Selection Criteria for Determination of Optimal Reconstruction Method for Cu-64 Trastuzumab Dosimetry on Siemens Inveon PET Scanner
Abstract
1. Introduction
2. Experimental Section
2.1. Ethical Statement
2.2. PET Scanner
2.3. Reconstruction Algorithms
2.4. Attenuation and Scatter Corrections
2.5. Phantom Studies
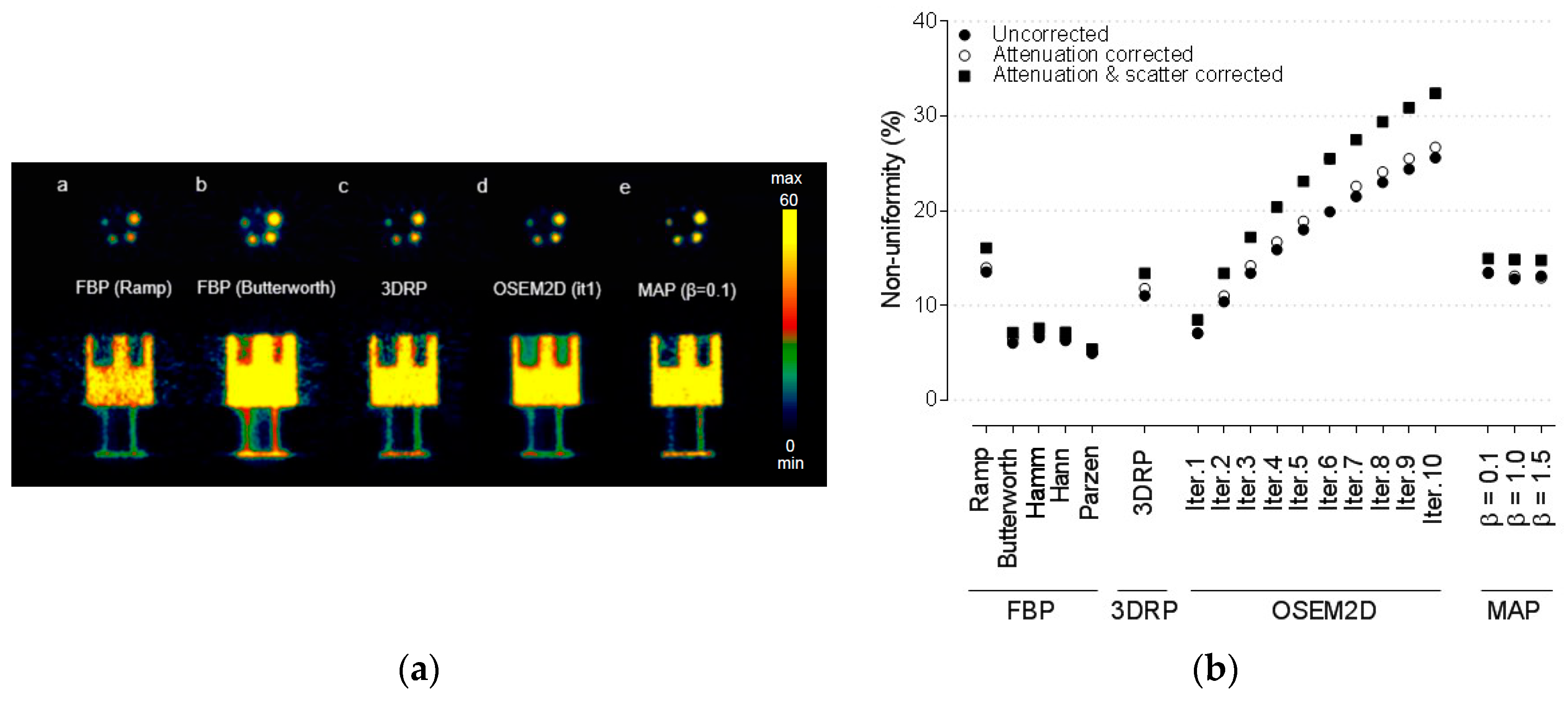
2.5.1. Non-Uniformity
2.5.2. Recovery Coefficient
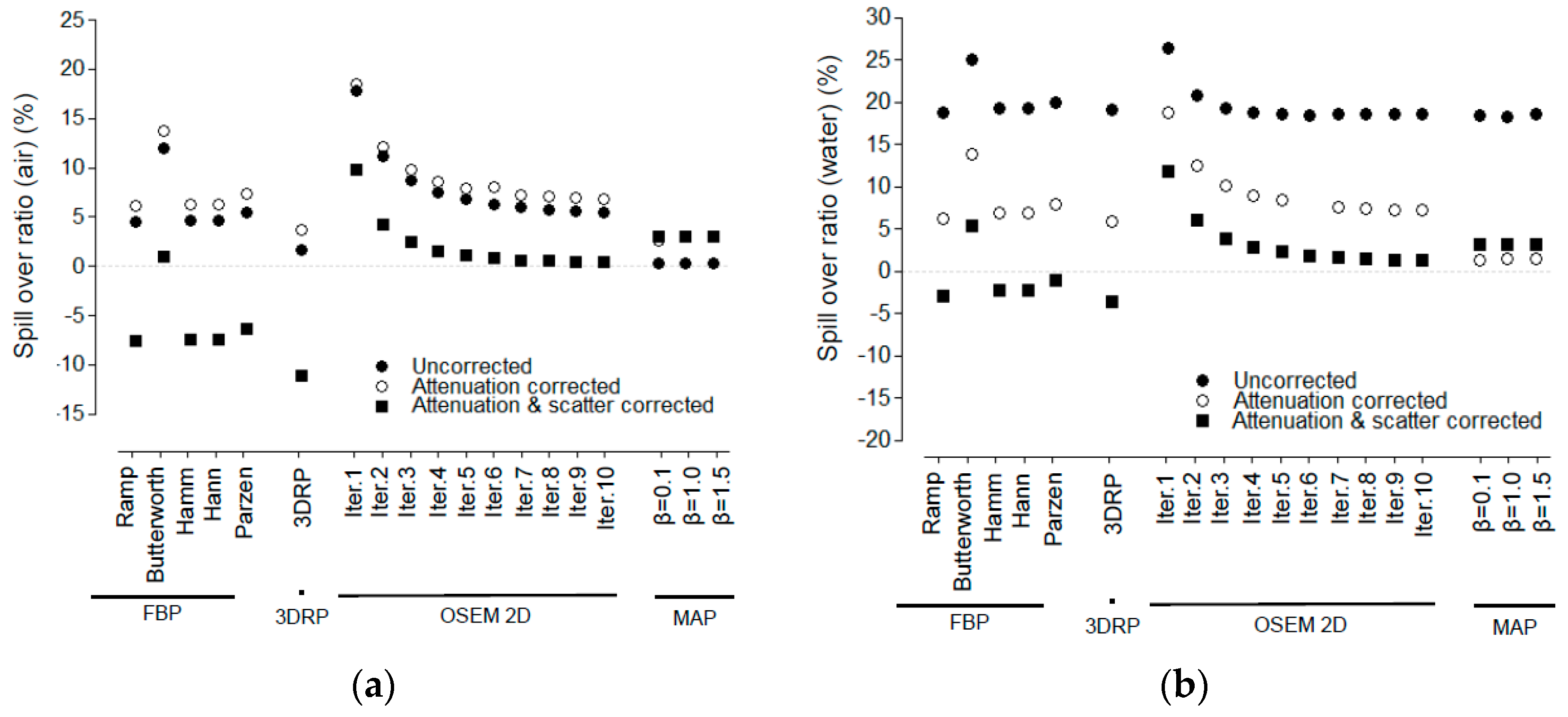
2.5.3. Spill-Over Ratio
2.6. Quantification of Cu-64 Trastuzumab PET
2.6.1. Cell Culture and Tumor Xenograft in Mice
2.6.2. Radiolabeling of Cu-64-DOTA-Trastuzumab
2.6.3. Cu-64 Trastuzumab PET
2.6.4. Dosimetry
3. Results
3.1. Non-Uniformity
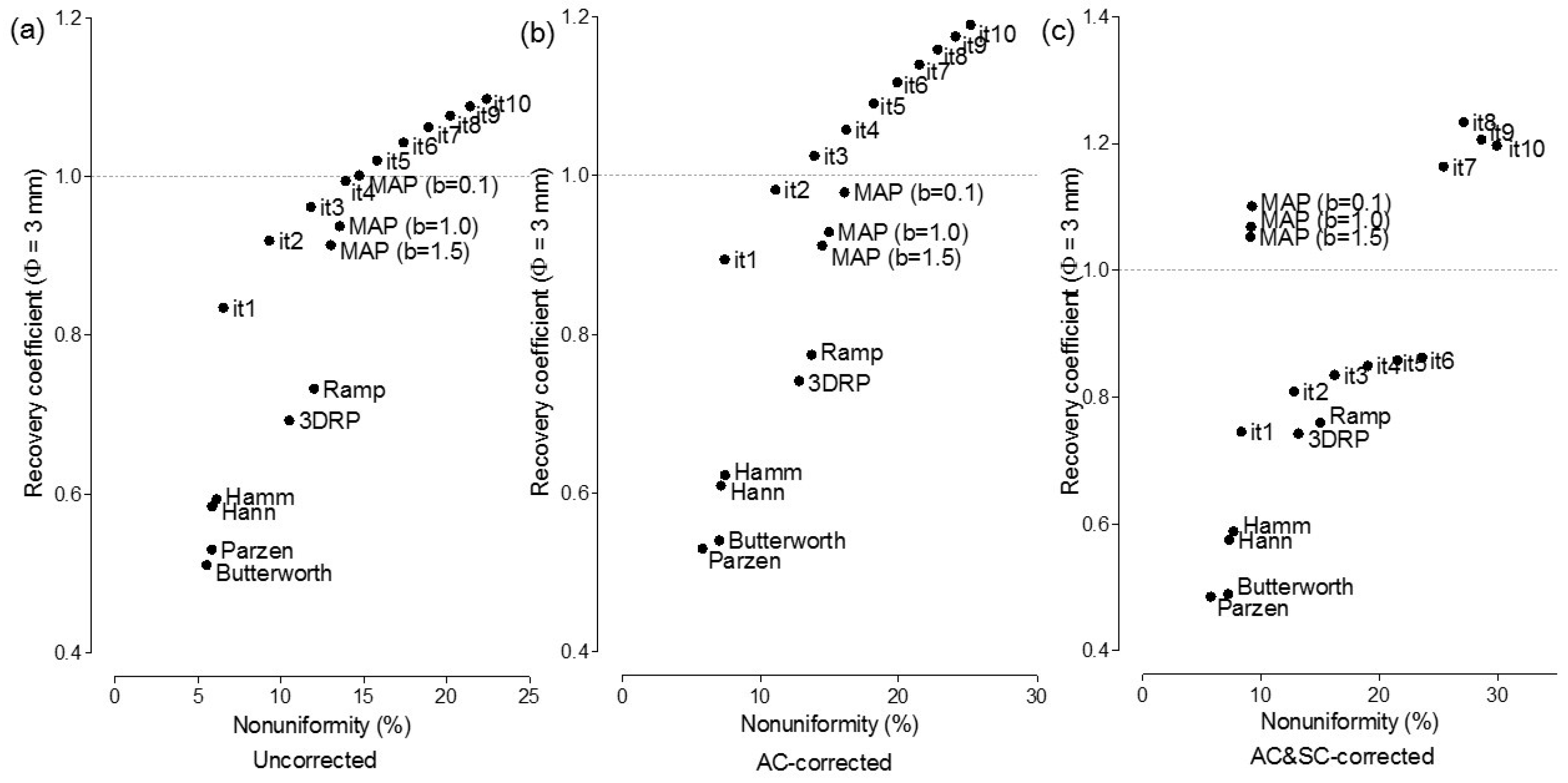
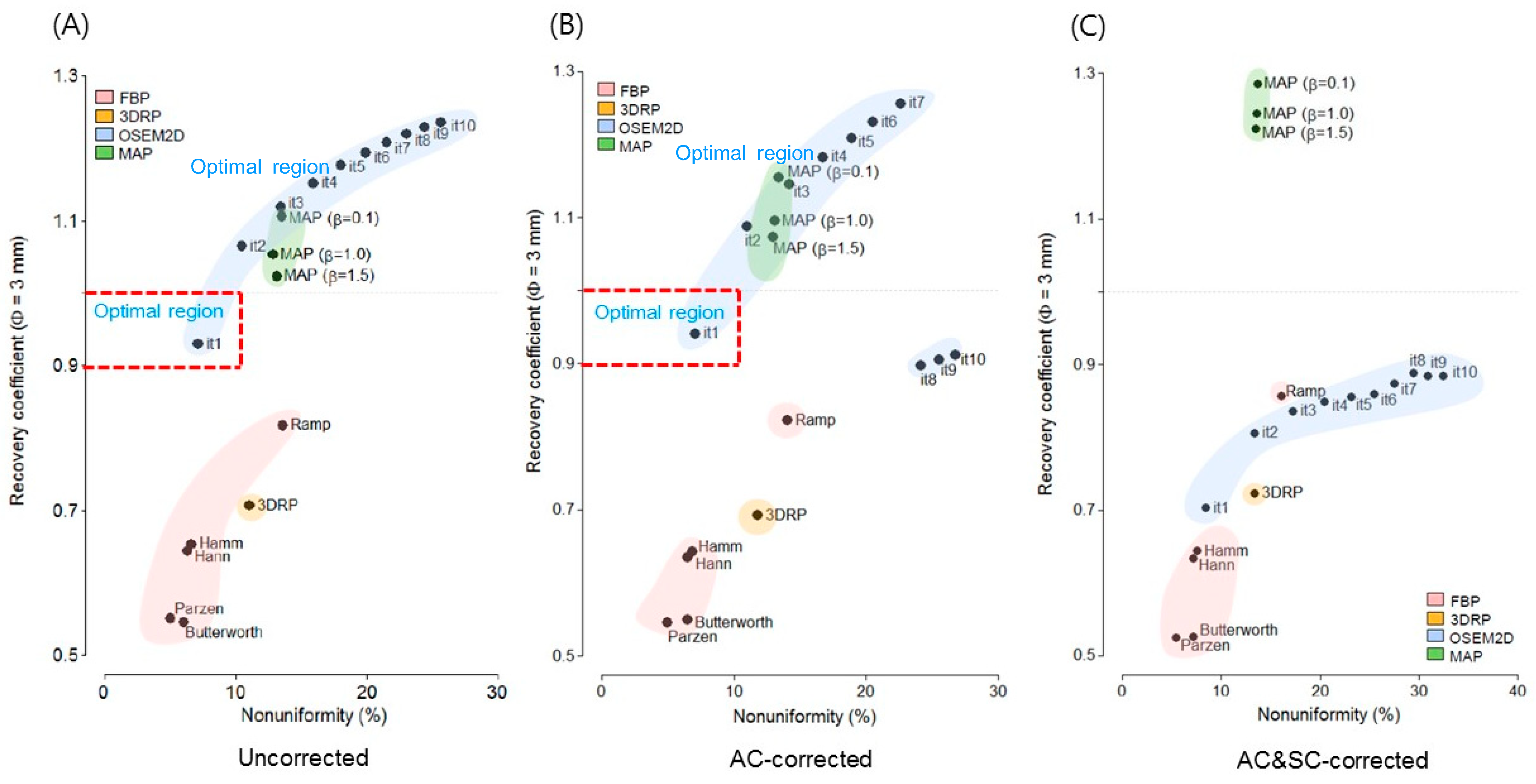
3.2. Recovery Coefficient and Non-Uniformity
3.3. Spill-Over Ratio
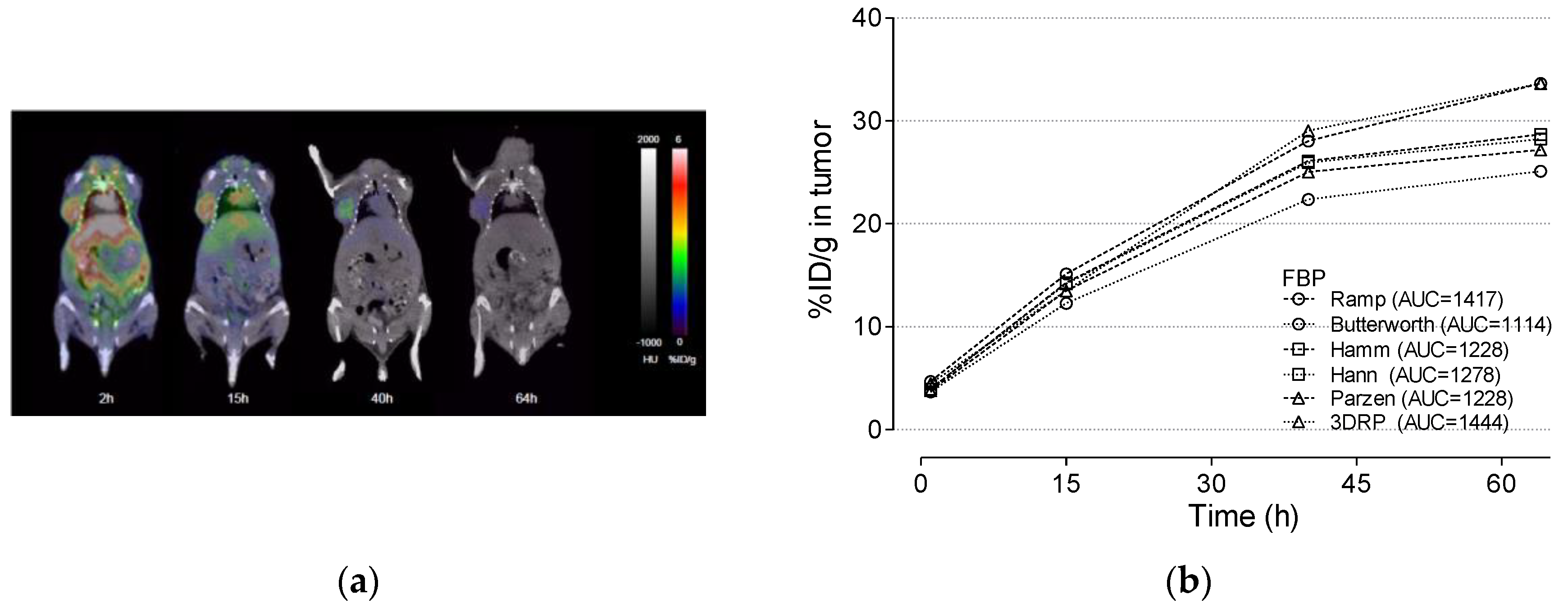
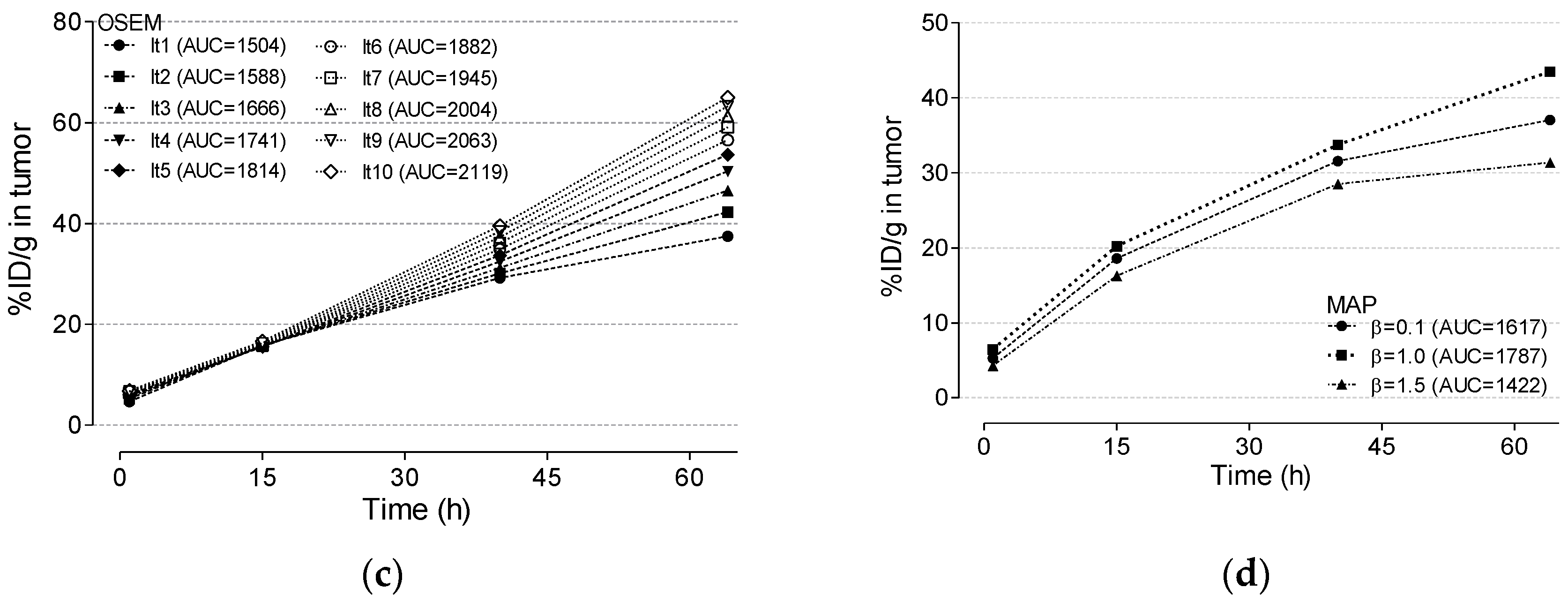
3.4. Cu-64 Trastuzumab PET
3.5. Radiation Dosimetry Using Various Reconstruction Algorithms and Filters
4. Discussion
4.1. Spill-Over Ratio
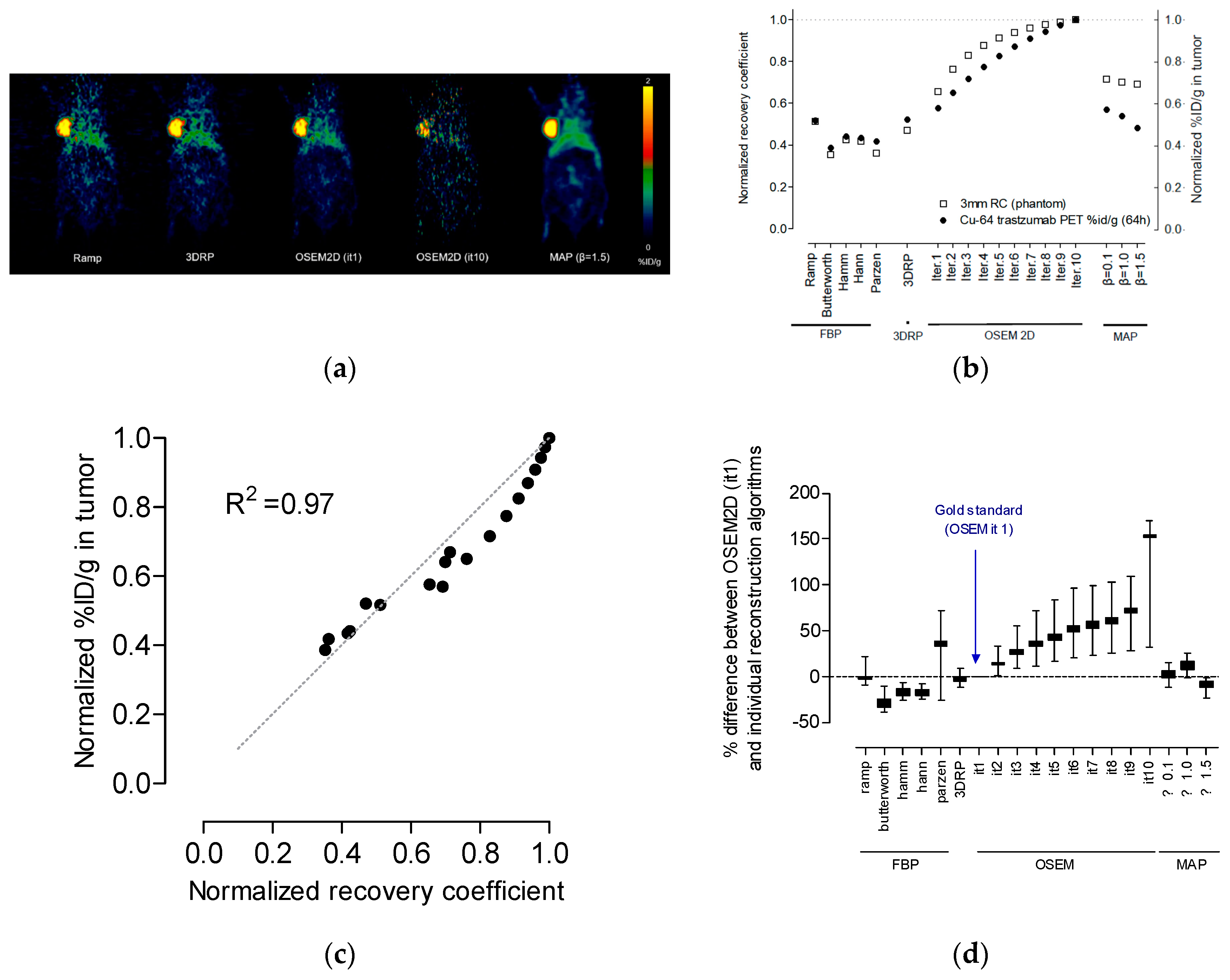
4.2. Selection of the Optimal Reconstruction Algorithm (a Trade-Off Relationship between Recovery Coefficients and Non-Uniformity)
4.3. Effect of Attenuation Correction and Scatter Correction
4.4. Correlation between Recovery Coefficient and % ID/g in the Tumor Region
4.5. Optimal Reconstruction Algorithm and Tumor Absorbed Dose
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
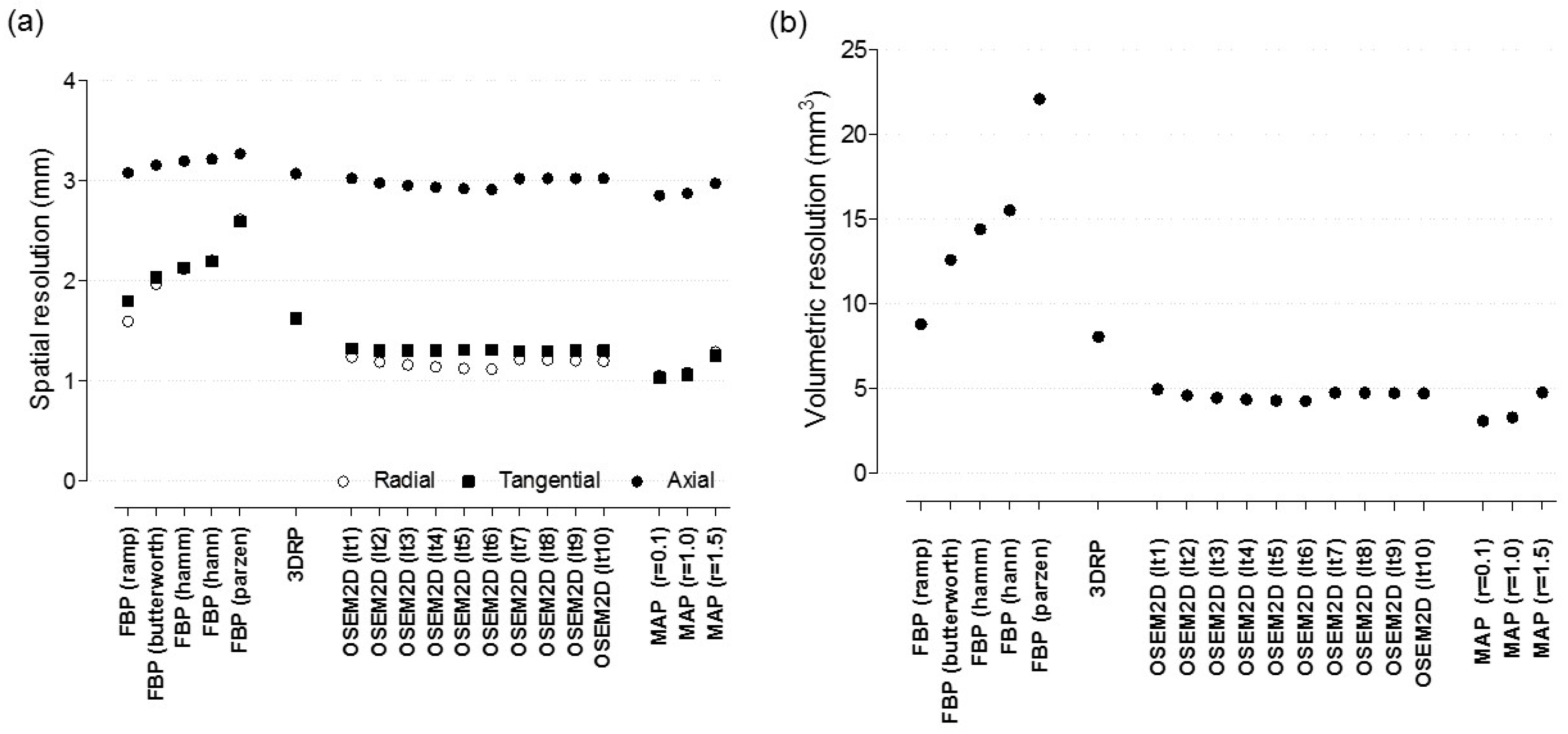
Appendix A.1. Spatial Resolution
Appendix A.2. The Effect of Reconstruction Algorithms with Various Filters
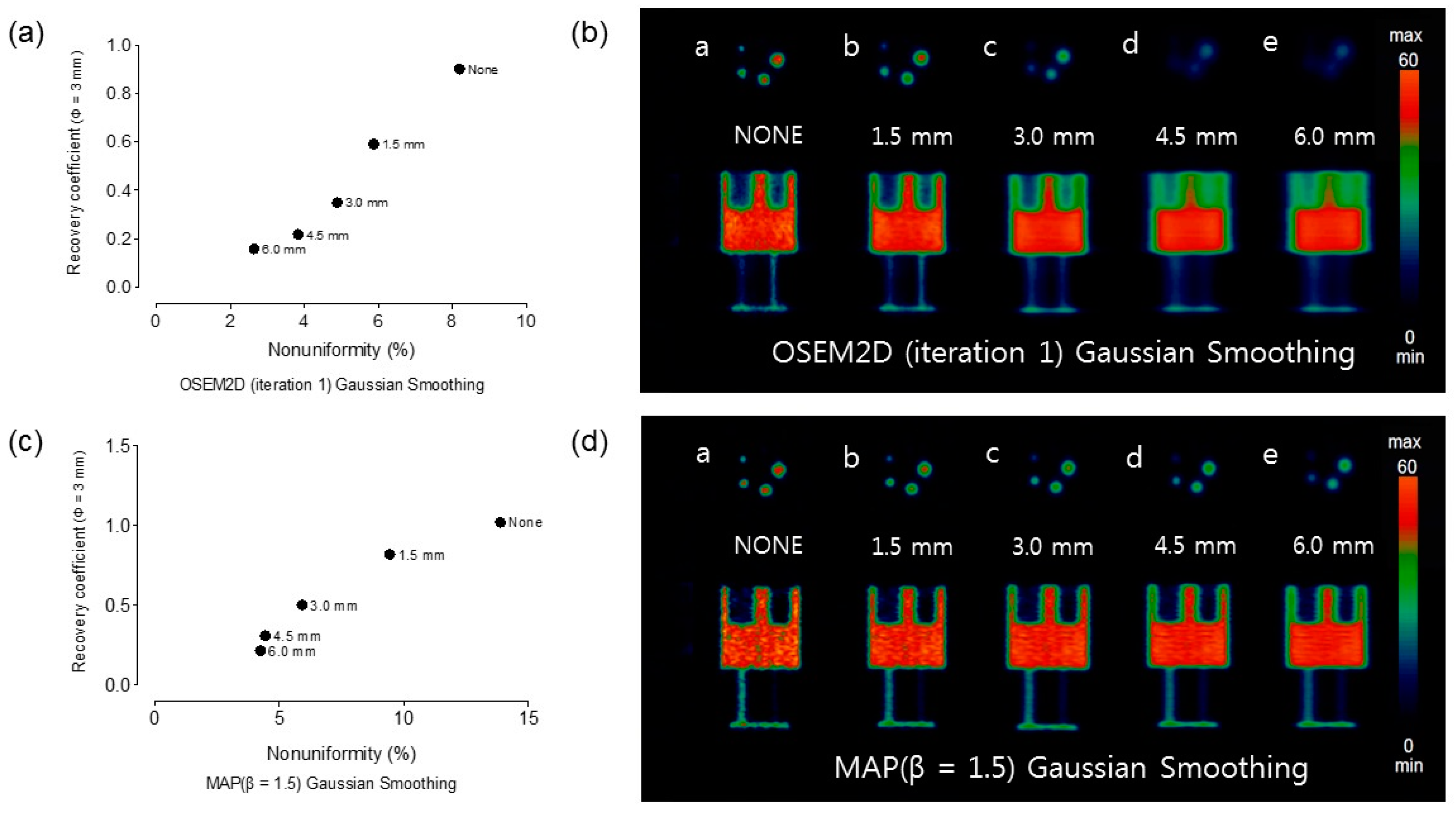
Appendix A.3. The Effect of Post-Smoothing with Various Gaussian Kernel
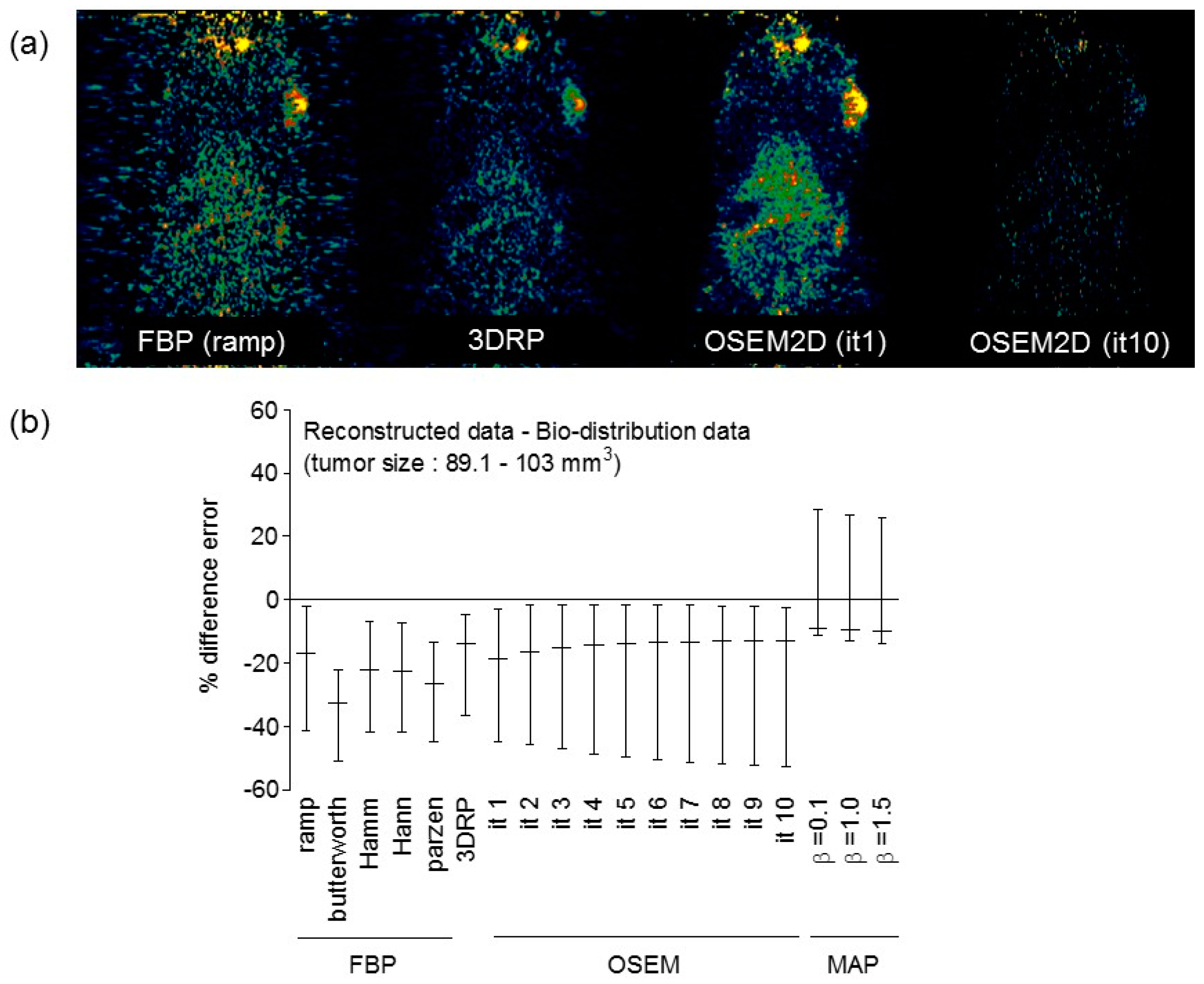
Appendix A.4. Comparison between Reconstructed Data and Bio-Distribution Data
Appendix A.5. Spill-Over Ratio vs. Recovery Coefficient

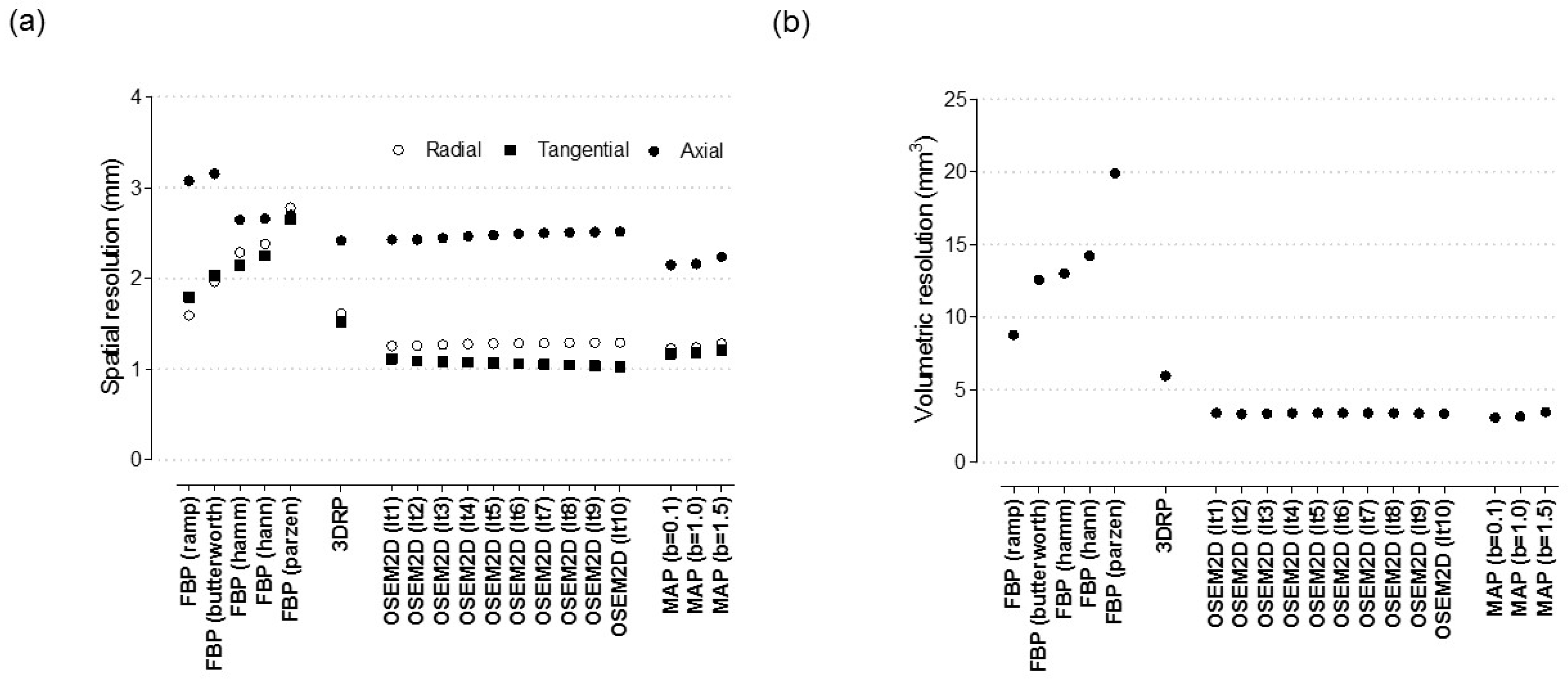
Appendix A.6. Assessment of Zr-89 PET Data in Terms of Spatial Resolution and SOR and NU
Appendix A.7. The effect of Reconstruction Algorithms with Various Filters
Appendix A.8. Spill-Over Ratio vs. Non-Uniformity
References
- Boerman, O.C.; Oyen, W.J. Immuno-PET of cancer: A revival of antibody imaging. J. Nucl. Med. 2011, 52, 1171–1172. [Google Scholar] [CrossRef]
- Kim, J.S. Combination Radioimmunotherapy Approaches and Quantification of Immuno-PET. Nucl. Med. Mol. Imaging 2016, 50, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Mestel, R. Cancer: Imaging with antibodies. Nature 2017, 543, 743–746. [Google Scholar] [CrossRef]
- van Dongen, G.A.; Visser, G.W.; Lub-de Hooge, M.N.; de Vries, E.G.; Perk, L.R. Immuno-PET: A navigator in monoclonal antibody development and applications. Oncologist 2007, 12, 1379–1389. [Google Scholar] [CrossRef]
- Tamura, K.; Kurihara, H.; Yonemori, K.; Tsuda, H.; Suzuki, J.; Kono, Y.; Honda, N.; Kodaira, M.; Yamamoto, H.; Yunokawa, M.; et al. Cu-64 DOTA-trastuzumab PET imaging in patients with HER2-positive breast cancer. J. Nucl. Med. 2013, 54, 1869–1875. [Google Scholar] [CrossRef]
- Mortimer, J.E.; Bading, J.R.; Colcher, D.M.; Conti, P.S.; Frankel, P.H.; Carroll, M.I.; Tong, S.; Poku, E.; Miles, J.K.; Shively, J.E.; et al. Functional imaging of human epidermal growth factor receptor 2-positive metastatic breast cancer using Cu-64 DOTA-trastuzumab PET. J. Nucl. Med. 2014, 55, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Kurihara, H.; Hamada, A.; Yoshida, M.; Shimma, S.; Hashimoto, J.; Yonemori, K.; Tani, H.; Miyakita, Y.; Kanayama, Y.; Wada, Y.; et al. Cu-64 DOTA-trastuzumab PET imaging and HER2 specificity of brain metastases in HER2-positive breast cancer patients. EJNMMI Res. 2015, 5, 8. [Google Scholar] [CrossRef]
- Woo, S.K.; Jang, S.J.; Seo, M.J.; Park, J.H.; Kim, B.S.; Kim, E.J.; Lee, Y.J.; Lee, T.S.; An, G.I.; Song, I.H.; et al. Development of Cu-64 -NOTA-Trastuzumab for HER2 targeting: Radiopharmaceutical with improved pharmacokinetics for human study. J. Nucl. Med. 2018, 60, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Stabin, M.G.; Sparks, R.B.; Crowe, E. OLINDA/EXM: The second-generation personal computer software for internal dose assessment in nuclear medicine. J. Nucl. Med. 2005, 46, 1023–1027. [Google Scholar]
- Lee, Y.S.; Kim, J.S.; Cho, K.D.; Kang, J.H.; Lim, S.M. Tumor dosimetry for I-131 trastuzumab therapy in a Her2+ NCI N87 xenograft mouse model using the Siemens SYMBIA E gamma camera with a pinhole collimator. J. INSTRUM 2015, 10, P07001. [Google Scholar] [CrossRef]
- Poston, J.W. Application of the effective dose equivalent to nuclear medicine patients. The MIRD Committee. J. Nucl. Med. 1993, 34, 714–716. [Google Scholar]
- Howell, R.W. The MIRD Schema: From organ to cellular dimensions. J. Nucl. Med. 1994, 35, 531–533. [Google Scholar]
- Boellaard, R.; Delgado-Bolton, R.; Oyen, W.J.; Giammarile, F.; Tatsch, K.; Eschner, W.; Verzijlbergen, F.J.; Barrington, S.F.; Pike, L.C.; Weber, W.A.; et al. FDG PET/CT: EANM procedure guidelines for tumour imaging: Version 2.0. Eur. J. Nucl. Med. Mol. Imaging 2015, 42, 328–354. [Google Scholar] [CrossRef]
- NEMA. Performance Measurements of Small Animal Positron Emission Tomographs (PETs). In NEMA Standards Publication NU 4-2008; NEMA: Rosslyn, VA, USA, 2008. [Google Scholar]
- Bahri, M.A.; Plenevaux, A.; Warnock, G.; Luxen, A.; Seret, A. NEMA NU4-2008 image quality performance report for the microPET focus 120 and for various transmission and reconstruction methods. J. Nucl. Med. 2009, 50, 1730–1738. [Google Scholar] [CrossRef][Green Version]
- Constantinescu, C.C.; Mukherjee, J. Performance evaluation of an Inveon PET preclinical scanner. Phys. Med. Biol. 2009, 54, 2885–2899. [Google Scholar] [CrossRef]
- Bao, Q.; Newport, D.; Chen, M.; Stout, D.B.; Chatziioannou, A.F. Performance evaluation of the inveon dedicated PET preclinical tomograph based on the NEMA NU-4 standards. J. Nucl. Med. 2009, 50, 401–408. [Google Scholar] [CrossRef]
- Lasnon, C.; Dugue, A.E.; Briand, M.; Blanc-Fournier, C.; Dutoit, S.; Louis, M.H.; Aide, N. NEMA NU 4-Optimized Reconstructions for Therapy Assessment in Cancer Research with the Inveon Small Animal PET/CT System. Mol. Imaging Biol. 2015, 17, 403–412. [Google Scholar] [CrossRef]
- Bradshaw, T.J.; Voorbach, M.J.; Reuter, D.R.; Giamis, A.M.; Mudd, S.R.; Beaver, J.D. Image quality of Zr-89 PET imaging in the Siemens microPET Focus 220 preclinical scanner. Mol. Imaging Biol. 2016, 18, 377–385. [Google Scholar] [CrossRef]
- Yu, A.R.; Kim, J.S. Effect of filters and reconstruction algrorithms on I-124 PET in Siemens Inveon PET scanner. J. INSTRUM 2015, 10, P10026. [Google Scholar] [CrossRef]
- Laforest, R.; Lapi, S.E.; Oyama, R.; Bose, R.; Tabchy, A.; Marquez-Nostra, B.V.; Burkemper, J.; Wright, B.D.; Frye, J.; Frye, S.; et al. Zr-89 Trastuzumab: Evaluation of Radiation Dosimetry, Safety, and Optimal Imaging Parameters in Women with HER2-Positive Breast Cancer. Mol. Imaging Biol. 2016, 18, 952–959. [Google Scholar] [CrossRef]
- O’Sullivan, F.; Pawitan, Y.; Haynor, D. Reducing negative artifact in emission tomography. IEEE Trans. Med. Imaging 1993, 12, 653–663. [Google Scholar] [CrossRef] [PubMed]
- Hudson, H.M.; Larkin, R.S. Accelerated image reconstruction using ordered subsets of projection data. IEEE Trans. Med. Imaging 1994, 13, 601–619. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Kim, J.S.; Kim, H.-J.; Woo, S.K.; Kim, J.G.; Park, J.A.; Choi, C.W.; Lim, S.M.; Kim, K.M. Imaging Characteristics of I-124 Between 3D and 2D on Siemens ECAT HR PET Scanner. IEEE Trans. Nucl. Sci. 2013, 60, 797–801. [Google Scholar] [CrossRef]
- Kidera, D.; Kihara, K.; Akamatsu, G.; Mikasa, S.; Taniguchi, T.; Tsutsui, Y.; Takeshita, T.; Maebatake, A.; Miwa, K.; Sasaki, M. The edge artifact in the point-spread function-based PET reconstruction at different sphere-to-background ratios of radioactivity. Ann. Nucl. Med. 2016, 30, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Mayer, A.T.; Natarajan, A.; Gordon, S.R.; Maute, R.L.; McCracken, M.N.; Ring, A.M.; Weissman, I.L.; Gambhir, S.S. Practical Immuno-PET Radiotracer Design Considerations for Human Immune Checkpoint Imaging. J. Nucl. Med. 2017, 58, 538–546. [Google Scholar] [CrossRef] [PubMed][Green Version]

| FBP | 3DRP | OSEM2D | OSEM3D-MAP | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ramp | Butterworth | Hamm | Hann | Parzen | Iter.1 | Iter.2 | Iter.3 | Iter.4 | Iter.5 | Iter.6 | Iter.7 | Iter.8 | Iter.9 | Iter.10 | β = 0.1 | β = 1.0 | β = 1.5 | ||
| Tumor (mGy/MBq) | 1200 | 923 | 1020 | 1060 | 1020 | 1250 | 1320 | 1340 | 1440 | 1480 | 1560 | 1620 | 1660 | 1710 | 1760 | 1830 | 1380 | 1500 | 1180 |
| Organ (mSv/MBq) | |||||||||||||||||||
| Adrenals | 0.0122 | 0.0094 | 0.0106 | 0.0104 | 0.0175 | 0.0121 | 0.0122 | 0.0140 | 0.0159 | 0.0175 | 0.0190 | 0.0206 | 0.0219 | 0.0227 | 0.0259 | 0.0911 | 0.0127 | 0.0141 | 0.0113 |
| Brain | 0.0327 | 0.0216 | 0.0222 | 0.0215 | 0.0556 | 0.0236 | 0.0263 | 0.0367 | 0.0466 | 0.0556 | 0.0642 | 0.0756 | 0.0775 | 0.0823 | 0.0899 | 0.3200 | 0.0233 | 0.0283 | 0.0207 |
| Breasts | 0.0036 | 0.0027 | 0.0031 | 0.0030 | 0.0053 | 0.0035 | 0.0037 | 0.0042 | 0.0048 | 0.0053 | 0.0057 | 0.0064 | 0.0066 | 0.0067 | 0.0077 | 0.0272 | 0.0036 | 0.0040 | 0.0033 |
| Gallbladder wall | 0.0107 | 0.0084 | 0.0093 | 0.0092 | 0.0147 | 0.0109 | 0.0105 | 0.0121 | 0.0135 | 0.0147 | 0.0158 | 0.0174 | 0.0180 | 0.0189 | 0.0211 | 0.0744 | 0.0110 | 0.0124 | 0.0096 |
| Lower large intestine wall | 0.0031 | 0.0023 | 0.0026 | 0.0026 | 0.0044 | 0.0031 | 0.0032 | 0.0036 | 0.0040 | 0.0044 | 0.0048 | 0.0053 | 0.0055 | 0.0058 | 0.0066 | 0.0231 | 0.0034 | 0.0038 | 0.0030 |
| Small intestine | 0.0041 | 0.0032 | 0.0036 | 0.0035 | 0.0057 | 0.0041 | 0.0041 | 0.0047 | 0.0052 | 0.0057 | 0.0062 | 0.0067 | 0.0071 | 0.0075 | 0.0084 | 0.0296 | 0.0043 | 0.0048 | 0.0038 |
| Stomach wall | 0.1060 | 0.0906 | 0.0947 | 0.0937 | 0.1230 | 0.1110 | 0.1010 | 0.1090 | 0.1160 | 0.1230 | 0.1300 | 0.1420 | 0.1440 | 0.1560 | 0.1820 | 0.6270 | 0.0993 | 0.1000 | 0.0925 |
| Upper lower intestine wall | 0.0046 | 0.0036 | 0.0040 | 0.0040 | 0.0063 | 0.0047 | 0.0046 | 0.0052 | 0.0058 | 0.0063 | 0.0069 | 0.0075 | 0.0078 | 0.0083 | 0.0093 | 0.0327 | 0.0048 | 0.0053 | 0.0043 |
| Heart wall | 0.0084 | 0.0064 | 0.0072 | 0.0071 | 0.0123 | 0.0084 | 0.0086 | 0.0097 | 0.0112 | 0.0123 | 0.0133 | 0.0148 | 0.0152 | 0.0155 | 0.0179 | 0.0629 | 0.0086 | 0.0094 | 0.0077 |
| Kidneys | 0.1890 | 0.1460 | 0.1650 | 0.1570 | 0.2680 | 0.1870 | 0.1800 | 0.2130 | 0.2420 | 0.2680 | 0.2930 | 0.2850 | 0.3390 | 0.3600 | 0.4040 | 1.4100 | 0.1830 | 0.2030 | 0.1720 |
| Liver | 0.0944 | 0.0746 | 0.0828 | 0.0821 | 0.1300 | 0.0979 | 0.0924 | 0.1070 | 0.1190 | 0.1300 | 0.1390 | 0.1550 | 0.1580 | 0.1660 | 0.1810 | 0.6440 | 0.0990 | 0.1130 | 0.0840 |
| Lungs | 0.1660 | 0.1240 | 0.1410 | 0.1390 | 0.2590 | 0.1630 | 0.1770 | 0.1980 | 0.2360 | 0.2590 | 0.2800 | 0.3140 | 0.3200 | 0.3220 | 0.3750 | 1.3200 | 0.1650 | 0.1790 | 0.1530 |
| Muscle | 0.0035 | 0.0027 | 0.0030 | 0.0030 | 0.0051 | 0.0035 | 0.0035 | 0.0041 | 0.0046 | 0.0051 | 0.0055 | 0.0061 | 0.0063 | 0.0066 | 0.0075 | 0.0264 | 0.0037 | 0.0040 | 0.0033 |
| Ovaries | 0.0031 | 0.0023 | 0.0027 | 0.0026 | 0.0045 | 0.0032 | 0.0032 | 0.0037 | 0.0041 | 0.0045 | 0.0049 | 0.0054 | 0.0057 | 0.0060 | 0.0067 | 0.0236 | 0.0035 | 0.0039 | 0.0031 |
| Pancreas | 0.0164 | 0.0127 | 0.0143 | 0.0141 | 0.0228 | 0.0162 | 0.0163 | 0.0187 | 0.0208 | 0.0228 | 0.0248 | 0.0276 | 0.0287 | 0.0297 | 0.0346 | 0.1210 | 0.0174 | 0.0190 | 0.0154 |
| Red. Marrow | 0.0038 | 0.0029 | 0.0033 | 0.0032 | 0.0056 | 0.0037 | 0.0038 | 0.0044 | 0.0051 | 0.0056 | 0.0062 | 0.0068 | 0.0071 | 0.0074 | 0.0084 | 0.0296 | 0.0039 | 0.0043 | 0.0035 |
| Osteogenic | 0.0031 | 0.0023 | 0.0026 | 0.0026 | 0.0047 | 0.0030 | 0.0031 | 0.0036 | 0.0042 | 0.0047 | 0.0051 | 0.0057 | 0.0059 | 0.0061 | 0.0070 | 0.0245 | 0.0031 | 0.0035 | 0.0028 |
| Skin | 0.0017 | 0.0013 | 0.0015 | 0.0014 | 0.0025 | 0.0017 | 0.0017 | 0.0020 | 0.0023 | 0.0025 | 0.0027 | 0.0030 | 0.0032 | 0.0033 | 0.0037 | 0.0131 | 0.0018 | 0.0019 | 0.0016 |
| Spleen | 0.4480 | 0.3260 | 0.3850 | 0.3820 | 0.6730 | 0.4120 | 0.4620 | 0.5380 | 0.6040 | 0.6730 | 0.7460 | 0.8620 | 0.8940 | 0.8990 | 1.0900 | 3.8000 | 0.5390 | 0.5960 | 0.4590 |
| Thymus | 0.0038 | 0.0029 | 0.0033 | 0.0032 | 0.0058 | 0.0038 | 0.0040 | 0.0045 | 0.0053 | 0.0058 | 0.0063 | 0.0070 | 0.0072 | 0.0073 | 0.0084 | 0.0296 | 0.0039 | 0.0042 | 0.0035 |
| Thyroid | 0.0014 | 0.0010 | 0.0011 | 0.0011 | 0.0021 | 0.0013 | 0.0014 | 0.0016 | 0.0019 | 0.0021 | 0.0023 | 0.0026 | 0.0027 | 0.0027 | 0.0031 | 0.0110 | 0.0013 | 0.0015 | 0.0012 |
| Urinary bladder | 0.1590 | 0.1140 | 0.1340 | 0.1320 | 0.2380 | 0.1600 | 0.1680 | 0.1940 | 0.2160 | 0.2380 | 0.2580 | 0.2890 | 0.2970 | 0.3160 | 0.3500 | 1.2300 | 0.1870 | 0.2120 | 0.1620 |
| Uterus | 0.0050 | 0.0037 | 0.0042 | 0.0042 | 0.0073 | 0.0050 | 0.0052 | 0.0060 | 0.0067 | 0.0073 | 0.0080 | 0.0089 | 0.0092 | 0.0097 | 0.0108 | 0.0381 | 0.0057 | 0.0064 | 0.0050 |
| FBP | 3DRP | OSEM2D | OSEM3D-MAP | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % Difference | Ramp | ButterWorth | Hamm | Hann | Parzen | 3DRP | Iter.1 * | Iter.2 | Iter.3 | Iter.4 | Iter.5 | Iter.6 | Iter.7 | Iter.8 | Iter.9 | Iter.10 | β = 0.1 | β = 1.0 | β = 1.5 |
| Tumor | −9.5 | −35.4 | −25.6 | −21.8 | −25.6 | −5.4 | - | 1.5 | 8.7 | 11.4 | 16.7 | 20.4 | 22.8 | 25.7 | 28.6 | 32.4 | 4.4 | 12.8 | −11.2 |
| Organ | |||||||||||||||||||
| Adrenals | 0.0 | −25.9 | −14.0 | −15.9 | 35.7 | −0.8 | - | 13.7 | 26.3 | 35.7 | 43.6 | 51.2 | 56.9 | 60.2 | 71.9 | 152.8 | 4.0 | 14.4 | −7.7 |
| Brain | 21.7 | −19.6 | −16.9 | −20.1 | 71.6 | −10.8 | - | 33.0 | 55.7 | 71.6 | 83.8 | 96.8 | 98.7 | 103.1 | 109.5 | 169.6 | −12.1 | 7.3 | −23.8 |
| Breasts | −2.7 | −31.3 | −17.6 | −20.9 | 35.6 | −5.6 | - | 12.7 | 25.9 | 35.6 | 42.6 | 53.5 | 56.3 | 57.7 | 70.2 | 152.1 | −2.7 | 7.8 | −11.4 |
| Gallbladder wall | 1.9 | −22.2 | −12.1 | −13.2 | 33.3 | 3.7 | - | 14.2 | 25.0 | 33.3 | 40.3 | 49.5 | 52.6 | 57.1 | 67.1 | 150.5 | 4.7 | 16.6 | −9.0 |
| Lower large intestine wall | −3.2 | −32.7 | −20.7 | −20.7 | 31.6 | −3.2 | - | 11.8 | 22.2 | 31.6 | 40.0 | 49.4 | 52.9 | 57.8 | 69.4 | 151.3 | 6.1 | 17.1 | −6.5 |
| Small intestine | 0.0 | −24.7 | −13.0 | −15.8 | 32.7 | 0.0 | - | 13.6 | 23.7 | 32.7 | 40.8 | 48.1 | 53.6 | 58.6 | 68.8 | 151.3 | 4.8 | 15.7 | −7.6 |
| Stomach wall | 4.8 | −10.9 | −6.4 | −7.5 | 19.6 | 9.4 | - | 7.6 | 13.8 | 19.6 | 25.1 | 33.7 | 35.1 | 42.8 | 57.2 | 144.5 | −1.7 | -1.0 | −8.8 |
| Upper lower intestine wall | 0.0 | −24.4 | −14.0 | −14.0 | 31.2 | 2.2 | - | 12.2 | 23.1 | 31.2 | 40.0 | 47.9 | 51.6 | 57.4 | 67.6 | 150.7 | 4.3 | 14.1 | −6.7 |
| Heart wall | −2.4 | −29.3 | −17.7 | −19.1 | 35.4 | −2.4 | - | 12.0 | 26.3 | 35.4 | 42.9 | 53.0 | 55.5 | 57.3 | 70.2 | 151.9 | 0.0 | 8.9 | −11.0 |
| Kidneys | 4.9 | −20.9 | −8.7 | −13.6 | 39.3 | 3.8 | - | 16.8 | 29.4 | 39.3 | 47.8 | 45.2 | 61.3 | 66.7 | 76.7 | 154.7 | 1.7 | 12.0 | −4.5 |
| Liver | 2.1 | −21.3 | −11.0 | −11.8 | 33.8 | 5.8 | - | 14.6 | 25.2 | 33.8 | 40.3 | 50.6 | 52.4 | 57.0 | 64.8 | 149.8 | 6.9 | 20.1 | −9.5 |
| Lungs | −6.4 | −35.2 | −22.6 | −24.1 | 37.6 | −8.2 | - | 11.2 | 28.6 | 37.6 | 45.1 | 55.8 | 57.5 | 58.1 | 71.7 | 152.7 | −7.0 | 1.1 | −14.5 |
| Muscle | 0.0 | −25.8 | −15.4 | −15.4 | 37.2 | 0.0 | - | 15.8 | 27.2 | 37.2 | 44.4 | 54.2 | 57.1 | 61.4 | 72.7 | 153.2 | 5.6 | 13.3 | −5.9 |
| Ovaries | −3.2 | −32.7 | −16.9 | −20.7 | 33.8 | 0.0 | - | 14.5 | 24.7 | 33.8 | 42.0 | 51.2 | 56.2 | 60.9 | 70.7 | 152.2 | 9.0 | 19.7 | −3.2 |
| Pancreas | 0.6 | −24.8 | −13.1 | −14.5 | 33.2 | −0.6 | - | 13.7 | 24.3 | 33.2 | 41.4 | 51.5 | 55.1 | 58.3 | 71.9 | 152.5 | 6.5 | 15.3 | −5.7 |
| Red. Marrow | 0.0 | −26.9 | −14.1 | −17.1 | 38.3 | −2.7 | - | 14.6 | 29.2 | 38.3 | 48.0 | 56.6 | 60.6 | 64.3 | 75.4 | 154.5 | 2.6 | 12.3 | −8.2 |
| Osteogenic | 0.0 | −29.6 | −17.5 | −17.5 | 41.0 | −3.3 | - | 14.9 | 30.1 | 41.0 | 48.8 | 59.1 | 62.2 | 65.2 | 77.2 | 155.1 | 0.0 | 12.1 | −10.2 |
| Skin | 0.0 | −26.7 | −12.5 | −19.4 | 38.1 | 0.0 | - | 16.2 | 30.0 | 38.1 | 45.5 | 55.3 | 61.2 | 64.0 | 74.1 | 154.1 | 5.7 | 11.1 | −6.1 |
| Spleen | −3.1 | −34.5 | −18.2 | −19.0 | 37.2 | −11.4 | - | 15.2 | 26.6 | 37.2 | 47.0 | 60.4 | 63.7 | 64.2 | 80.9 | 156.6 | 15.4 | 25.3 | −0.7 |
| Thymus | −5.1 | −31.9 | −19.2 | −22.2 | 36.7 | −5.1 | - | 11.8 | 28.0 | 36.7 | 44.7 | 54.5 | 57.1 | 58.4 | 71.0 | 152.4 | −2.5 | 4.9 | −13.3 |
| Thyroid | 0.0 | −33.3 | −24.0 | −24.0 | 40.0 | −7.4 | - | 13.3 | 30.3 | 40.0 | 48.6 | 60.0 | 63.4 | 63.4 | 75.6 | 154.8 | −7.4 | 6.9 | −15.4 |
| Urinary bladder | −5.5 | −38.3 | −22.5 | −24.0 | 34.5 | −4.9 | - | 14.4 | 25.0 | 34.5 | 42.3 | 53.0 | 55.5 | 61.2 | 70.3 | 151.9 | 10.7 | 23.2 | −3.6 |
| Uterus | −3.9 | −33.7 | −21.3 | −21.3 | 33.6 | −3.9 | - | 14.3 | 25.2 | 33.6 | 42.4 | 52.5 | 55.6 | 60.4 | 70.0 | 152.0 | 9.2 | 20.7 | −3.8 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.; Kim, H.; Kang, Y.-r.; Kim, H.; Kim, J.Y.; Lee, Y.-J.; Kim, J.M.; Kim, J.S. Selection Criteria for Determination of Optimal Reconstruction Method for Cu-64 Trastuzumab Dosimetry on Siemens Inveon PET Scanner. J. Clin. Med. 2019, 8, 512. https://doi.org/10.3390/jcm8040512
Lee S, Kim H, Kang Y-r, Kim H, Kim JY, Lee Y-J, Kim JM, Kim JS. Selection Criteria for Determination of Optimal Reconstruction Method for Cu-64 Trastuzumab Dosimetry on Siemens Inveon PET Scanner. Journal of Clinical Medicine. 2019; 8(4):512. https://doi.org/10.3390/jcm8040512
Chicago/Turabian StyleLee, Seonhwa, Hyeongi Kim, Ye-rin Kang, Hyungwoo Kim, Jung Young Kim, Yong-Jin Lee, Jung Min Kim, and Jin Su Kim. 2019. "Selection Criteria for Determination of Optimal Reconstruction Method for Cu-64 Trastuzumab Dosimetry on Siemens Inveon PET Scanner" Journal of Clinical Medicine 8, no. 4: 512. https://doi.org/10.3390/jcm8040512
APA StyleLee, S., Kim, H., Kang, Y.-r., Kim, H., Kim, J. Y., Lee, Y.-J., Kim, J. M., & Kim, J. S. (2019). Selection Criteria for Determination of Optimal Reconstruction Method for Cu-64 Trastuzumab Dosimetry on Siemens Inveon PET Scanner. Journal of Clinical Medicine, 8(4), 512. https://doi.org/10.3390/jcm8040512





