New Chondrosarcoma Cell Lines with Preserved Stem Cell Properties to Study the Genomic Drift During In Vitro/In Vivo Growth
Abstract
:1. Introduction
2. Experimental Section
2.1. Establishment of Cell Lines
2.2. Tumorsphere Culture
2.3. Western Blotting
2.4. Aldefluor Assay and Cell Sorting
2.5. Three-Dimensional Spheroid Invasion Assay
2.6. In Vivo Tumor Growth
2.7. Histological Analysis
2.8. MDM2 and CDKN2A Gene Copy Number Analysis
2.9. Mutational Analysis of TP53, IDH1, IDH2, PI3KCA
2.10. Library Construction and WES
3. Results
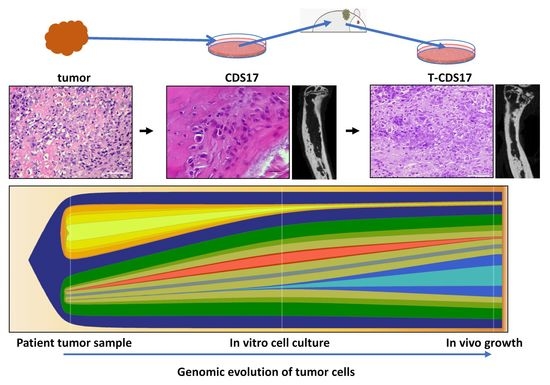
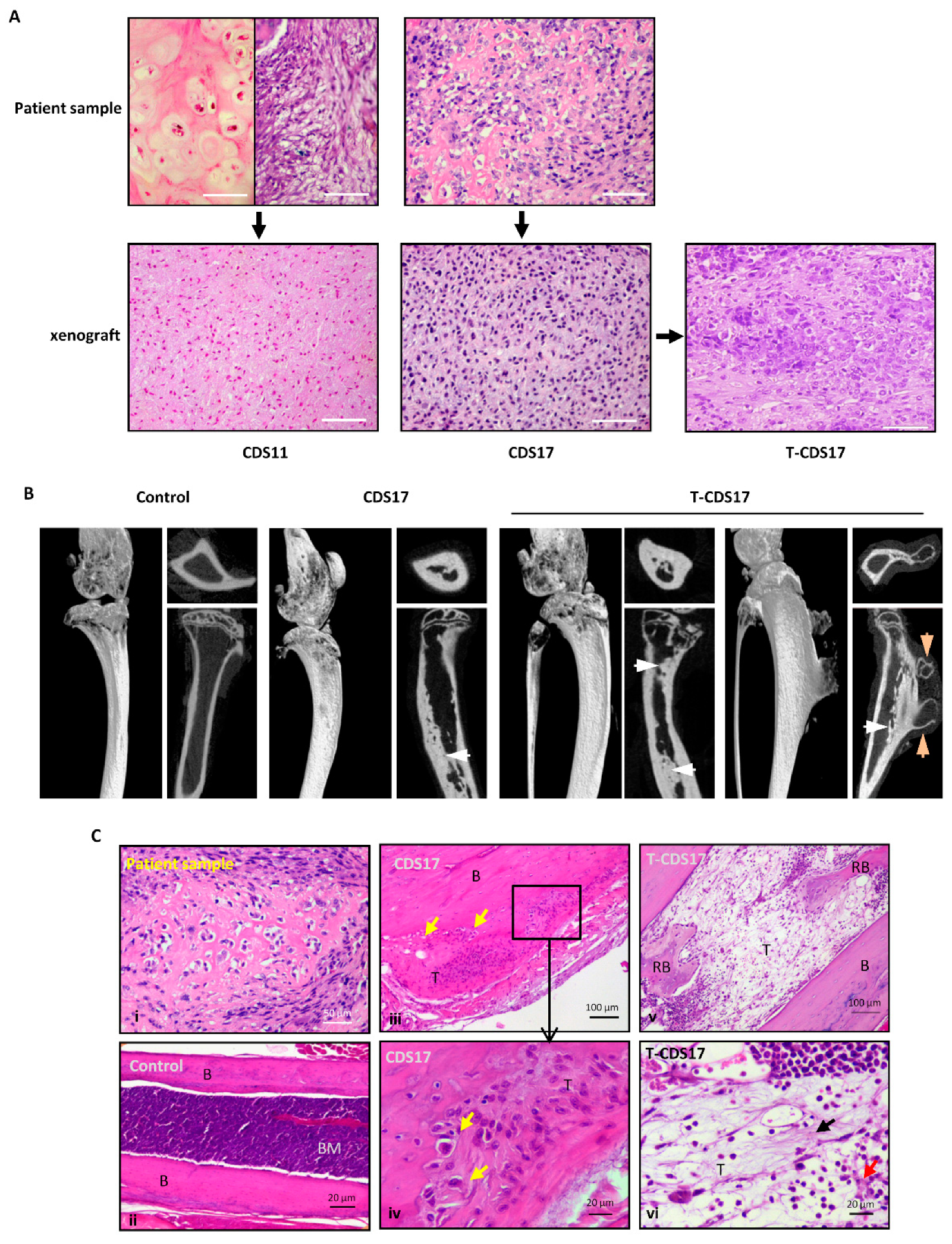
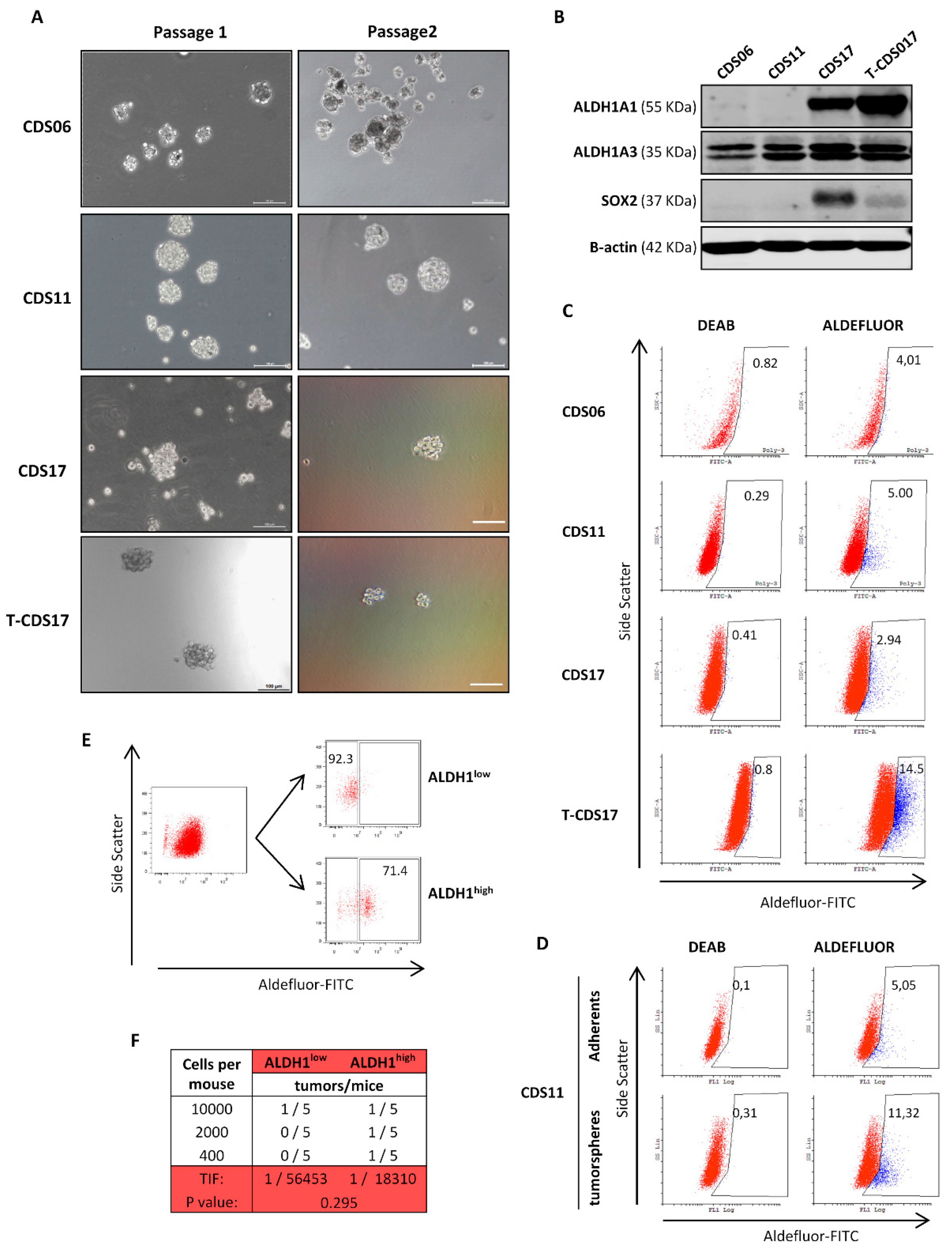
3.1. Establishment of Patient-Derived Chondrosarcoma Cell Lines and Analysis of In Vivo Tumorigenic Potential
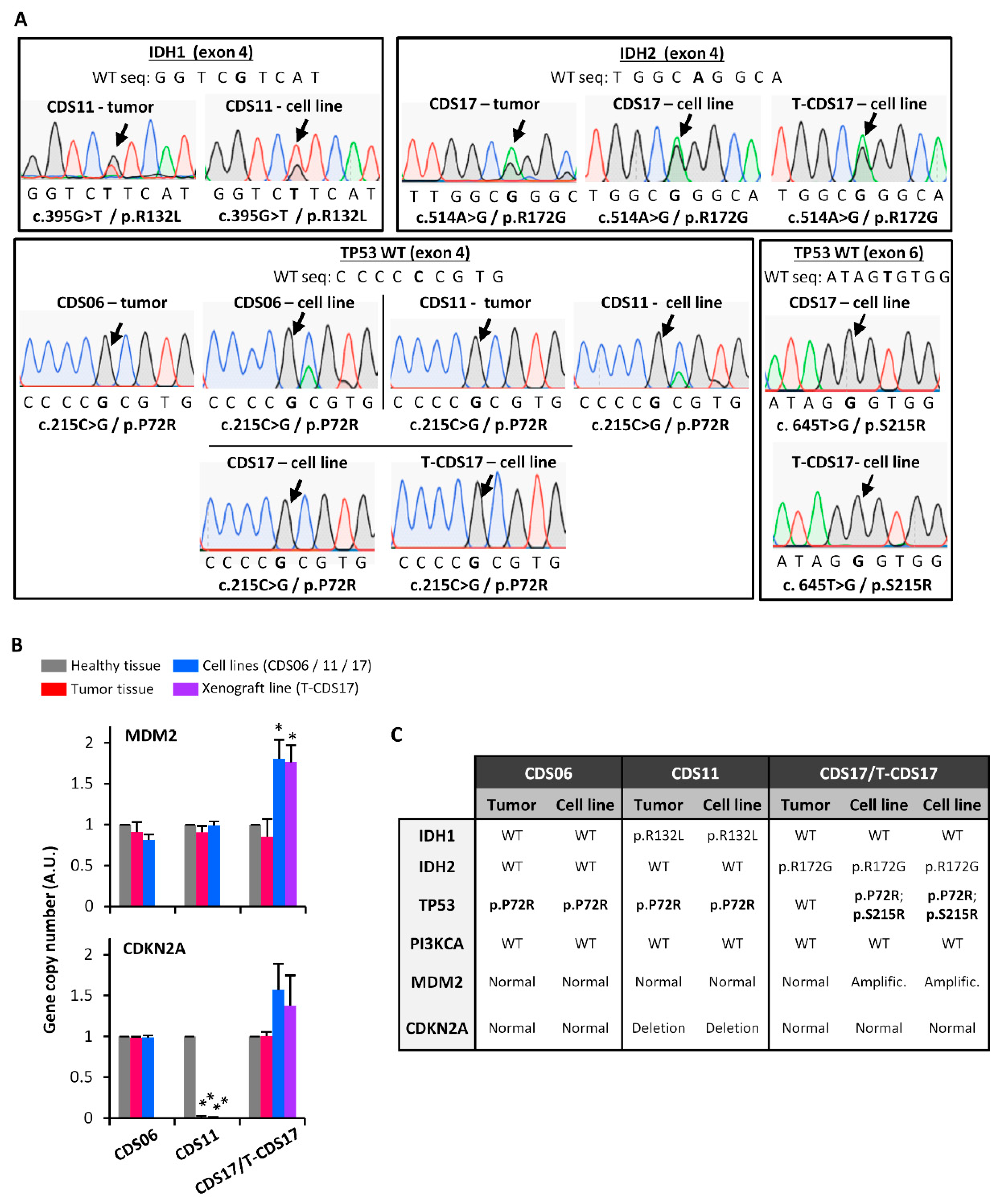
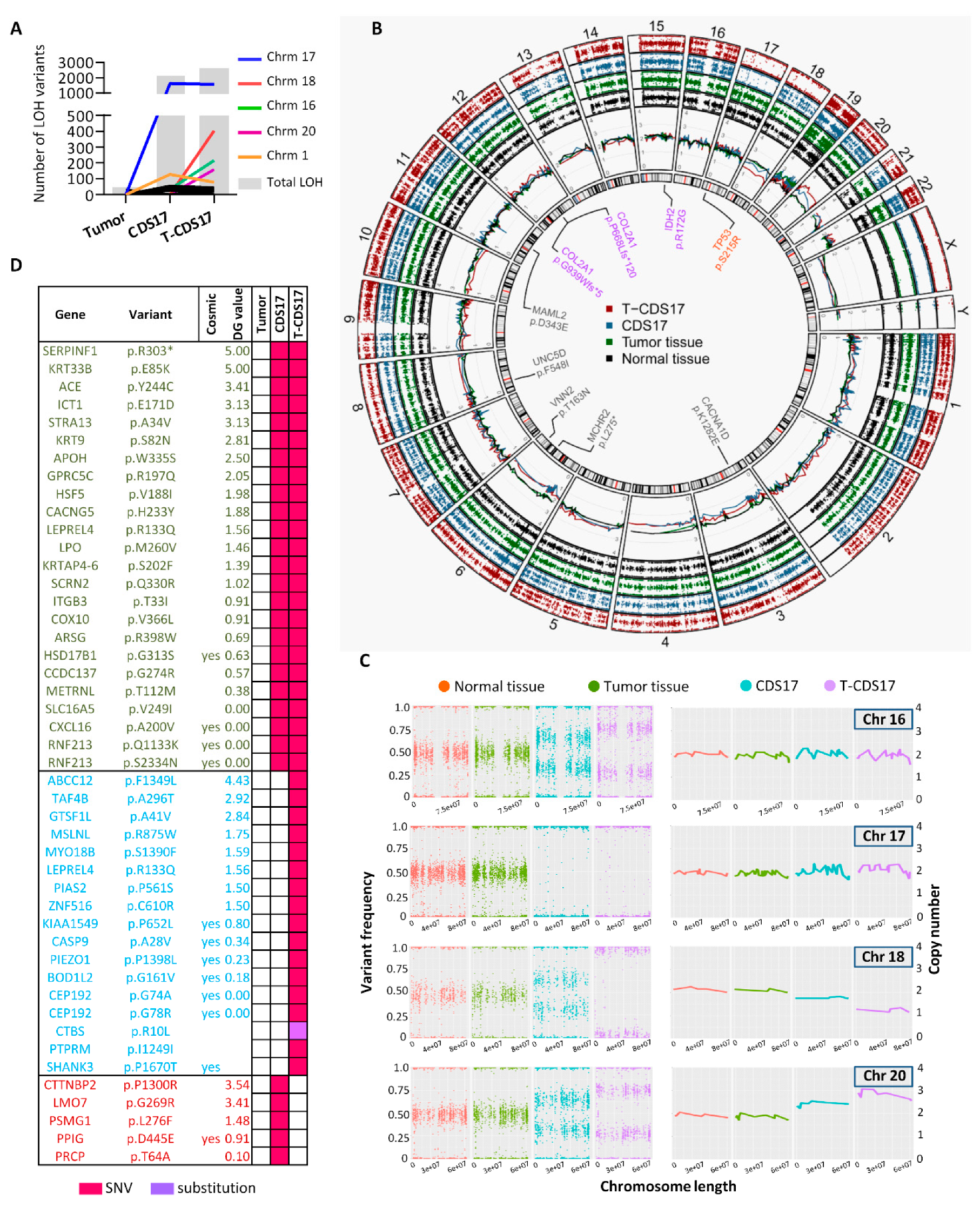
3.2. Genetic Characterization of Chondrosarcoma Cell Lines
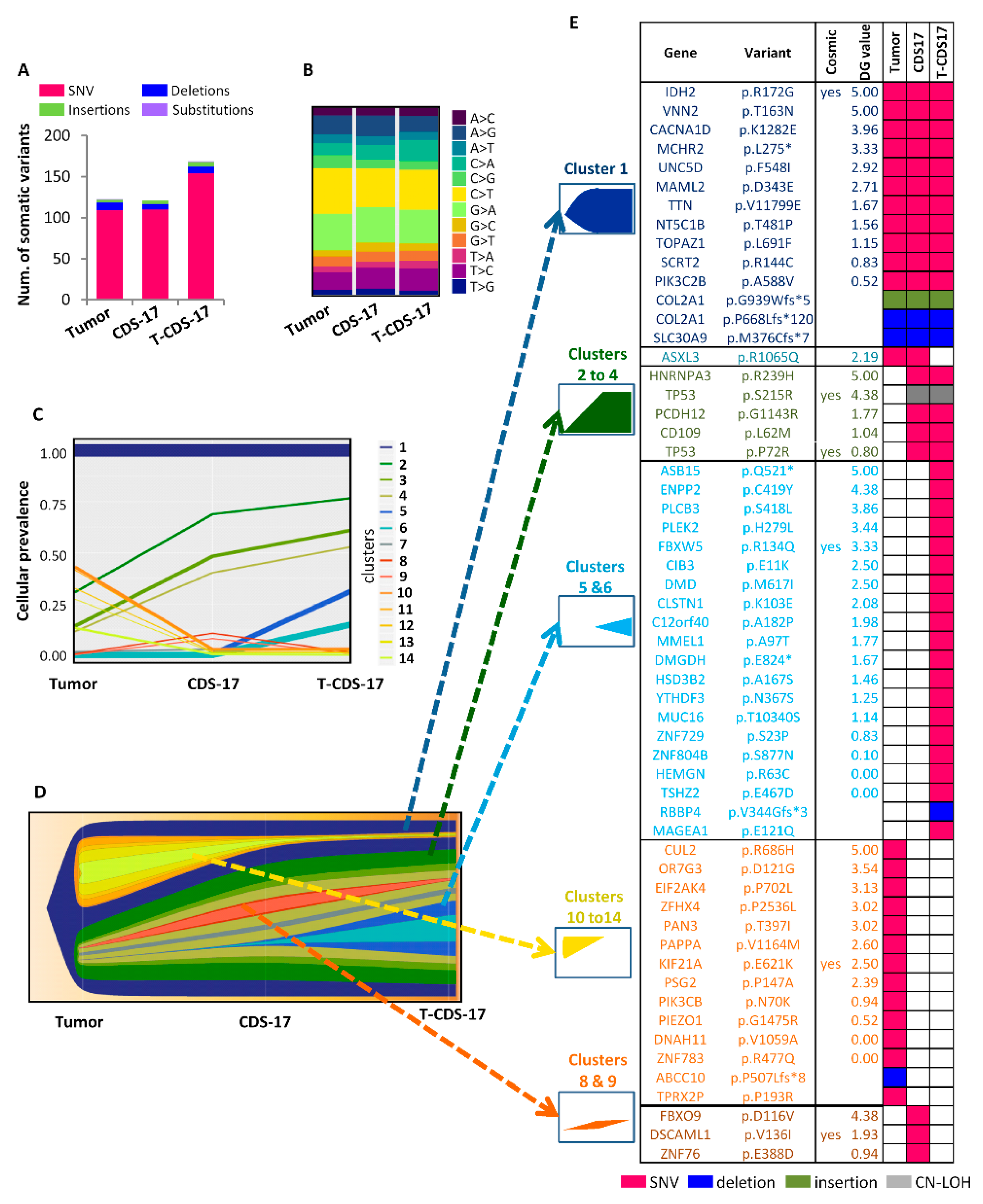
3.3. WES of Chondrosarcoma Cell Lines and Clonal Evolution after In Vitro and In Vivo Growth
3.4. Analysis of CSC Subpopulations in New Chondrosarcoma Cell Lines
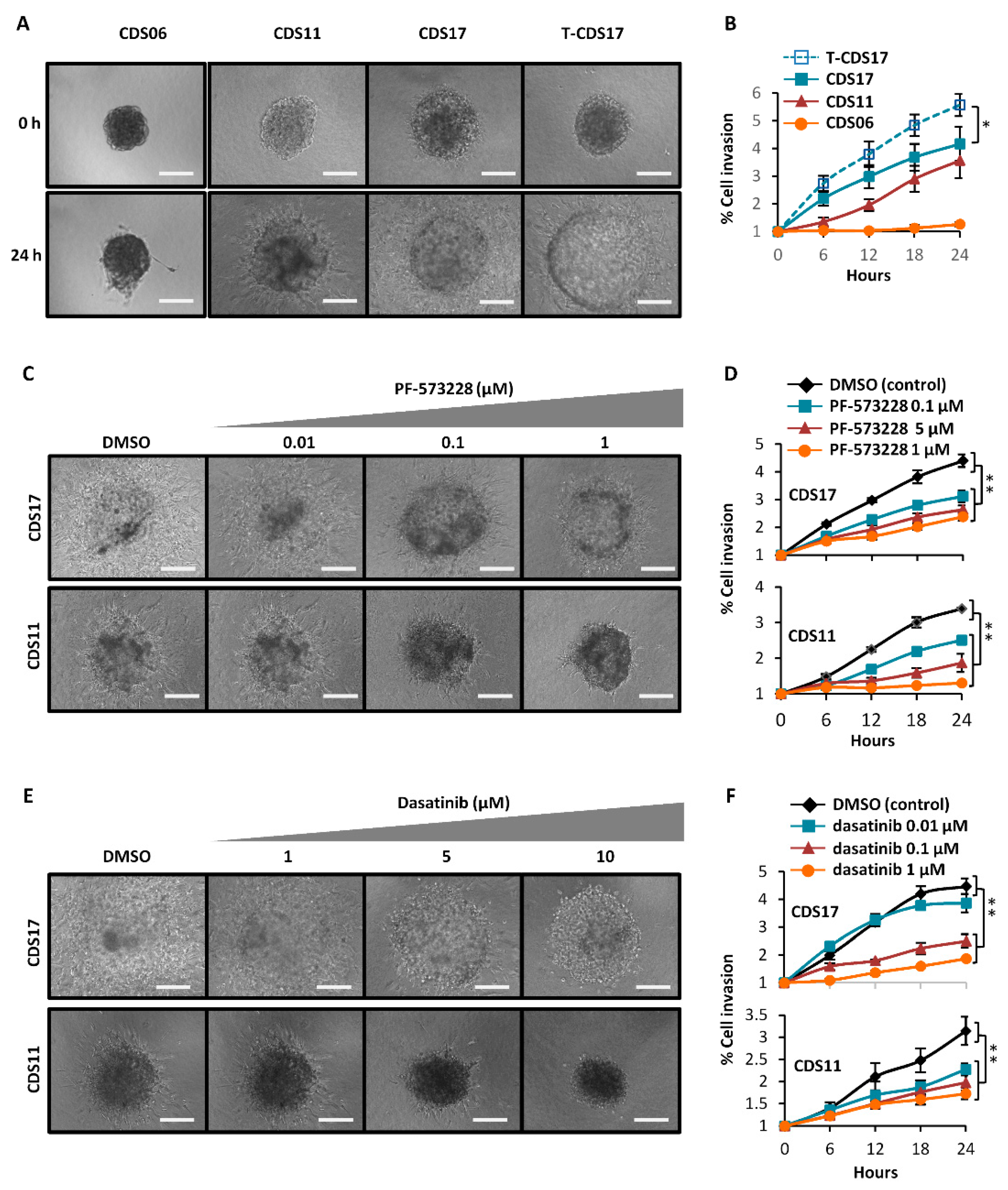
3.5. Invasion Ability of Chondrosarcoma Cell Lines
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fletcher, C.; Bridge, J.; Hogendoorn, P.; Mertens, F. (Eds.) WHO Classification of Tumours of Soft Tissue and Bone. Pathology and Genetics of Tumours of Soft Tissue and Bone, 4th ed.; IARC Press: Lyon, France, 2013. [Google Scholar]
- Amary, M.F.; Bacsi, K.; Maggiani, F.; Damato, S.; Halai, D.; Berisha, F.; Pollock, R.; O’Donnell, P.; Grigoriadis, A.; Diss, T.; et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J. Pathol. 2011, 224, 334–343. [Google Scholar] [CrossRef]
- Lu, C.; Venneti, S.; Akalin, A.; Fang, F.; Ward, P.S.; Dematteo, R.G.; Intlekofer, A.M.; Chen, C.; Ye, J.; Hameed, M.; et al. Induction of sarcomas by mutant IDH2. Genes Dev. 2013, 27, 1986–1998. [Google Scholar] [CrossRef]
- Tinoco, G.; Wilky, B.A.; Paz-Mejia, A.; Rosenberg, A.; Trent, J.C. The biology and management of cartilaginous tumors: A role for targeting isocitrate dehydrogenase. Am. Soc. Clin. Oncol. Educ. Book 2015, e648–e655. [Google Scholar] [CrossRef]
- Tarpey, P.S.; Behjati, S.; Cooke, S.L.; Van Loo, P.; Wedge, D.C.; Pillay, N.; Marshall, J.; O’Meara, S.; Davies, H.; Nik-Zainal, S.; et al. Frequent mutation of the major cartilage collagen gene COL2A1 in chondrosarcoma. Nat. Genet. 2013, 45, 923–926. [Google Scholar] [CrossRef]
- Totoki, Y.; Yoshida, A.; Hosoda, F.; Nakamura, H.; Hama, N.; Ogura, K.; Fujiwara, T.; Arai, Y.; Toguchida, J.; Tsuda, H.; et al. Unique mutation portraits and frequent COL2A1 gene alteration in chondrosarcoma. Genome Res. 2014, 24, 1411–1420. [Google Scholar] [CrossRef] [PubMed]
- Schrage, Y.M.; Lam, S.; Jochemsen, A.G.; Cleton-Jansen, A.M.; Taminiau, A.H.; Hogendoorn, P.C.; Bovee, J.V. Central chondrosarcoma progression is associated with pRb pathway alterations: CDK4 down-regulation and p16 overexpression inhibit cell growth in vitro. J. Cell. Mol. Med. 2009, 13, 2843–2852. [Google Scholar] [CrossRef] [PubMed]
- Boehme, K.A.; Schleicher, S.B.; Traub, F.; Rolauffs, B. Chondrosarcoma: A Rare Misfortune in Aging Human Cartilage? The Role of Stem and Progenitor Cells in Proliferation, Malignant Degeneration and Therapeutic Resistance. Int. J. Mol. Sci. 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Bovee, J.V.; Cleton-Jansen, A.M.; Taminiau, A.H.; Hogendoorn, P.C. Emerging pathways in the development of chondrosarcoma of bone and implications for targeted treatment. Lancet Oncol. 2005, 6, 599–607. [Google Scholar] [CrossRef]
- Brown, H.K.; Schiavone, K.; Gouin, F.; Heymann, M.F.; Heymann, D. Biology of Bone Sarcomas and New Therapeutic Developments. Calcif. Tissue Int. 2018, 102, 174–195. [Google Scholar] [CrossRef]
- David, E.; Blanchard, F.; Heymann, M.F.; De Pinieux, G.; Gouin, F.; Redini, F.; Heymann, D. The Bone Niche of Chondrosarcoma: A Sanctuary for Drug Resistance, Tumour Growth and also a Source of New Therapeutic Targets. Sarcoma 2011, 2011, 932451. [Google Scholar] [CrossRef]
- Polychronidou, G.; Karavasilis, V.; Pollack, S.M.; Huang, P.H.; Lee, A.; Jones, R.L. Novel therapeutic approaches in chondrosarcoma. Future Oncol. 2017, 13, 637–648. [Google Scholar] [CrossRef]
- Martinez-Cruzado, L.; Tornin, J.; Santos, L.; Rodriguez, A.; Garcia-Castro, J.; Moris, F.; Rodriguez, R. Aldh1 Expression and Activity Increase During Tumor Evolution in Sarcoma Cancer Stem Cell Populations. Sci. Rep. 2016, 6, 27878. [Google Scholar] [CrossRef]
- Tirino, V.; Desiderio, V.; Paino, F.; De Rosa, A.; Papaccio, F.; Fazioli, F.; Pirozzi, G.; Papaccio, G. Human primary bone sarcomas contain CD133+ cancer stem cells displaying high tumorigenicity in vivo. FASEB J. 2011, 25, 2022–2030. [Google Scholar] [CrossRef]
- Goodspeed, A.; Heiser, L.M.; Gray, J.W.; Costello, J.C. Tumor-Derived Cell Lines as Molecular Models of Cancer Pharmacogenomics. Mol. Cancer Res. 2016, 14, 3–13. [Google Scholar] [CrossRef]
- McDermott, U. Cancer cell lines as patient avatars for drug response prediction. Nat. Genet. 2018, 50, 1350–1351. [Google Scholar] [CrossRef]
- Wilding, J.L.; Bodmer, W.F. Cancer cell lines for drug discovery and development. Cancer Res. 2014, 74, 2377–2384. [Google Scholar] [CrossRef]
- Calabuig-Farinas, S.; Benso, R.G.; Szuhai, K.; Machado, I.; Lopez-Guerrero, J.A.; de Jong, D.; Peydro, A.; San Miguel, T.; Navarro, L.; Pellin, A.; et al. Characterization of a new human cell line (CH-3573) derived from a grade II chondrosarcoma with matrix production. Pathol. Oncol. Res. 2012, 18, 793–802. [Google Scholar] [CrossRef]
- Farges, M.; Mazeau, C.; Gioanni, J.; Ettore, F.; Denovion, H.; Schneider, M. Establishment and characterization of a new cell line derived from a human chondrosarcoma. Oncol. Rep. 1997, 4, 697–700. [Google Scholar] [CrossRef]
- Gil-Benso, R.; Lopez-Gines, C.; Lopez-Guerrero, J.A.; Carda, C.; Callaghan, R.C.; Navarro, S.; Ferrer, J.; Pellin, A.; Llombart-Bosch, A. Establishment and characterization of a continuous human chondrosarcoma cell line, ch-2879: Comparative histologic and genetic studies with its tumor of origin. Lab. Investig. 2003, 83, 877–887. [Google Scholar] [CrossRef]
- Jagasia, A.A.; Block, J.A.; Qureshi, A.; Diaz, M.O.; Nobori, T.; Gitelis, S.; Iyer, A.P. Chromosome 9 related aberrations and deletions of the CDKN2 and MTS2 putative tumor suppressor genes in human chondrosarcomas. Cancer Lett. 1996, 105, 91–103. [Google Scholar] [CrossRef]
- Kalinski, T.; Krueger, S.; Pelz, A.F.; Wieacker, P.; Hartig, R.; Ropke, M.; Schneider-Stock, R.; Dombrowski, F.; Roessner, A. Establishment and characterization of the permanent human cell line C3842 derived from a secondary chondrosarcoma in Ollier’s disease. Virchows Arch. 2005, 446, 287–299. [Google Scholar] [CrossRef]
- Kudawara, I.; Araki, N.; Myoui, A.; Kato, Y.; Uchida, A.; Yoshikawa, H. New cell lines with chondrocytic phenotypes from human chondrosarcoma. Virchows Arch. 2004, 444, 577–586. [Google Scholar] [CrossRef]
- Kudo, N.; Ogose, A.; Hotta, T.; Kawashima, H.; Gu, W.; Umezu, H.; Toyama, T.; Endo, N. Establishment of novel human dedifferentiated chondrosarcoma cell line with osteoblastic differentiation. Virchows Arch. 2007, 451, 691–699. [Google Scholar] [CrossRef]
- Kunisada, T.; Miyazaki, M.; Mihara, K.; Gao, C.; Kawai, A.; Inoue, H.; Namba, M. A new human chondrosarcoma cell line (OUMS-27) that maintains chondrocytic differentiation. Int. J. Cancer 1998, 77, 854–859. [Google Scholar] [CrossRef]
- Monderer, D.; Luseau, A.; Bellec, A.; David, E.; Ponsolle, S.; Saiagh, S.; Bercegeay, S.; Piloquet, P.; Denis, M.G.; Lode, L.; et al. New chondrosarcoma cell lines and mouse models to study the link between chondrogenesis and chemoresistance. Lab. Investig. 2013, 93, 1100–1114. [Google Scholar] [CrossRef]
- van Oosterwijk, J.G.; de Jong, D.; van Ruler, M.A.; Hogendoorn, P.C.; Dijkstra, P.D.; van Rijswijk, C.S.; Machado, I.; Llombart-Bosch, A.; Szuhai, K.; Bovee, J.V. Three new chondrosarcoma cell lines: One grade III conventional central chondrosarcoma and two dedifferentiated chondrosarcomas of bone. BMC Cancer 2012, 12, 375. [Google Scholar] [CrossRef]
- Tornin, J.; Martinez-Cruzado, L.; Santos, L.; Rodriguez, A.; Nunez, L.E.; Oro, P.; Hermosilla, M.A.; Allonca, E.; Fernandez-Garcia, M.T.; Astudillo, A.; et al. Inhibition of SP1 by the mithramycin analog EC-8042 efficiently targets tumor initiating cells in sarcoma. Oncotarget 2016, 7, 30935–30950. [Google Scholar] [CrossRef]
- Martinez-Cruzado, L.; Tornin, J.; Rodriguez, A.; Santos, L.; Allonca, E.; Fernandez-Garcia, M.T.; Astudillo, A.; Garcia-Pedrero, J.M.; Rodriguez, R. Trabectedin and Campthotecin Synergistically Eliminate Cancer Stem Cells in Cell-of-Origin Sarcoma Models. Neoplasia 2017, 19, 460–470. [Google Scholar] [CrossRef]
- Tornin, J.; Hermida-Prado, F.; Padda, R.S.; Gonzalez, M.V.; Alvarez-Fernandez, C.; Rey, V.; Martinez-Cruzado, L.; Estupinan, O.; Menendez, S.T.; Fernandez-Nevado, L.; et al. FUS-CHOP Promotes Invasion in Myxoid Liposarcoma through a SRC/FAK/RHO/ROCK-Dependent Pathway. Neoplasia 2018, 20, 44–56. [Google Scholar] [CrossRef]
- Estupinan, O.; Santos, L.; Rodriguez, A.; Fernandez-Nevado, L.; Costales, P.; Perez-Escuredo, J.; Hermosilla, M.A.; Oro, P.; Rey, V.; Tornin, J.; et al. The multikinase inhibitor EC-70124 synergistically increased the antitumor activity of doxorubicin in sarcomas. Int. J. Cancer 2018. [Google Scholar] [CrossRef]
- Rubio, R.; Abarrategi, A.; Garcia-Castro, J.; Martinez-Cruzado, L.; Suarez, C.; Tornin, J.; Santos, L.; Astudillo, A.; Colmenero, I.; Mulero, F.; et al. Bone environment is essential for osteosarcoma development from transformed mesenchymal stem cells. Stem Cells 2014, 32, 1136–1148. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Cabanillas, R.; Dineiro, M.; Castillo, D.; Pruneda, P.C.; Penas, C.; Cifuentes, G.A.; de Vicente, A.; Duran, N.S.; Alvarez, R.; Ordonez, G.R.; et al. A novel molecular diagnostics platform for somatic and germline precision oncology. Mol. Genet. Genomic Med. 2017, 5, 336–359. [Google Scholar] [CrossRef]
- Choi, Y.; Sims, G.E.; Murphy, S.; Miller, J.R.; Chan, A.P. Predicting the functional effect of amino acid substitutions and indels. PLoS ONE 2012, 7, e46688. [Google Scholar] [CrossRef]
- Chun, S.; Fay, J.C. Identification of deleterious mutations within three human genomes. Genome Res. 2009, 19, 1553–1561. [Google Scholar] [CrossRef]
- Davydov, E.V.; Goode, D.L.; Sirota, M.; Cooper, G.M.; Sidow, A.; Batzoglou, S. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput. Biol. 2010, 6, e1001025. [Google Scholar] [CrossRef]
- Deshwar, A.G.; Vembu, S.; Yung, C.K.; Jang, G.H.; Stein, L.; Morris, Q. PhyloWGS: Reconstructing subclonal composition and evolution from whole-genome sequencing of tumors. Genome Biol. 2015, 16, 35. [Google Scholar] [CrossRef]
- Dong, C.; Wei, P.; Jian, X.; Gibbs, R.; Boerwinkle, E.; Wang, K.; Liu, X. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum. Mol. Genet. 2015, 24, 2125–2137. [Google Scholar] [CrossRef]
- Jagadeesh, K.A.; Wenger, A.M.; Berger, M.J.; Guturu, H.; Stenson, P.D.; Cooper, D.N.; Bernstein, J.A.; Bejerano, G. M-CAP eliminates a majority of variants of uncertain significance in clinical exomes at high sensitivity. Nat. Genet. 2016, 48, 1581–1586. [Google Scholar] [CrossRef]
- Kumar, P.; Henikoff, S.; Ng, P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009, 4, 1073–1081. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.A.; McMichael, J.; Dang, H.X.; Maher, C.A.; Ding, L.; Ley, T.J.; Mardis, E.R.; Wilson, R.K. Visualizing tumor evolution with the fishplot package for R. BMC Genom. 2016, 17, 880. [Google Scholar] [CrossRef] [PubMed]
- Puente, X.S.; Pinyol, M.; Quesada, V.; Conde, L.; Ordonez, G.R.; Villamor, N.; Escaramis, G.; Jares, P.; Bea, S.; Gonzalez-Diaz, M.; et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature 2011, 475, 101–105. [Google Scholar] [CrossRef]
- Reva, B.; Antipin, Y.; Sander, C. Predicting the functional impact of protein mutations: Application to cancer genomics. Nucleic Acids Res. 2011, 39, e118. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, J.M.; Cooper, D.N.; Schuelke, M.; Seelow, D. MutationTaster2: Mutation prediction for the deep-sequencing age. Nat. Methods 2014, 11, 361–362. [Google Scholar] [CrossRef] [PubMed]
- Shihab, H.A.; Gough, J.; Mort, M.; Cooper, D.N.; Day, I.N.; Gaunt, T.R. Ranking non-synonymous single nucleotide polymorphisms based on disease concepts. Hum. Genom. 2014, 8, 11. [Google Scholar] [CrossRef]
- European Nucleotide Archive repository. Available online: http://www.ebi.ac.uk/ena/data/view/PRJEB31233 (accessed on 28 February 2019).
- Suijker, J.; Oosting, J.; Koornneef, A.; Struys, E.A.; Salomons, G.S.; Schaap, F.G.; Waaijer, C.J.; Wijers-Koster, P.M.; Briaire-de Bruijn, I.H.; Haazen, L.; et al. Inhibition of mutant IDH1 decreases D-2-HG levels without affecting tumorigenic properties of chondrosarcoma cell lines. Oncotarget 2015, 6, 12505–12519. [Google Scholar] [CrossRef]
- Campbell, V.T.; Nadesan, P.; Ali, S.A.; Wang, C.Y.; Whetstone, H.; Poon, R.; Wei, Q.; Keilty, J.; Proctor, J.; Wang, L.W.; et al. Hedgehog pathway inhibition in chondrosarcoma using the smoothened inhibitor IPI-926 directly inhibits sarcoma cell growth. Mol. Cancer Ther. 2014, 13, 1259–1269. [Google Scholar] [CrossRef]
- Heymann, D.; Redini, F. Targeted therapies for bone sarcomas. Bonekey Rep. 2013, 2, 378. [Google Scholar] [CrossRef]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehar, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef]
- Iorio, F.; Knijnenburg, T.A.; Vis, D.J.; Bignell, G.R.; Menden, M.P.; Schubert, M.; Aben, N.; Goncalves, E.; Barthorpe, S.; Lightfoot, H.; et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell 2016, 166, 740–754. [Google Scholar] [CrossRef]
- Lee, J.K.; Liu, Z.; Sa, J.K.; Shin, S.; Wang, J.; Bordyuh, M.; Cho, H.J.; Elliott, O.; Chu, T.; Choi, S.W.; et al. Pharmacogenomic landscape of patient-derived tumor cells informs precision oncology therapy. Nat. Genet. 2018, 50, 1399–1411. [Google Scholar] [CrossRef]
- Ledford, H. US cancer institute to overhaul tumour cell lines. Nature 2016, 530, 391. [Google Scholar] [CrossRef]
- Golan, H.; Shukrun, R.; Caspi, R.; Vax, E.; Pode-Shakked, N.; Goldberg, S.; Pleniceanu, O.; Bar-Lev, D.D.; Mark-Danieli, M.; Pri-Chen, S.; et al. In Vivo Expansion of Cancer Stemness Affords Novel Cancer Stem Cell Targets: Malignant Rhabdoid Tumor as an Example. Stem Cell Rep. 2018, 11, 795–810. [Google Scholar] [CrossRef]
- Ben-David, U.; Ha, G.; Tseng, Y.Y.; Greenwald, N.F.; Oh, C.; Shih, J.; McFarland, J.M.; Wong, B.; Boehm, J.S.; Beroukhim, R.; et al. Patient-derived xenografts undergo mouse-specific tumor evolution. Nat. Genet. 2017, 49, 1567–1575. [Google Scholar] [CrossRef]
- Oshiro, Y.; Chaturvedi, V.; Hayden, D.; Nazeer, T.; Johnson, M.; Johnston, D.A.; Ordonez, N.G.; Ayala, A.G.; Czerniak, B. Altered p53 is associated with aggressive behavior of chondrosarcoma: A long term follow-up study. Cancer 1998, 83, 2324–2334. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Toguchida, J.; Wadayama, B.; Kanoe, H.; Nakayama, T.; Ishizaki, K.; Ikenaga, M.; Kotoura, Y.; Sasaki, M.S. Loss of heterozygosity and tumor suppressor gene mutations in chondrosarcomas. Anticancer Res. 1996, 16, 2009–2015. [Google Scholar]
| Cell Line | Chondrosar. Subtype | Anchorage Independ. Growth § | Passage * | Tumorsphere Growth (% Frequency ± SD) | In Vivo Tumor Growth ** | Aldefluor Assay (%) | Invasion ‡ | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Subcutaneous | Intra-Bone | |||||||||
| 1st Tumorsph. Passage | 2nd Tumorsph. Passage | Tumor Growth (Tumors/Mice) | Mean Volume (mm3 ± SD) | Tumor Growth (Tumors/Mice) | ||||||
| CDS06 | Secondary | Yes | 20 | Yes (0.24 ± 0.10) | Yes (0.10 ± 0.05) | n/a | n/a | n/a | Yes (4.01) | No |
| CDS11 | Secondary | Yes | 25 | Yes (0.28 ± 0.11) | Yes (0.16 ± 0.06) | Yes (3/3) | 77.00 ± 10.91 | n/a | Yes (5.05) | Yes |
| CDS17 | Dedifferent. | Yes | <35 | Yes (0.20 ± 0.08) | Yes (0.10 ± 0.01) | Yes (3/3) | 198.27 ± 5.11 | Yes (1/2) | Yes (2.94) | Yes |
| T-CDS17 | Dedifferent. | Yes | <35 | Yes (0.26 ± 0.08) | Yes (0.11 ± 0.01) | Yes (3/3) | 350.32 ± 39.45 ¶ | Yes (2/2) | Yes (14.50) | Yes |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rey, V.; Menendez, S.T.; Estupiñan, O.; Rodriguez, A.; Santos, L.; Tornin, J.; Martinez-Cruzado, L.; Castillo, D.; Ordoñez, G.R.; Costilla, S.; et al. New Chondrosarcoma Cell Lines with Preserved Stem Cell Properties to Study the Genomic Drift During In Vitro/In Vivo Growth. J. Clin. Med. 2019, 8, 455. https://doi.org/10.3390/jcm8040455
Rey V, Menendez ST, Estupiñan O, Rodriguez A, Santos L, Tornin J, Martinez-Cruzado L, Castillo D, Ordoñez GR, Costilla S, et al. New Chondrosarcoma Cell Lines with Preserved Stem Cell Properties to Study the Genomic Drift During In Vitro/In Vivo Growth. Journal of Clinical Medicine. 2019; 8(4):455. https://doi.org/10.3390/jcm8040455
Chicago/Turabian StyleRey, Veronica, Sofia T. Menendez, Oscar Estupiñan, Aida Rodriguez, Laura Santos, Juan Tornin, Lucia Martinez-Cruzado, David Castillo, Gonzalo R. Ordoñez, Serafin Costilla, and et al. 2019. "New Chondrosarcoma Cell Lines with Preserved Stem Cell Properties to Study the Genomic Drift During In Vitro/In Vivo Growth" Journal of Clinical Medicine 8, no. 4: 455. https://doi.org/10.3390/jcm8040455
APA StyleRey, V., Menendez, S. T., Estupiñan, O., Rodriguez, A., Santos, L., Tornin, J., Martinez-Cruzado, L., Castillo, D., Ordoñez, G. R., Costilla, S., Alvarez-Fernandez, C., Astudillo, A., Braña, A., & Rodriguez, R. (2019). New Chondrosarcoma Cell Lines with Preserved Stem Cell Properties to Study the Genomic Drift During In Vitro/In Vivo Growth. Journal of Clinical Medicine, 8(4), 455. https://doi.org/10.3390/jcm8040455