Deep Convolutional Neural Network-Based Positron Emission Tomography Analysis Predicts Esophageal Cancer Outcome
Abstract
1. Introduction
2. Methods
2.1. Data Source
2.2. Convolution Neural Network Model Setup and Training
2.3. Statistics
3. Results
3.1. Patients
3.2. CNN Performance
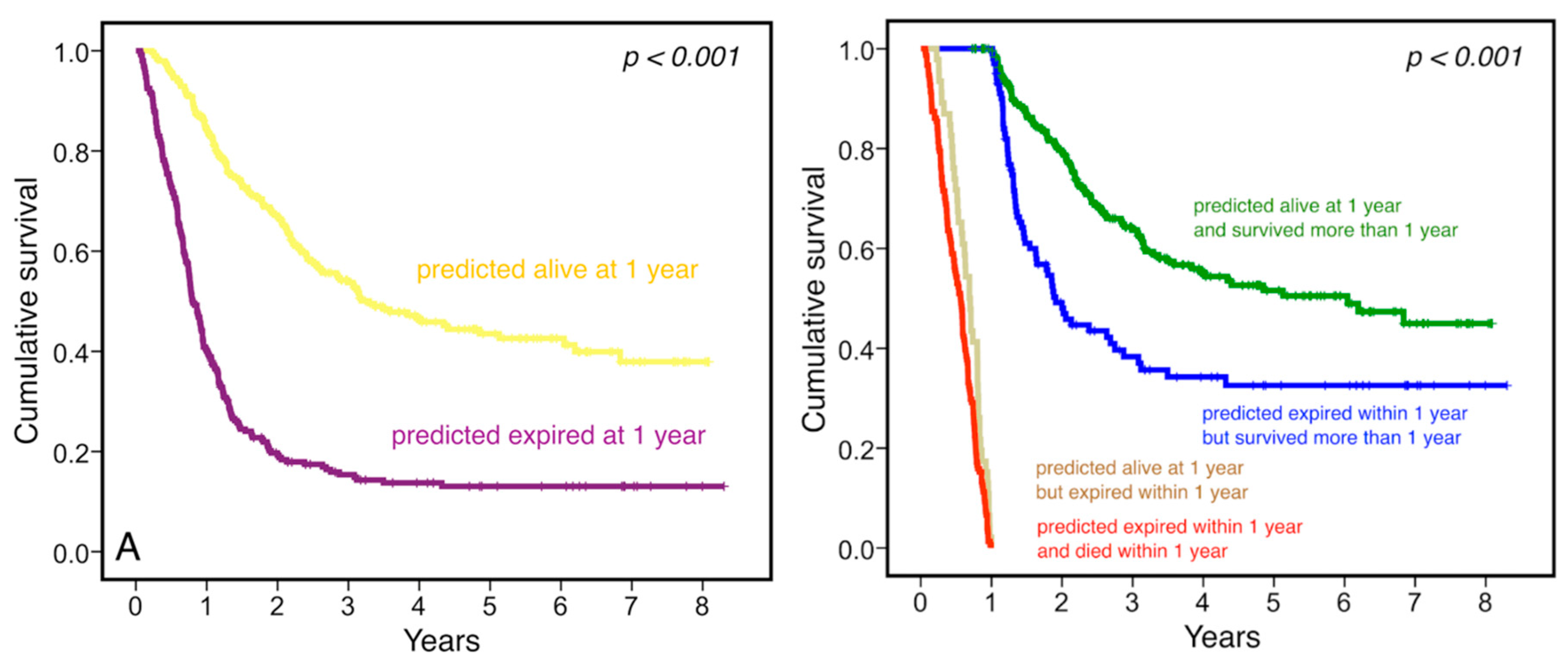
3.3. Clinical Relevance
4. Discussion
Supplementary Materials
Author Contributions
Conflicts of Interest
References
- Gupta, V.; Coburn, N.; Kidane, B.; Hess, K.R.; Compton, C.; Ringash, J.; Darling, G.; Mahar, A.L. Survival prediction tools for esophageal and gastroesophageal junction cancer: A systematic review. J. Thorac. Cardiovasc. Surg. 2018, 156, 847–856. [Google Scholar] [CrossRef]
- Aerts, H.J.; Velazquez, E.R.; Leijenaar, R.T.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef]
- Chen, R.; Zhang, R.; Gan, Y.; Yang, L.; Li, W. Development and clinical application of radiomics in lung cancer. Radiat. Oncol. 2017, 12, 154. [Google Scholar] [CrossRef] [PubMed]
- Sollini, M.; Cozzi, L.; Antunovic, L.; Chiti, A.; Kirienko, M. PET Radiomics in NSCLC: State of the art and a proposal for harmonization of methodology. Sci. Rep. 2017, 7, 358. [Google Scholar] [CrossRef] [PubMed]
- Lovinfosse, P.; Polus, M.; Van Daele, D.; Martinive, P.; Daenen, F.; Hatt, M.; Visvikis, D.; Koopmansch, B.; Lambert, F.; Coimbra, C.; et al. FDG PET/CT radiomics for predicting the outcome of locally advanced rectal cancer. Eur. J. Nucl. Med. Mol. Imaging 2018, 45, 365–375. [Google Scholar] [CrossRef]
- Beukinga, R.J.; Hulshoff, J.B.; Mul, V.E.M.; Noordzij, W.; Kats-Ugurlu, G.; Slart, R.H.; Plukker, J.T. Prediction of response to neoadjuvant chemotherapy and radiation therapy with baseline and restaging (18)F-FDG PET imaging biomarkers in patients with esophageal cancer. Radiology 2018, 287, 983–992. [Google Scholar] [CrossRef]
- Suzuki, K. Overview of deep learning in medical imaging. Radiol. Phys. Technol. 2017, 10, 257–273. [Google Scholar] [CrossRef]
- Shin, H.C.; Roth, H.R.; Gao, M.; Lu, L.; Xu, Z.; Nogues, I.; Yao, J.; Mollura, D.; Summers, R.M. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer Learning. IEEE Trans. Med. Imaging 2016, 35, 1285–1298. [Google Scholar] [CrossRef]
- Yamashita, R.; Nishio, M.; Do, R.K.G.; Togashi, K. Convolutional neural networks: An overview and application in radiology. Insights Imaging 2018, 9, 611–629. [Google Scholar] [CrossRef] [PubMed]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Gulshan, V.; Peng, L.; Coram, M.; Stumpe, M.C.; Wu, D.; Narayanaswamy, A.; Venugopalan, S.; Widner, K.; Madams, T.; Cuadros, J.; et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 2016, 316, 2402–2410. [Google Scholar] [CrossRef] [PubMed]
- Bejnordi, B.E.; Veta, M.; Van Diest, P.J.; Van Ginneken, B.; Karssemeijer, N.; Litjens, G.; Van Der Laak, J.A.; Hermsen, M.; Manson, Q.F.; Balkenhol, M.; et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 2017, 318, 2199–2210. [Google Scholar] [CrossRef] [PubMed]
- Maxmen, A. Deep learning sharpens views of cells and genes. Nature 2018, 553, 9–10. [Google Scholar] [CrossRef] [PubMed]
- Poplin, R.; Varadarajan, A.V.; Blumer, K.; Liu, Y.; McConnell, M.V.; Corrado, G.S.; Peng, L.; Webster, D.R. Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nat. Biomed. Eng. 2018, 2, 158–164. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. December 2015. Available online: https://arxiv.org/pdf/1512.03385.pdf (accessed on 3 August 2018).
- Parkinson, C.; Foley, K.; Whybra, P.; Hills, R.; Roberts, A.; Marshall, C.; Staffurth, J.; Spezi, E. Evaluation of prognostic models developed using standardised image features from different PET automated segmentation methods. EJNMMI Res. 2018, 8, 29. [Google Scholar] [CrossRef] [PubMed]
- Goel, R.; Subramaniam, R.M.; Wachsmann, J.W. PET/computed tomography scanning and precision medicine: Esophageal cancer. PET Clin. 2017, 12, 373–391. [Google Scholar] [CrossRef] [PubMed]
- Van de Wiele, C.; Kruse, V.; Smeets, P.; Sathekge, M.; Maes, A. Predictive and prognostic value of metabolic tumour volume and total lesion glycolysis in solid tumours. Eur. J. Nucl. Med. Mol. Imaging 2013, 40, 290–301. [Google Scholar] [CrossRef]
- Cerfolio, R.J.; Bryant, A.S. Maximum standardized up-take values on positron emission tomography of esophageal cancer predicts stage, tumor biology, and survival. Ann. Thorac. Surg. 2006, 82, 391–394. [Google Scholar] [CrossRef]
- Ott, K.; Weber, W.A.; Lordick, F.; Becker, K.; Busch, R.; Herrmann, K.; Wieder, H.; Fink, U.; Schwaiger, M.; Siewert, J.R. Metabolic imaging predicts response, survival, and recurrence in adenocarcinomas of the esophagogastric junction. J. Clin. Oncol. 2006, 24, 4692–4698. [Google Scholar] [CrossRef]
- Omloo, J.M.; van Heijl, M.; Hoekstra, O.S.; van Berge Henegouwen, M.I.; Van Lanschot, J.J.B.; Sloof, G.W. FDG-PET parameters as prognostic factor in esopha-geal cancer patients: A review. Ann. Surg. Oncol. 2011, 18, 3338–3352. [Google Scholar] [CrossRef]
- Zaidi, H.; Karakatsanis, N. Towards enhanced PET quantification in clinical oncology. Br. J. Radiol. 2018, 91, 20170508. [Google Scholar] [CrossRef] [PubMed]
- Chicklore, S.; Goh, V.; Siddique, M.; Roy, A.; Marsden, P.K.; Cook, G.J. Quantifying tumour heterogeneity in 18F-FDG PET/CT imaging by texture analysis. Eur. J. Nucl. Med. Mol. Imaging 2013, 40, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; Van Stiphout, R.G.; Granton, P.; Zegers, C.M.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Larue, R.T.H.M.; Klaassen, R.; Jochems, A.; Leijenaar, R.T.; Hulshof, M.C.; van Berge Henegouwen, M.I.; Schreurs, W.M.; Sosef, M.N.; van Elmpt, W.; van Laarhoven, H.W.; et al. Pre-treatment CT radiomics to predict 3-year overall survival following chemoradiotherapy of esophageal cancer. Acta Oncol. 2018, 57, 1475–1481. [Google Scholar] [CrossRef] [PubMed]
- Foley, K.G.; Shi, Z.; Whybra, P.; Kalendralis, P.; Larue, R.; Berbee, M.; Sosef, M.N.; Parkinson, C.; Staffurth, J.; Crosby, T.D.; et al. External validation of a prognostic model incorporating quantitative PET image features in oesophageal cancer. Radiother. Oncol. 2018, 133, 205–212. [Google Scholar] [CrossRef]
- Hosny, A.; Parmar, C.; Coroller, T.P.; Grossmann, P.; Zeleznik, R.; Kumar, A.; Bussink, J.; Gillies, R.J.; Mak, R.H.; Aerts, H.J. Deep learning for lung cancer prognostication: A retrospective multi-cohort radiomics study. PLoS Med. 2018, 15, e1002711. [Google Scholar] [CrossRef] [PubMed]
- Haenssle, H.A.; Fink, C.; Schneiderbauer, R.; Toberer, F.; Buhl, T.; Blum, A.; Kalloo, A.; Hassen, A.B.H.; Thomas, L.; Enk, A.; et al. Man against machine: Diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann. Oncol. 2018, 29, 1836–1842. [Google Scholar] [CrossRef]
- Takiyama, H.; Ozawa, T.; Ishihara, S.; Fujishiro, M.; Shichijo, S.; Nomura, S.; Miura, M.; Tada, T. Automatic anatomical classification of esophagogastroduodenoscopy images using deep convolutional neural networks. Sci. Rep. 2018, 8, 7497. [Google Scholar] [CrossRef]
- Wang, H.; Zhou, Z.; Li, Y.; Chen, Z.; Lu, P.; Wang, W.; Liu, W.; Yu, L. Comparison of machine learning methods for classifying mediastinal lymph node metastasis of non-small cell lung cancer from 18F-FDG PET/CT images. EJNMMI Res. 2017, 7, 11. [Google Scholar] [CrossRef]
- Ypsilantis, P.P.; Siddique, M.; Sohn, H.M.; Davies, A.; Cook, G.; Goh, V.; Montana, G. Predicting response to neoadjuvant chemotherapy with pet imaging using convolutional neural networks. PLoS ONE 2015, 10, e0137036. [Google Scholar] [CrossRef]
- Sheetz, K.H.; Zhao, L.; Holcombe, S.A.; Wang, S.C.; Reddy, R.M.; Lin, J.; Orringer, M.B.; Chang, A.C. Decreased core muscle size is associated with worse patient survival following esophagectomy for cancer. Dis. Esophagus 2013, 26, 716–722. [Google Scholar] [CrossRef] [PubMed]
- Murimwa, G.Z.; Venkat, P.S.; Jin, W.; Leuthold, S.; Latifi, K.; Almhanna, K.; Pimiento, J.M.; Fontaine, J.P.; Hoffe, S.E.; Frakes, J.M. Impact of sarcopenia on outcomes of locally advanced esophageal cancer patients treated with neoadjuvant chemoradiation followed by surgery. J. Gastrointest. Oncol. 2017, 8, 808–815. [Google Scholar] [CrossRef] [PubMed]

| Total | Correct Prediction | Incorrect Prediction | p | |
|---|---|---|---|---|
| n = 548 | n = 401 | n = 147 | ||
| Age, (years, mean ± SD) | 61.4 ± 12.4 | 61.7 ± 12.7 | 60.5 ± 11.7 | 0.290 |
| Gender (%) | 0.320 | |||
| Male | 504 (92.0) | 366 (91.3) | 138 (93.9) | |
| Female | 44 (8.0) | 35 (8.7) | 9 (6.1) | |
| Clinical T stage (%) | 0.109 | |||
| T1 | 65 (11.9) | 55 (13.7) | 10 (6.8) | |
| T2 | 118 (21.5) | 89 (22.2) | 29 (19.7) | |
| T3 | 325 (59.3) | 229 (57.1) | 96 (65.3) | |
| T4 | 40 (7.3) | 28 (7.0) | 12 (8.2) | |
| Clinical N stage (%) | 0.016 | |||
| N0 | 178 (32.5) | 141 (35.2) | 37 (25.2) | |
| N1 | 196 (35.8) | 128 (31.9) | 68 (46.3) | |
| N2 | 102 (18.6) | 76 (19.0) | 26 (17.7) | |
| N3 | 72 (13.1) | 56 (14.0) | 16 (10.9) | |
| Clinical M stage (%) | 0.970 | |||
| M0 | 458 (83.6) | 335 (83.5) | 123 (83.7) | |
| M1 | 90 (16.4) | 66 (16.5) | 24 (16.3) | |
| Tumor location (%) | 0.349 | |||
| Upper third | 161 (29.4) | 111 (27.7) | 50 (34.0) | |
| Middle third | 242 (44.2) | 182 (45.4) | 60 (40.8) | |
| Lower third | 145 (26.5) | 108 (26.9) | 37 (25.2) | |
| Tumor size, (cm, mean ± SD) | 4.4 ± 3.4 | 4.3 ± 3.3 | 4.5 ± 3.5 | 0.730 |
| SUVMAX, (mean ± SD) | 12.5 ± 6.0 | 12.3 ± 6.2 | 12.9 ± 5.4 | 0.308 |
| Tumor markers, (mean ± SD) | ||||
| SCC, ng/ml | 2.6 ± 5.3 | 2.6 ± 6.0 | 2.6 ± 2.8 | 0.987 |
| Cyfra 21-1, ng/ml | 4.2 ± 5.9 | 4.4 ± 7.2 | 4.1 ± 5.9 | 0.879 |
| Treatments (%) | 0.758 | |||
| Primary resection | 141 (25.7) | 101 (25.2) | 40 (27.2) | |
| Trimodal treatments | 160 (29.2) | 115 (28.7) | 45 (30.6) | |
| Medical treatment | 208 (38.0) | 154 (38.4) | 54 (36.7) | |
| Supportive care | 39 (7.1) | 31 (7.7) | 8 (5.4) |
| Models | AUC | 95% CI | ||
|---|---|---|---|---|
| Pretrain | Optimizer | Layers | ||
| No | Adam | 18 | 0.710 | 0.688–0.733 |
| No | SGD | 18 | 0.724 | 0.714–0.734 |
| No | Adam | 34 | 0.740 | 0.704–0.776 |
| No | SGD | 34 | 0.722 | 0.697–0.747 |
| Yes | Adam | 18 | 0.717 | 0.693–0.741 |
| Yes | SGD | 18 | 0.709 | 0.678–0.741 |
| Yes | Adam | 34 | 0.738 | 0.714–0.761 |
| Yes | SGD | 34 | 0.720 | 0.694–0.746 |
| HR | 95% CI | p | |
|---|---|---|---|
| Age | 1.018 | 1.010–1.027 | <0.001 |
| Gender | |||
| Male | 1 | - | - |
| Female | 0.426 | 0.252–0.720 | 0.001 |
| Tumor location | |||
| Lower third | 1 | - | - |
| Middle third | 1.297 | 0.960–1.752 | 0.090 |
| Upper third | 1.333 | 1.007–1.763 | 0.044 |
| Clinical T stage | |||
| T1/2 | 1 | - | - |
| T3/4 | 1.209 | 0.946–1.545 | 0.130 |
| Clinical N stage | |||
| N(−) | 1 | - | - |
| N(+) | 1.417 | 1.082–1.856 | 0.011 |
| Clinical M stage | |||
| M0 | 1 | - | - |
| M1 | 2.041 | 1.571–2.651 | <0.001 |
| Prediction results | |||
| Alive at one year | 1 | - | - |
| Expired within one year | 2.830 | 2.252–3.555 | <0.001 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, C.-K.; Yeh, J.C.-Y.; Yu, W.-H.; Chien, L.-I.; Lin, K.-H.; Huang, W.-S.; Hsu, P.-K. Deep Convolutional Neural Network-Based Positron Emission Tomography Analysis Predicts Esophageal Cancer Outcome. J. Clin. Med. 2019, 8, 844. https://doi.org/10.3390/jcm8060844
Yang C-K, Yeh JC-Y, Yu W-H, Chien L-I, Lin K-H, Huang W-S, Hsu P-K. Deep Convolutional Neural Network-Based Positron Emission Tomography Analysis Predicts Esophageal Cancer Outcome. Journal of Clinical Medicine. 2019; 8(6):844. https://doi.org/10.3390/jcm8060844
Chicago/Turabian StyleYang, Cheng-Kun, Joe Chao-Yuan Yeh, Wei-Hsiang Yu, Ling-I. Chien, Ko-Han Lin, Wen-Sheng Huang, and Po-Kuei Hsu. 2019. "Deep Convolutional Neural Network-Based Positron Emission Tomography Analysis Predicts Esophageal Cancer Outcome" Journal of Clinical Medicine 8, no. 6: 844. https://doi.org/10.3390/jcm8060844
APA StyleYang, C.-K., Yeh, J. C.-Y., Yu, W.-H., Chien, L.-I., Lin, K.-H., Huang, W.-S., & Hsu, P.-K. (2019). Deep Convolutional Neural Network-Based Positron Emission Tomography Analysis Predicts Esophageal Cancer Outcome. Journal of Clinical Medicine, 8(6), 844. https://doi.org/10.3390/jcm8060844





