infections, n (%) | 10 (20.0) | 0 (0) | 0.004 | ||
| No of antibiotic therapies (per patient) | 3.7 ± 1.8 | 4.0 (1–8) | 0.16 ± 0.37 | 0 (0–1) | <0.0001 |
| Mean ± SD | Median (Range) | |
|---|---|---|
| URTIs (%), including: | ||
| - Pharyngitis | 40 (80.0) | |
| - Laryngitis | 7 (14.0) | |
| - Otitis | 19 (38.0) | |
| LRTIs (%), including: | ||
| - Bronchitis | 22 (44.0) | |
| - Pneumonia | 13 (26.0) | |
| Genitourinary infections (%) | 11 (22.0) | |
| Skin/soft tissue infections (bacterial/fungal) (%) | 10 (20.0) | |
| The Rai stage, n (%): 0 | 16 (32.0) | |
| 1 | 18 (36.0) | |
| 2 | 16 (32.0) | |
| Binet stage, n (%): A | 16 (32.0) | |
| B | 34 (68.0) | |
| Splenomegaly, n (%) | 14 (28.0) | |
| Hepatomegaly, n (%) | 8 (16.0) | |
| Institution of treatment, n (%) | 21 (42.0) | |
| Remission, n (%) | 5 (10.0) | |
| Progression, n (%) | 16 (32.0) | |
| EBV(+) patients, n (%) | 25 (50.0) | |
| EBV EA IgA (U/mL) EBV(+)/EBV(-) | 59.0 ± 125.4/6.3 ± 6.0 | 4.8 (2.4–580.4)/5.1 (1.2–33.3) |
| EBV EA IgG (U/mL) EBV(+)/EBV(-) | 63.4 ± 89.0/14.0 ± 19.8 | 23.7 (3.9–381.1)/8.2 (2.0–94.8) |
| EBV EA IgM (U/mL) EBV(+)/EBV(-) | 5.4 ± 4.3/4.3 ± 2.8 | 4.1 (1.4–19.2)/3.6 (0.8–10.5) |
| EBV EBNA-1 IgA (U/mL) EBV(+)/EBV(-) | 12.0 ± 11.5/5.7 ± 3.6 | 8.3 (1.3–38.3)/5.1 (1.5–15.3) |
| EBV EBNA-1 IgG (U/mL) EBV(+)/EBV(-) | 132.6 ± 141.8/64.8 ± 41.0 | 78.5 (22.5–659.3)/55.7 (23.5–181.7) |
| EBV EBNA-1 IgM (U/mL) EBV(+)/EBV(-) | 6.9 ± 4.5/4.8 ± 2.3 | 6.2 (1.8–19.1)/4.8 (0.9–10.5) |
| EBV VCA IgA (U/mL) EBV(+)/EBV(-) | 10.7 ± 9.3/7.7 ± 4.3 | 6.1 (1.9–34.0)/5.5 (2.3–19.8) |
| EBV VCA IgG (U/mL) EBV(+)/EBV(-) | 203.2 ± 82.6/162.0 ± 87.8 | 209.2 (40.6–352.8)/149.4 (25.2–358.0) |
| Duplication of lymphocytosis, n (%) | 26 (52.0) | |
| Leukocytosis (G/L) | 41.5 ± 27.5 | 30.9 (12.7–128.0) |
| Lymphocytosis (G/L) | 35.1 ± 26.3 | 25.5 (8.4–124.0) |
| Hemoglobin (g/dL) | 13.4 ± 1.6 | 13.4 (10.0-16.6) |
| Platelets (G/L) | 183.5 ± 55.6 | 177.5 (101.0–299.0) |
| Lactate dehydrogenase, LDH (U/L) | 313.7 ± 130.8 | 297.0 (115.0–705.0) |
| Beta-2 microglobulin, B2M (mg/L) | 2.7 ± 1.0 | 2.5 (1.1–4.9) |
| IgA (g/L) | 1.4 ± 1.2 | 1.1 (0.14–5.4) |
| IgG (g/L) | 8.7 ± 3.7 | 8.5 (3.2–18.6) |
| IgM (g/L) | 0.65 ± 0.5 | 0.5 (0.04–2.0) |
| T CD3+cells (%) | 10.8 ± 7.4 | 9.7 (1.2–30.6) |
| B CD19+cells (%) | 84.25 ± 9.5 | 85.3 (51.5–97.8) |
| CD5+CD19+cells (%) | 84.4 ± 10.3 | 84.2 (54.9–99.3) |
| CD19+ZAP70+cells (%) | 16.3 ± 12.0 | 14.0 (0.5–47.0) |
| Negative (<20%) | 30 (60.0) | |
| Positive (>20%) | 20 (40.0) | |
| CD19+CD38+cells (%) | 25.2 ± 25.3 | 16.4 (0.2–87.5) |
| Negative (<30%), n (%) | 29 (58.0) | |
| Positive (>30%), n (%) | 21 (42.0) | |
| ZAP70 in relation to CD38, n (%): | ||
| ZAP70(-) CD38(-) | 20 (40.0) | |
| ZAP70(+) CD38(+) | 14 (28.0) | |
| ZAP70(+) CD38(-) | 7 (14.0) | |
| ZAP70(-) CD38(+) | 9 (18.0) | |
| CD19+CD5+CD23+(%) | 79.0 ± 11.2 | 79.9 (54.1–95.2) |
| CD19+CD25+cells (%) | 52.4 ± 23.1 | 52.3 (6.7–93.0) |
| CD19+CD69+cells (%) | 33.5 ± 19.3 | 30.5 (3.9–77.6) |
| CD3+CD25+cells (%) | 21.95 ± 13.9 | 20.4 (1.2–57.6) |
| CD3+CD69+cells (%) | 4.7 ± 4.8 | 3.2 (0.1–21.8) |
| CD4+/PD-1+ (among CD4+) (%) | 19.6 ± 9.8 | 16.4 (8.5–49.8) |
| CD8+/PD-1+(among CD8+) (%) | 14.6 ± 7.1 | 14.1 (1.6–33.2) |
| CD19+/PD-1+(among CD19+) (%) | 19.1 ± 7.1 | 19.7 (6.5–35.65) |
| Factors | LRTIs (n = 34) | No LRTIs (n = 16) | OR (95%) | p Value |
|---|---|---|---|---|
| Age | 63.7 ± 9.9 | 61.6 ± 12.4 | 1.01 (0.96–1.1) | 0.66 |
| Male gender: | 13 (38.2) | 6 (37.5) | 1.03 (0.3–3.5) | 1.0 |
| The Rai stage: 0 | 6 (17.7) | 10 (62.5) | referent | |
| 1 | 14 (41.2) | 4 (25.0) | 5.8 (1.3–26.2) | 0.022 |
| 2 | 14 (41.2) | 5 (12.5) | 11.7 (1.9–70.2) | 0.007 |
| Binet stage B | 28 (82.4) | 6 (37.5) | 7.8 (2.0–29.8) | 0.0029 |
| Splenomegaly | 12 (35.3) | 2 (12.5) | 3.8 (0.7–19.7) | 0.087 |
| Hepatomegaly | 8 (23.5) | 0 (0) | 10.6 (0.6–159.9) | 0.034 |
| EBV positive | 23 (67.7) | 2 (12.5) | 14.6 (2.8–75.9) | 0.0006 |
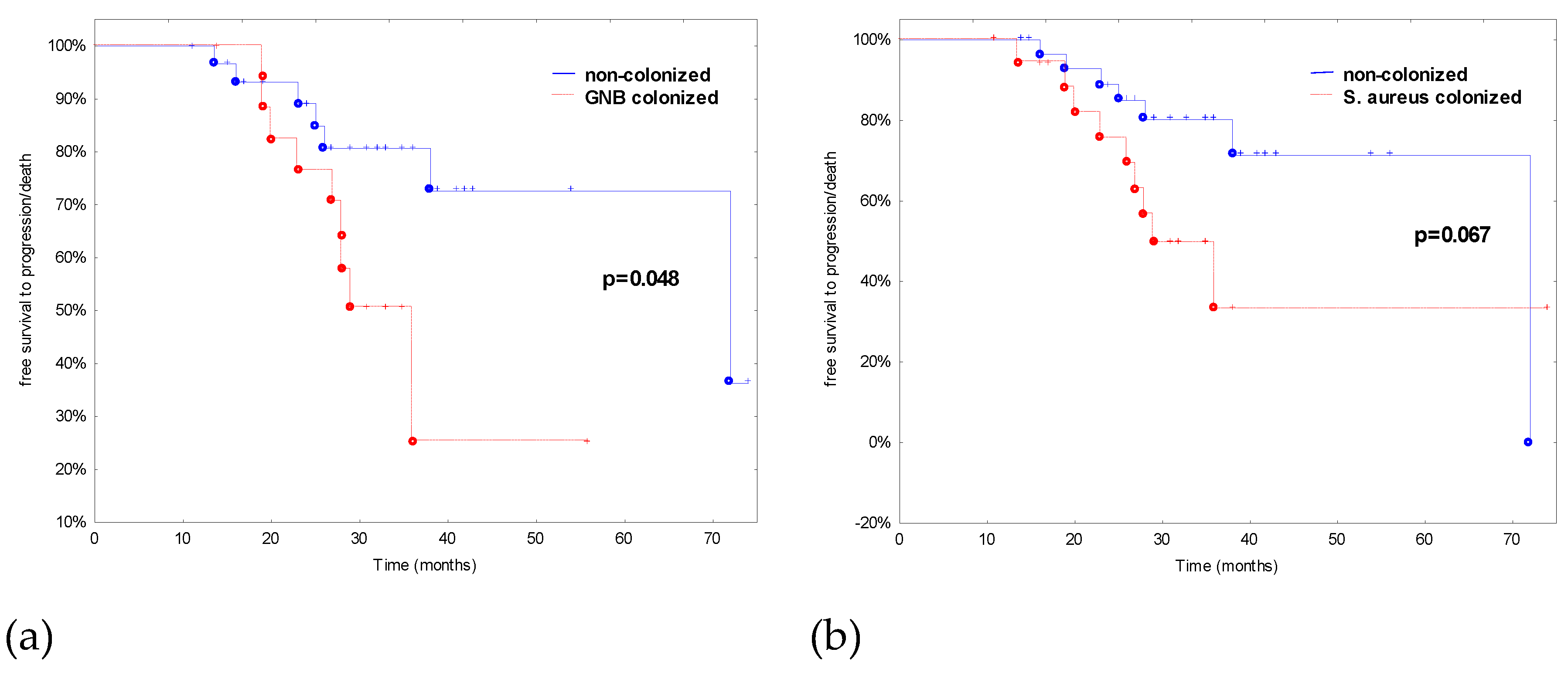
| S. aureus colonization | 15 (44.1) | 4 (25.0) | 3.8 (0.7–19.7) | 0.11 |
| GNB colonization | 15 (44.1) | 4 (25.0) | 0.5 (0.1–2.3) | 0.23 |
| URT pathogens | 6 (17.7) | 3 (18.8) | 0.9 (0.2–4.3) | 1.0 |
| Duplication of lymphocytosis | 23 (67.7) | 3 (18.8) | 9.1 (2.1–38.5) | 0.002 |
| Institution of treatment | 21 (61.8) | 0 (0) | 52.6 (2.9–950.7) | <0.0001 |
| Lactate dehydrogenase (U/L) | 339.7 ± 137.5 | 258.4 ± 97.6 | 1.01 (1.0–1.012) | 0.04 |
| Beta-2 microglobulin (mg/L) | 3.0 ± 1.0 | 2.03 ± 0.7 | 3.4 (1.4–8.0) | 0.001 |
| IgA (g/L) | 1.5 ± 1.3 | 1.2 ± 0.99 | 1.3 (0.8–2.3) | 0.38 |
| IgG (g/L) | 9.1 ± 3.6 | 7.8 ± 3.8 | 1.1 (0.9–1.3) | 0.18 |
| IgM (g/L) | 0.6 ± 0.5 | 0.7 ± 0.6 | 0.7 (0.2–2.2) | 0.72 |
| CD19+ZAP70+cells (%) | 18.4 ± 12.1 | 11.9 ± 10.8 | 1.1 (1.0–1.12) | 0.098 |
| CD19+CD38+cells (%) | 30.5 ± 25.8 | 13.8 ± 20.7 | 1.03 (1.0–1.06) | 0.012 |
| CD4+/PD-1+ (within CD4+) (%) | 16.1 ± 7.7 | 11.3 ± 4.5 | 1.2 (1.0–1.3) | 0.001 |
| CD8+/PD-1+ (within CD8+) (%) | 21.2 ± 6.2 | 14.5 ± 6.7 | 1.1 (1.0–1.3) | 0.030 |
| CD19+/PD-1+ (within CD19+) (%) | 22.2 ± 10.0 | 14.0 ± 6.6 | 1.2 (1.1–1.3) | 0.001 |
| S. aureus | GNB | |||||
|---|---|---|---|---|---|---|
| Factors | Colonized (n = 19) | Non-Colonized (n = 31) | OR (95%CI) | Colonized (n = 19) | Non-Colonized (n = 31) | OR (95%CI) |
| Age | 71 (38–79) | 62.0 (44–80) | 1.0 (0.98–1.1) | 63 (45–79) | 63 (38-80) | 1.0 (0.95–1.1) |
| Male gender: | 11 (57.9%) | 8 (25.8%) | 3.9 (1.2–13.3) * | 10 (52.6%) | 9 (29.0%) | 2.7 (0.8–8.9) |
| The Rai stage: 0 | 3 (15.8%) | 13 (41.9%) | reference | 6 (31.6%) | 10 (32.3%) | reference |
| 1 | 10 (52.6%) | 8 (25.8%) | 5.4 (1.1–25.8) * | 8 (42.1%) | 10 (32.3%) | 1.3 (0.3–5.3) |
| 2 | 6 (31.6%) | 10 (32.3%) | 2.6 (0.5–13.0) | 5 (26.3%) | 11 (35.5%) | 0.7 (0.2–3.3) |
| Binet stage B | 16 (84.2%) | 18 (58.1%) | 3.8 (0.9–16.0) | 13 (68.4%) | 21 (67.7%) | 1.03 (0.3–3.5) |
| Splenomegaly | 6 (31.6%) | 8 (25.8%) | 1.3 (0.4–4.7) | 4 (21.1%) | 10 (32.3%) | 0.6 (0.1–2.1) |
| Hepatomegaly | 4 (21.0%) | 4 (12.9%) | 1.8 (0.4–8.3) | 1 (5.3%) | 7 (22.6%) | 0.2 (0.02–1.7) |
| EBV positive | 14 (73.7%) | 11 (35.5%) | 5.1 (1.4–17.9) * | 10 (52.6%) | 15 (48.4%) | 1.2 (0.4–3.7) |
| URTIs | 18 (94.7%) | 25 (80.7%) | 1.5 (1.0–2.2) | 17 (89.5%) | 26 (83.9%) | 1.2 (0.7–2.0) |
| LRTIs | 15 (79.0%) | 19 (61.3%) | 1.3 (0.9–2.0) | 15 (79.0%) | 19 (61.3%) | 1.3 (0.9–2.0) |
| UTIs | 2 (10.5%) | 8 (25.8%) | 0.7 (0.5–1.1) | 3 (15.8%) | 7 (22.6%) | 0.9 (0.5–1.4) |
| Skin infections | 5 (26.3%) | 5 (16.1%) | 1.8 (0.5–7.5) | 3 (15.8%) | 7 (22.6%) | 0.9 (0.5–1.4) |
| Duplication of lymphocytosis | 14 (73.7%) | 12 (38.7%) | 4.4 (1.3–15.5) * | 10 (52.6%) | 16 (51.6%) | 1.04 (0.3–3.3) |
| Institution of treatment | 10 (52.6%) | 11 (35.5%) | 2.02 (0.6–6.5) | 11 (57.9%) | 10 (32.3%) | 2.9 (0.9–9.4) |
| Lactate dehydrogenase (U/L) | 346 (179–625) | 245 (115–705) | 1.0 (0.99–1.0) | 311 (115–595) | 285 (153–705) | 1.0 (0.99–1.0) |
| Beta-2 microglobulin (mg/L) | 3.1 (1–5) | 2.1(1–4.5) | 2.3 (1.2–4.3) * | 2.3 (1–4) | 2.6 (1–5) | 1.2 (0.7–2.2) |
| IgA (g/L) | 1.5 (0.1–5) | 1.1 (0.2–5.4) | 1.1 (0.7–1.8) | 1.2 (0.2–5) | 1.0 (0.1–5) | 1.3 (0.8–2.2) |
| IgG (g/L) | 8.7 (4–17) | 7 (3–19) | 1.05 (0.9–1.2) | 8.3 (4–17) | 8.7 (3–19) | 1.03 (0.88–1.2) |
| IgM (g/L) | 0.6 (0.1–2) | 0.45 (0.04–2) | 1.0 (0.3–3.2) | 0.6 (0.1–2) | 0.45 (0.04–2) | 1.3 (0.4–4.2) |
| CD19+ZAP70+cells (%) | 20.6 (2–47) | 12.4 (0.5–38) | 1.05 (1.0–1.1) | 18.3 (0.5–47) | 13.1 (0.7–45) | 1.01 (0.96–1.06) |
| CD19+CD38+cells (%) | 30.6 (0.4–87) | 8.9 (0.2–5) | 1.02 (1.0–1.05) | 11.8 (0.2–75) | 20.5 (0.4–87) | 1.0 (0.98–1.02) |
| CD4+/PD-1+ (within CD4+) (%) | 20.4 (9–35) | 14.4 (8–50) | 1.0 (0.95–1.07) | 15.9 (9–50) | 16.8 (8–43) | 0.98 (0.92–1.04) |
| CD8+/PD-1+ (within CD8+) (%) | 18.3 (2.5–33) | 12.6 (1.6–29) | 1.1 (1.01–1.2) * | 13.2 (5–33 ) | 14.5 (1–29) | 1.03 (0.95–1.1) |
| CD19+/PD-1+ (within CD19+) (%) | 20.7 (7–30) | 18.3 (6–36) | 1.06 (0.97–1.2) | 20.6(8–36) | 19.7 (6.5–30) | 1.02 (0.9–1.1) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Korona-Glowniak, I.; Grywalska, E.; Grzegorczyk, A.; Roliński, J.; Glowniak, A.; Malm, A. Bacterial Colonization in Patients with Chronic Lymphocytic Leukemia and Factors Associated with Infections and Colonization. J. Clin. Med. 2019, 8, 861. https://doi.org/10.3390/jcm8060861
Korona-Glowniak I, Grywalska E, Grzegorczyk A, Roliński J, Glowniak A, Malm A. Bacterial Colonization in Patients with Chronic Lymphocytic Leukemia and Factors Associated with Infections and Colonization. Journal of Clinical Medicine. 2019; 8(6):861. https://doi.org/10.3390/jcm8060861
Chicago/Turabian StyleKorona-Glowniak, Izabela, Ewelina Grywalska, Agnieszka Grzegorczyk, Jacek Roliński, Andrzej Glowniak, and Anna Malm. 2019. "Bacterial Colonization in Patients with Chronic Lymphocytic Leukemia and Factors Associated with Infections and Colonization" Journal of Clinical Medicine 8, no. 6: 861. https://doi.org/10.3390/jcm8060861
APA StyleKorona-Glowniak, I., Grywalska, E., Grzegorczyk, A., Roliński, J., Glowniak, A., & Malm, A. (2019). Bacterial Colonization in Patients with Chronic Lymphocytic Leukemia and Factors Associated with Infections and Colonization. Journal of Clinical Medicine, 8(6), 861. https://doi.org/10.3390/jcm8060861








